Available In Vitro Models for Human Satellite Cells from Skeletal Muscle
Abstract
1. Introduction
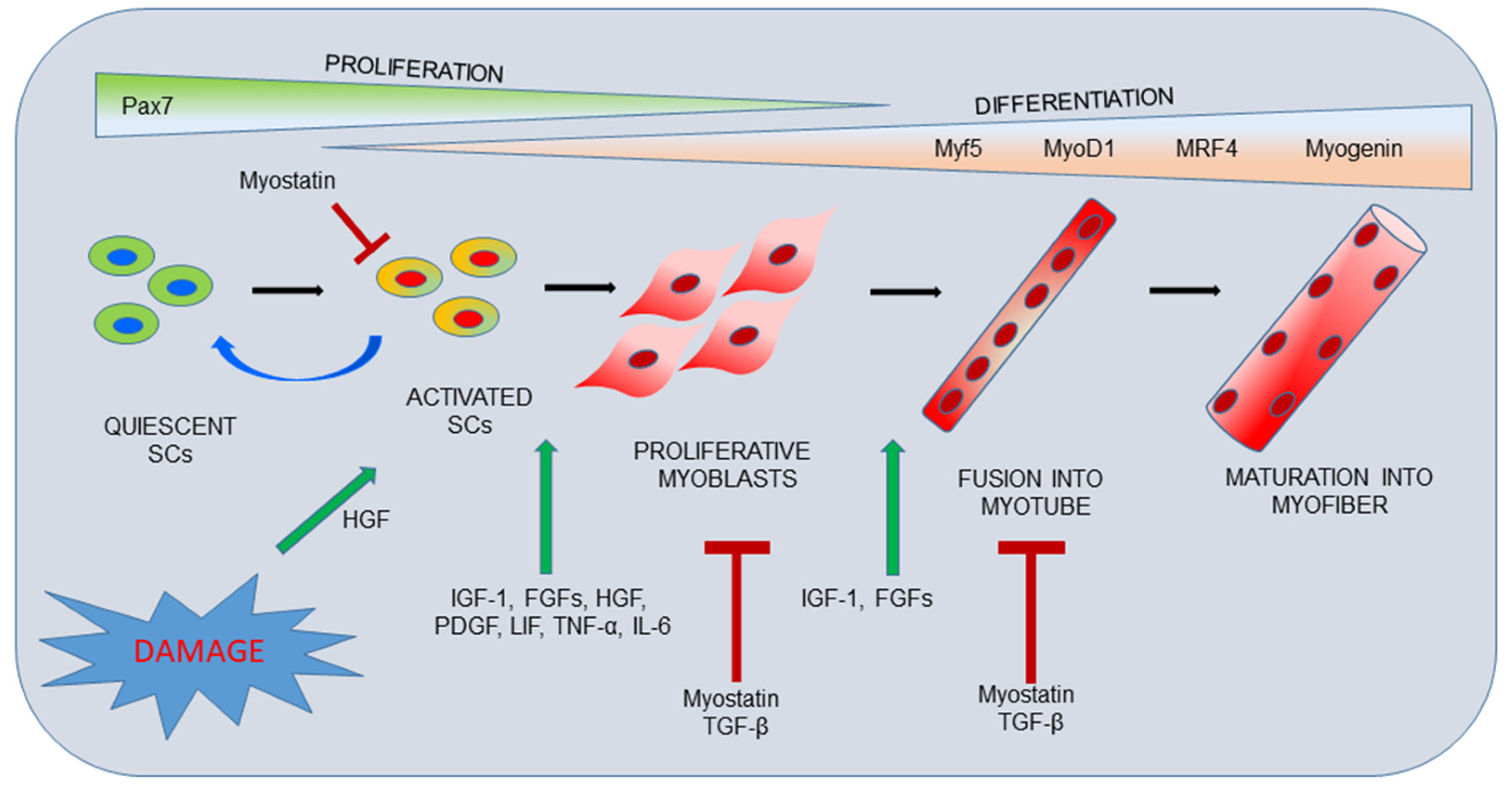
2. Skeletal Muscle Organization and Repair Process
3. Models of Culture for Skeletal Muscle Study
3.1. Skeletal Muscle Explants
3.2. In Vitro 2D Models for Skeletal Muscle
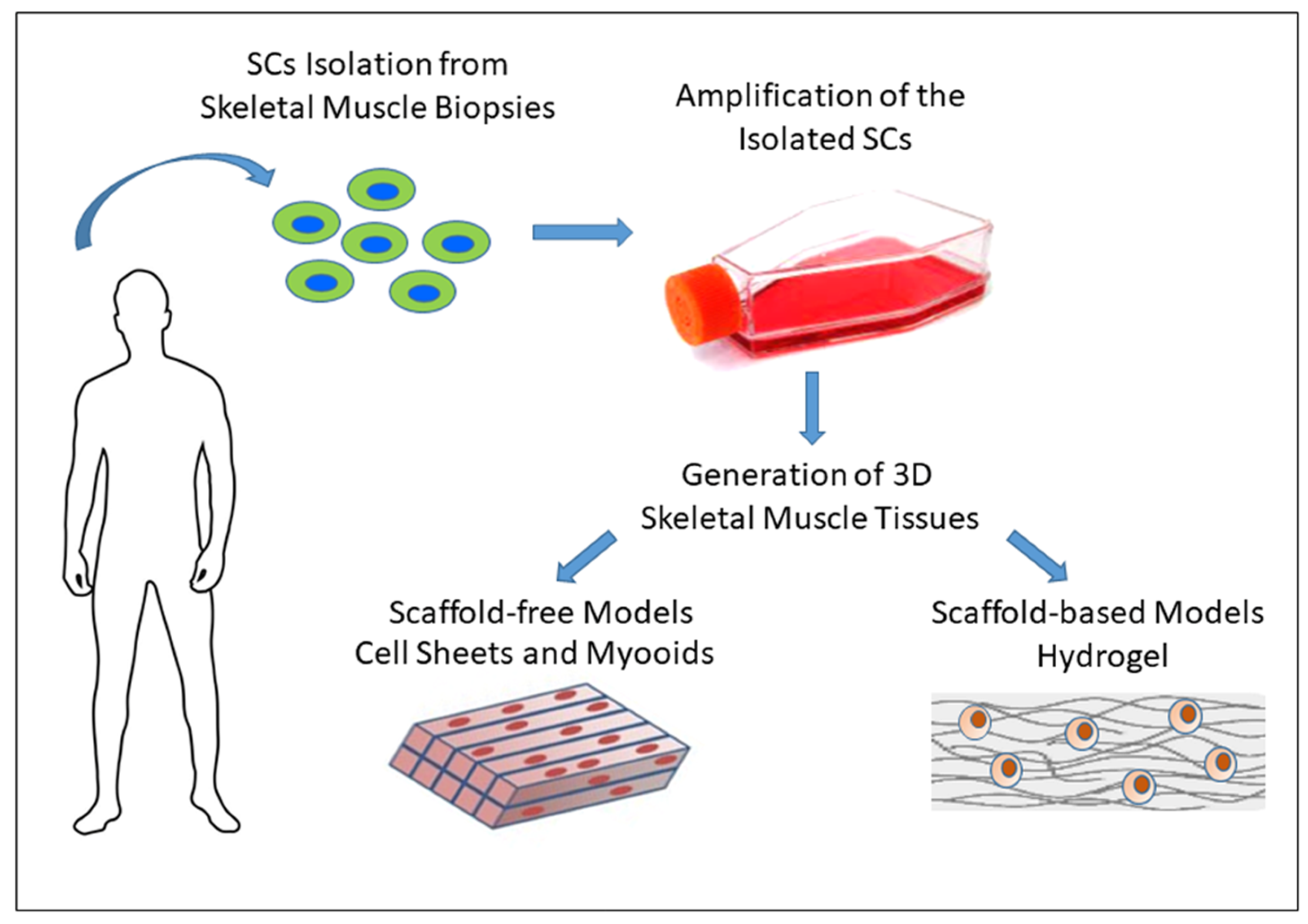
3.3. In Vitro 3D Models of Skeletal Muscle
3.3.1. Scaffold-Free Approaches
3.3.2. Scaffold-Based Approaches
4. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frontera, W.R.; Ochala, J. Skeletal muscle: A brief review of structure and function. Calcif. Tissue Int. 2015, 96, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Das, D.K.; Graham, Z.A.; Cardozo, C.P. Myokines in skeletal muscle physiology and metabolism: Recent advances and future perspectives. Acta Physiol. 2020, 228, e13367. [Google Scholar] [CrossRef]
- Chang, N.C.; Chevalier, F.P.; Rudnicki, M.A. Satellite Cells in Muscular Dystrophy-Lost in Polarity. Trends Mol. Med. 2016, 22, 479–496. [Google Scholar] [CrossRef] [PubMed]
- Sousa-Victor, P.; Muñoz-Cánoves, P. Regenerative decline of stem cells in sarcopenia. Mol. Aspects Med. 2016, 50, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Marchildon, F.; Lamarche, É.; Lala-Tabbert, N.; St-Louis, C.; Wiper-Bergeron, N. Expression of CCAAT/Enhancer Binding Protein Beta in Muscle Satellite Cells Inhibits Myogenesis in Cancer Cachexia. PLoS ONE 2015, 10, e0145583. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Partridge, T.A. The mdx mouse model as a surrogate for Duchenne muscular dystrophy. FEBS J. 2013, 280, 4177–4186. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.; Echigoya, Y.; Fukada, S.I.; Yokota, T. Current Translational Research and Murine Models For Duchenne Muscular Dystrophy. J. Neuromuscul. Dis. 2016, 3, 29–48. [Google Scholar] [CrossRef]
- Heron, M.I.; Richmond, F.J. In-series fiber architecture in long human muscles. J. Morphol. 1993, 216, 35–45. [Google Scholar] [CrossRef]
- Wang, J.; Khodabukus, A.; Rao, L.; Vandusen, K.; Abutaleb, N.; Bursac, N. Engineered skeletal muscles for disease modeling and drug discovery. Biomaterials 2019, 221, 119416. [Google Scholar] [CrossRef]
- Feher, J. Quantitative Human Physiology; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar] [CrossRef]
- Barone, V.; Randazzo, D.; Del Re, V.; Sorrentino, V.; Rossi, D. Organization of junctional sarcoplasmic reticulum proteins in skeletal muscle fibers. J. Muscle Res. Cell Motil. 2015, 36, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Mauro, A. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 1961, 9, 493–495. [Google Scholar] [CrossRef]
- Danoviz, M.E.; Yablonka-Reuveni, Z. Skeletal muscle satellite cells: Background and methods for isolation and analysis in a primary culture system. Methods Mol. Biol. 2012, 798, 21–52. [Google Scholar] [CrossRef]
- Tedesco, F.S.; Dellavalle, A.; Diaz-Manera, J.; Messina, G.; Cossu, G. Repairing skeletal muscle: Regenerative potential of skeletal muscle stem cells. J. Clin. Investig. 2010, 120, 11–19. [Google Scholar] [CrossRef]
- Chen, B.; Shan, T. The role of satellite and other functional cell types in muscle repair and regeneration. J. Muscle Res. Cell Motil. 2019, 40, 1–8. [Google Scholar] [CrossRef]
- Dumont, N.A.; Bentzinger, C.F.; Sincennes, M.C.; Rudnicki, M.A. Satellite Cells and Skeletal Muscle Regeneration. Compr. Physiol. 2015, 5, 1027–1059. [Google Scholar] [CrossRef] [PubMed]
- Seale, P.; Sabourin, L.A.; Girgis-Gabardo, A.; Mansouri, A.; Gruss, P.; Rudnicki, M.A. Pax7 is required for the specification of myogenic satellite cells. Cell 2000, 102, 777–786. [Google Scholar] [CrossRef]
- Zammit, P.S. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin. Cell Dev. Biol. 2017, 72, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Doynova, M.D.; Markworth, J.F.; Cameron-Smith, D.; Vickers, M.H.; O’Sullivan, J.M. Linkages between changes in the 3D organization of the genome and transcription during myotube differentiation in vitro. Skelet. Muscle 2017, 7, 5. [Google Scholar] [CrossRef]
- Karalaki, M.; Fili, S.; Philippou, A.; Koutsilieris, M. Muscle regeneration: Cellular and molecular events. In Vivo 2009, 23, 779–796. [Google Scholar]
- Floss, T.; Arnold, H.H.; Braun, T. A role for FGF-6 in skeletal muscle regeneration. Genes Dev. 1997, 11, 2040–2051. [Google Scholar] [CrossRef]
- Yoshida, T.; Delafontaine, P. Mechanisms of IGF-1-Mediated Regulation of Skeletal Muscle Hypertrophy and Atrophy. Cells 2020, 9, E1970. [Google Scholar] [CrossRef]
- Li, Y.P. TNF-alpha is a mitogen in skeletal muscle. Am. J. Physiol. Cell Physiol. 2003, 285, C370–C376. [Google Scholar] [CrossRef] [PubMed]
- Serrano, A.L.; Baeza-Raja, B.; Perdiguero, E.; Jardí, M.; Muñoz-Cánoves, P. Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. Cell Metab. 2008, 7, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Kurek, J.B.; Bower, J.J.; Romanella, M.; Koentgen, F.; Murphy, M.; Austin, L. The role of leukemia inhibitory factor in skeletal muscle regeneration. Muscle Nerve 1997, 20, 815–822. [Google Scholar] [CrossRef]
- Allouh, M.Z.; Yablonka-Reuveni, Z.; Rosser, B.W.C. Pax7 reveals a greater frequency and concentration of satellite cells at the ends of growing skeletal muscle fibers. J. Histochem. Cytochem. 2008, 56, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Blau, H.M.; Cosgrove, B.D.; Ho, A.T.V. The central role of muscle stem cells in regenerative failure with aging. Nat. Med. 2015, 21, 854–862. [Google Scholar] [CrossRef]
- Day, K.; Shefer, G.; Shearer, A.; Yablonka-Reuveni, Z. The depletion of skeletal muscle satellite cells with age is concomitant with reduced capacity of single progenitors to produce reserve progeny. Dev. Biol. 2010, 340, 330–343. [Google Scholar] [CrossRef]
- Shefer, G.; Rauner, G.; Yablonka-Reuveni, Z.; Benayahu, D. Reduced satellite cell numbers and myogenic capacity in aging can be alleviated by endurance exercise. PLoS ONE 2010, 5, e13307. [Google Scholar] [CrossRef]
- Tierney, M.T.; Stec, M.J.; Rulands, S.; Simons, B.D.; Sacco, A. Muscle Stem Cells Exhibit Distinct Clonal Dynamics in Response to Tissue Repair and Homeostatic Aging. Cell Stem. Cell 2018, 22, 119–127. [Google Scholar] [CrossRef]
- Blau, H.M.; Webster, C.; Pavlath, G.K. Defective myoblasts identified in Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. USA 1983, 80, 4856–4860. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.R.; Meyer, G.A. Skeletal muscle explants: Ex-vivo models to study cellular behavior in a complex tissue environment. Connect. Tissue Res. 2020, 61, 248–261. [Google Scholar] [CrossRef]
- Park, K.H.; Brotto, L.; Lehoang, O.; Brotto, M.; Ma, J.; Zhao, X. Ex vivo assessment of contractility, fatigability and alternans in isolated skeletal muscles. J. Vis. Exp. 2012, 69, e4198. [Google Scholar] [CrossRef]
- Bekoff, A.; Betz, W. Properties of isolated adult rat muscle fibres maintained in tissue culture. J. Physiol. 1977, 271, 537–547. [Google Scholar] [CrossRef]
- Bischoff, R. Regeneration of single skeletal muscle fibers in vitro. Anat. Rec. 1975, 182, 215–235. [Google Scholar] [CrossRef]
- Pasut, A.; Jones, A.E.; Rudnicki, M.A. Isolation and culture of individual myofibers and their satellite cells from adult skeletal muscle. J. Vis. Exp. 2013, 73, e50074. [Google Scholar] [CrossRef]
- Keire, P.; Shearer, A.; Shefer, G.; Yablonka-Reuveni, Z. Isolation and culture of skeletal muscle myofibers as a means to analyze satellite cells. Methods Mol. Biol. 2013, 946, 431–468. [Google Scholar] [CrossRef] [PubMed]
- Stuelsatz, P.; Keire, P.; Yablonka-Reuveni, Z. Isolation, Culture, and Immunostaining of Skeletal Muscle Myofibers from Wildtype and Nestin-GFP Mice as a Means to Analyze Satellite Cell. Methods Mol. Biol. 2017, 1556, 51–102. [Google Scholar] [CrossRef] [PubMed]
- Brun, C.E.; Wang, Y.X.; Rudnicki, M.A. Single EDL Myofiber Isolation for Analyses of Quiescent and Activated Muscle Stem Cells. Methods Mol. Biol. 2018, 1686, 149–159. [Google Scholar] [CrossRef]
- Renzini, A.; Benedetti, A.; Bouchè, M.; Silvestroni, L.; Adamo, S.; Moresi, V. Culture conditions influence satellite cell activation and survival of single myofibers. Eur. J. Transl. Myol. 2018, 28, 7567. [Google Scholar] [CrossRef] [PubMed]
- Kjøbsted, R.; Kido, K.; Larsen, J.K.; Jørgensen, N.O.; Birk, J.B.; Hellsten, Y.; Wojtaszewski, J.F.P. Measurement of Insulin- and Contraction-Stimulated Glucose Uptake in Isolated and Incubated Mature Skeletal Muscle from Mice. J. Vis. Exp. 2021, 171, e61398. [Google Scholar] [CrossRef] [PubMed]
- Olsson, K.; Cheng, A.J.; Alam, S.; Al-Ameri, M.; Rullman, E.; Westerblad, H.; Lanner, J.T.; Bruton, J.D.; Gustafsson, T. Intracellular Ca2+-handling differs markedly between intact human muscle fibers and myotubes. Skelet. Muscle 2015, 5, 26. [Google Scholar] [CrossRef] [PubMed]
- Olsson, K.; Cheng, A.J.; Al-Ameri, M.; Wyckelsma, V.L.; Rullman, E.; Westerblad, H.; Lanner, J.T.; Gustafsson, T.; Bruton, J. Impaired sarcoplasmic reticulum Ca2+ release is the major cause of fatigue-induced force loss in intact single fibres from human intercostal muscle. J. Physiol. 2020, 598, 773–787. [Google Scholar] [CrossRef]
- Tabakov, V.Y.; Zinov’eva, O.E.; Voskresenskaya, O.N.; Skoblov, M.Y. Isolation and Characterization of Human Myoblast Culture In Vitro for Technologies of Cell and Gene Therapy of Skeletal Muscle Pathologies. Bull Exp. Biol. Med. 2018, 164, 536–542. [Google Scholar] [CrossRef]
- Uezumi, A.; Nakatani, M.; Ikemoto-Uezumi, M.; Yamamoto, N.; Morita, M.; Yamaguchi, A.; Yamada, H.; Kasai, T.; Masuda, S.; Narita, A.; et al. Cell-Surface Protein Profiling Identifies Distinctive Markers of Progenitor Cells in Human Skeletal Muscle. Stem. Cell Rep. 2016, 7, 263–278. [Google Scholar] [CrossRef]
- Logan, M.S.; Propst, J.T.; Nottingham, J.M.; Goodwin, R.L.; Pabon, D.F.; Terracio, L.; Yost, M.J.; Fann, S.A. Human satellite progenitor cells for use in myofascial repair: Isolation and characterization. Ann. Plast. Surg. 2010, 64, 794–799. [Google Scholar] [CrossRef]
- Čamernik, K.; Marc, J.; Zupan, J. Human Skeletal Muscle-Derived Mesenchymal Stem/Stromal Cell Isolation and Growth Kinetics Analysis. Methods Mol. Biol. 2019, 2045, 119–129. [Google Scholar] [CrossRef]
- Soriano-Arroquia, A.; Clegg, P.D.; Molloy, A.P.; Goljanek-Whysall, K. Preparation and Culture of Myogenic Precursor Cells/Primary Myoblasts from Skeletal Muscle of Adult and Aged Humans. J. Vis. Exp. 2017, 120, e55047. [Google Scholar] [CrossRef] [PubMed]
- Agley, C.C.; Rowlerson, A.M.; Velloso, C.P.; Lazarus, N.L.; Harridge, S.D.R. Isolation and quantitative immunocytochemical characterization of primary myogenic cells and fibroblasts from human skeletal muscle. J. Vis. Exp. 2015, 95, 52049. [Google Scholar] [CrossRef] [PubMed]
- Lecourt, S.; Marolleau, J.P.; Fromigué, O.; Vauchez, K.; Andriamanalijaona, R.; Ternaux, B.; Lacassagne, M.-N.; Robert, I.; Boumédiene, K.; Chéreau, F.; et al. Characterization of distinct mesenchymal-like cell populations from human skeletal muscle in situ and in vitro. Exp. Cell Res. 2010, 316, 2513–2526. [Google Scholar] [CrossRef] [PubMed]
- Charville, G.W.; Cheung, T.H.; Yoo, B.; Santos, P.J.; Lee, G.K.; Shrager, J.B.; Rando, T.A. Ex Vivo Expansion and In Vivo Self-Renewal of Human Muscle Stem Cells. Stem. Cell Rep. 2015, 5, 621–632. [Google Scholar] [CrossRef]
- Garcia, S.M.; Tamaki, S.; Lee, S.; Wong, A.; Jose, A.; Dreux, J.; Kouklis, G.; Sbitany, H.; Seth, R.; Knott, P.D.; et al. High-Yield Purification, Preservation, and Serial Transplantation of Human Satellite Cells. Stem. Cell Rep. 2018, 10, 1160–1174. [Google Scholar] [CrossRef]
- Hartley, R.S.; Yablonka-Reuveni, Z. Long-term maintenance of primary myogenic cultures on a reconstituted basement membrane. In Vitro Cell Dev. Biol. 1990, 26, 955–961. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.R.; Binti Ismail, A.; Chee, S.C.; Bin Laupa, M.S.; Jaffri, F.B.; Saberi, S.E.M.; Idrus, R.B.H. One-Step Purification of Human Skeletal Muscle Myoblasts and Subsequent Expansion Using Laminin-Coated Surface. Tissue Eng. Part C Methods 2015, 21, 1135–1142. [Google Scholar] [CrossRef] [PubMed]
- Kühl, U.; Ocalan, M.; Timpl, R.; von der Mark, K. Role of laminin and fibronectin in selecting myogenic versus fibrogenic cells from skeletal muscle cells in vitro. Dev. Biol. 1986, 117, 628–635. [Google Scholar] [CrossRef]
- Gaster, M.; Beck-Nielsen, H.; Schrøder, H.D. Proliferation conditions for human satellite cells. The fractional content of satellite cells. APMIS 2001, 109, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Romagnoli, C.; Zonefrati, R.; Sharma, P.; Innocenti, M.; Cianferotti, L.; Brandi, M.L. Characterization of Skeletal Muscle Endocrine Control in an In Vitro Model of Myogenesis. Calcif. Tissue Int. 2020, 107, 18–30. [Google Scholar] [CrossRef]
- Gharaibeh, B.; Lu, A.; Tebbets, J.; Zheng, B.; Feduska, J.; Crisan, M.; Péault, B.; Cummins, J.; Huard, J. Isolation of a slowly adhering cell fraction containing stem cells from murine skeletal muscle by the preplate technique. Nat. Protoc. 2008, 3, 1501–1509. [Google Scholar] [CrossRef]
- Gaster, M.; Kristensen, S.R.; Beck-Nielsen, H.; Schrøder, H.D. A cellular model system of differentiated human myotubes. APMIS 2001, 109, 735–744. [Google Scholar] [CrossRef]
- Kleinman, H.K.; Martin, G.R. Matrigel: Basement membrane matrix with biological activity. Semin. Cancer Biol. 2005, 15, 378–386. [Google Scholar] [CrossRef]
- Passaniti, A.; Kleinman, H.K.; Martin, G.R. Matrigel: History/background, uses, and future applications. J. Cell Commun. Signal. 2021. [Google Scholar] [CrossRef]
- Funanage, V.L.; Smith, S.M.; Minnich, M.A. Entactin promotes adhesion and long-term maintenance of cultured regenerated skeletal myotubes. J. Cell Physiol. 1992, 150, 251–257. [Google Scholar] [CrossRef]
- Grefte, S.; Vullinghs, S.; Kuijpers-Jagtman, A.M.; Torensma, R.; Von den Hoff, J.W. Matrigel, but not collagen I, maintains the differentiation capacity of muscle derived cells in vitro. Biomed. Mater. 2012, 7, 055004. [Google Scholar] [CrossRef]
- Blau, H.M.; Webster, C. Isolation and characterization of human muscle cells. Proc. Natl. Acad. Sci. USA 1981, 78, 5623–5627. [Google Scholar] [CrossRef] [PubMed]
- Pavlath, G.K. Isolation, purification, and growth of human skeletal muscle cells. Methods Mol. Med. 1996, 2, 307–317. [Google Scholar] [CrossRef]
- Bareja, A.; Holt, J.A.; Luo, G.; Chang, C.; Lin, J.; Hinken, A.C.; Freudenberg, J.; Kraus, W.E.; Evans, W.J.; Billin, A.N. Human and mouse skeletal muscle stem cells: Convergent and divergent mechanisms of myogenesis. PLoS ONE 2014, 9, e90398. [Google Scholar] [CrossRef]
- Castiglioni, A.; Hettmer, S.; Lynes, M.D.; Rao, T.N.; Tchessalova, D.; Sinha, I.; Lee, B.; Tseng, Y.-H.; Wagers, A.J. Isolation of progenitors that exhibit myogenic/osteogenic bipotency in vitro by fluorescence-activated cell sorting from human fetal muscle. Stem. Cell Rep. 2014, 2, 92–106. [Google Scholar] [CrossRef]
- Webster, C.; Pavlath, G.K.; Parks, D.R.; Walsh, F.S.; Blau, H.M. Isolation of human myoblasts with the fluorescence-activated cell sorter. Exp. Cell Res. 1988, 174, 252–265. [Google Scholar] [CrossRef]
- Fukada, S.I.; Higuchi, S.; Segawa, M.; Koda, K.-I.; Yamamoto, Y.; Tsujikawa, K.; Kohama, Y.; Uezumi, A.; Imamura, M.; Miyagoe-Suzuki, Y.; et al. Purification and cell-surface marker characterization of quiescent satellite cells from murine skeletal muscle by a novel monoclonal antibody. Exp. Cell Res. 2004, 296, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Montarras, D.; Morgan, J.; Collins, C.; Relaix, F.; Zaffran, S.; Cumano, A.; Partridge, T.; Buckingham, M. Direct isolation of satellite cells for skeletal muscle regeneration. Science 2005, 309, 2064–2067. [Google Scholar] [CrossRef]
- Tsuchiya, Y.; Kitajima, Y.; Masumoto, H.; Ono, Y. Damaged Myofiber-Derived Metabolic Enzymes Act as Activators of Muscle Satellite Cells. Stem. Cell Rep. 2020, 15, 926–940. [Google Scholar] [CrossRef] [PubMed]
- Sacco, A.; Doyonnas, R.; Kraft, P.; Vitorovic, S.; Blau, H.M. Self-renewal and expansion of single transplanted muscle stem cells. Nature 2008, 456, 502–506. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, B.D.; Sacco, A.; Gilbert, P.M.; Blau, H.M. A home away from home: Challenges and opportunities in engineering in vitro muscle satellite cell niches. Differentiation 2009, 78, 185–194. [Google Scholar] [CrossRef]
- Machado, L.; Esteves de Lima, J.; Fabre, O.; Proux, C.; Legendre, R.; Szegedi, A.; Varet, H.; Ingerslev, L.R.; Barres, R.; Relaix, F.; et al. In Situ Fixation Redefines Quiescence and Early Activation of Skeletal Muscle Stem Cells. Cell Rep. 2017, 21, 1982–1993. [Google Scholar] [CrossRef]
- Chowdhury, S.R.; Muneyuki, Y.; Takezawa, Y.; Kino-Oka, M.; Saito, A.; Sawa, Y.; Taya, M. Growth and differentiation potentials in confluent state of culture of human skeletal muscle myoblasts. J. Biosci. Bioeng. 2010, 109, 310–313. [Google Scholar] [CrossRef] [PubMed]
- Krauss, R.S.; Cole, F.; Gaio, U.; Takaesu, G.; Zhang, W.; Kang, J.S. Close encounters: Regulation of vertebrate skeletal myogenesis by cell-cell contact. J. Cell Sci. 2005, 118 Pt 11, 2355–2362. [Google Scholar] [CrossRef]
- Bigot, A.; Jacquemin, V.; Debacq-Chainiaux, F.; Butler-Browne, G.S.; Toussaint, O.; Furling, D.; Mouly, V. Replicative aging down-regulates the myogenic regulatory factors in human myoblasts. Biol. Cell 2008, 100, 189–199. [Google Scholar] [CrossRef]
- Alsharidah, M.; Lazarus, N.R.; George, T.E.; Agley, C.C.; Velloso, C.P.; Harridge, S.D.R. Primary human muscle precursor cells obtained from young and old donors produce similar proliferative, differentiation and senescent profiles in culture. Aging Cell 2013, 12, 333–344. [Google Scholar] [CrossRef]
- Renault, V.; Thornell, L.E.; Eriksson, P.O.; Butler-Browne, G.; Mouly, V.; Thorne, L.E. Regenerative potential of human skeletal muscle during aging. Aging Cell 2002, 1, 132–139. [Google Scholar] [CrossRef]
- Shay, J.W.; Wright, W.E. Senescence and immortalization: Role of telomeres and telomerase. Carcinogenesis 2005, 26, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.H.; Mouly, V.; Cooper, R.N.; Mamchaoui, K.; Bigot, A.; Shay, J.W.; Di Santo, J.; Butler-Browne, G.; Wright, W.E. Cellular senescence in human myoblasts is overcome by human telomerase reverse transcriptase and cyclin-dependent kinase 4: Consequences in aging muscle and therapeutic strategies for muscular dystrophies. Aging Cell 2007, 6, 515–523. [Google Scholar] [CrossRef]
- Mamchaoui, K.; Trollet, C.; Bigot, A.; Negroni, E.; Chaouch, S.; Wolff, A.; Kandalla, P.K.; Marie, S.; Di Santo, J.; Guily, J.L.S.; et al. Immortalized pathological human myoblasts: Towards a universal tool for the study of neuromuscular disorders. Skelet. Muscle 2011, 1, 34. [Google Scholar] [CrossRef]
- Chal, J.; Oginuma, M.; Al Tanoury, Z.; Gobert, B.; Sumara, O.; Hick, A.; Bousson, F.; Zidouni, Y.; Mursch, C.; Moncuquet, P.; et al. Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat. Biotechnol. 2015, 33, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Shelton, M.; Metz, J.; Liu, J.; Carpenedo, R.L.; Demers, S.-P.; Stanford, W.L.; Skerjanc, I.S. Derivation and expansion of PAX7-positive muscle progenitors from human and mouse embryonic stem cells. Stem. Cell Rep. 2014, 3, 516–529. [Google Scholar] [CrossRef] [PubMed]
- Darabi, R.; Gehlbach, K.; Bachoo, R.M.; Kamath, S.; Osawa, M.; Kamm, K.E.; Kyba, M.; Perlingeiro, R.C.R. Functional skeletal muscle regeneration from differentiating embryonic stem cells. Nat. Med. 2008, 14, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Magli, A.; Chan, S.S.K.; Oliveira, V.K.; Wu, J.; Darabi, R.; Kyba, M.; Perlingeiro, R.C. Expansion and Purification Are Critical for the Therapeutic Application of Pluripotent Stem Cell-Derived Myogenic Progenitors. Stem. Cell Rep. 2017, 9, 12–22. [Google Scholar] [CrossRef]
- Rao, L.; Qian, Y.; Khodabukus, A.; Ribar, T.; Bursac, N. Engineering human pluripotent stem cells into a functional skeletal muscle tissue. Nat. Commun. 2018, 9, 126. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef]
- Kwon, J.B.; Vankara, A.; Ettyreddy, A.R.; Bohning, J.D.; Gersbach, C.A. Myogenic Progenitor Cell Lineage Specification by CRISPR/Cas9-Based Transcriptional Activators. Stem. Cell Rep. 2020, 14, 755–769. [Google Scholar] [CrossRef]
- Lieber, R.L.; Fridén, J. Clinical significance of skeletal muscle architecture. Clin. Orthop. Relat. Res. 2001, 383, 140–151. [Google Scholar] [CrossRef]
- Juhas, M.; Ye, J.; Bursac, N. Design, evaluation, and application of engineered skeletal muscle. Methods 2016, 99, 81–90. [Google Scholar] [CrossRef]
- Strohman, R.C.; Bayne, E.; Spector, D.; Obinata, T.; Micou-Eastwood, J.; Maniotis, A. Myogenesis and histogenesis of skeletal muscle on flexible membranes in vitro. In Vitro Cell Dev. Biol. 1990, 26, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.T.; Huang, Y.C.; Birla, R.K.; Takayama, S. Microfeature guided skeletal muscle tissue engineering for highly organized 3-dimensional free-standing constructs. Biomaterials 2009, 30, 1150–1155. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Dickinson, C.E.; Finkelstein, E.B.; Neville, C.M.; Sundback, C.A. The role of fibroblasts in self-assembled skeletal muscle. Tissue Eng. Part A. 2011, 17, 2641–2650. [Google Scholar] [CrossRef]
- Dennis, R.G.; Kosnik, P.E.; Gilbert, M.E.; Faulkner, J.A. Excitability and contractility of skeletal muscle engineered from primary cultures and cell lines. Am. J. Physiol. Cell Physiol. 2001, 280, C288–C295. [Google Scholar] [CrossRef] [PubMed]
- Kosnik, P.E.; Faulkner, J.A.; Dennis, R.G. Functional development of engineered skeletal muscle from adult and neonatal rats. Tissue Eng. 2001, 7, 573–584. [Google Scholar] [CrossRef]
- Dennis, R.G.; Kosnik, P.E. Excitability and isometric contractile properties of mammalian skeletal muscle constructs engineered in vitro. In Vitro Cell Dev. Biol. Anim. 2000, 36, 327–335. [Google Scholar] [CrossRef]
- Larkin, L.M.; Calve, S.; Kostrominova, T.Y.; Arruda, E.M. Structure and functional evaluation of tendon-skeletal muscle constructs engineered in vitro. Tissue Eng. 2006, 12, 3149–3158. [Google Scholar] [CrossRef]
- Nagamori, E.; Ngo, T.X.; Takezawa, Y.; Saito, A.; Sawa, Y.; Shimizu, T.; Okano, T.; Taya, M.; Kino-Oka, M. Network formation through active migration of human vascular endothelial cells in a multilayered skeletal myoblast sheet. Biomaterials 2013, 34, 662–668. [Google Scholar] [CrossRef]
- Takahashi, H.; Shimizu, T.; Nakayama, M.; Yamato, M.; Okano, T. The use of anisotropic cell sheets to control orientation during the self-organization of 3D muscle tissue. Biomaterials 2013, 34, 7372–7380. [Google Scholar] [CrossRef]
- Takahashi, H.; Shimizu, T.; Nakayama, M.; Yamato, M.; Okano, T. Anisotropic cellular network formation in engineered muscle tissue through the self-organization of neurons and endothelial cells. Adv. Healthc. Mater. 2015, 4, 356–360. [Google Scholar] [CrossRef]
- Takahashi, H.; Shimizu, T.; Okano, T. Engineered Human Contractile Myofiber Sheets as a Platform for Studies of Skeletal Muscle Physiology. Sci. Rep. 2018, 8, 13932. [Google Scholar] [CrossRef]
- Arsic, N.; Mamaeva, D.; Lamb, N.J.; Fernandez, A. Muscle-derived stem cells isolated as non-adherent population give rise to cardiac, skeletal muscle and neural lineages. Exp. Cell Res. 2008, 314, 1266–1280. [Google Scholar] [CrossRef]
- Westerman, K.A.; Penvose, A.; Yang, Z.; Allen, P.D.; Vacanti, C.A. Adult muscle “stem” cells can be sustained in culture as free-floating myospheres. Exp. Cell Res. 2010, 316, 1966–1976. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Li, Y.; Chen, C.; Stoelzel, K.; Kaufmann, A.M.; Albers, A.E. Human skeletal muscle-derived stem cells retain stem cell properties after expansion in myosphere culture. Exp. Cell Res. 2011, 317, 1016–1027. [Google Scholar] [CrossRef] [PubMed]
- Westerman, K.A. Myospheres are composed of two cell types: One that is myogenic and a second that is mesenchymal. PLoS ONE 2015, 10, e0116956. [Google Scholar] [CrossRef]
- Bian, W.; Bursac, N. Engineered skeletal muscle tissue networks with controllable architecture. Biomaterials 2009, 30, 1401–1412. [Google Scholar] [CrossRef] [PubMed]
- Lev, R.; Seliktar, D. Hydrogel biomaterials and their therapeutic potential for muscle injuries and muscular dystrophies. J. R. Soc. Interface 2018, 15, 20170380. [Google Scholar] [CrossRef] [PubMed]
- Okano, T.; Matsuda, T. Hybrid muscular tissues: Preparation of skeletal muscle cell-incorporated collagen gels. Cell Transplant. 1997, 6, 109–118. [Google Scholar] [CrossRef]
- Huang, Y.C.; Dennis, R.G.; Larkin, L.; Baar, K. Rapid formation of functional muscle in vitro using fibrin gels. J. Appl. Physiol. 2005, 98, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Liao, I.C.; Liu, J.B.; Bursac, N.; Leong, K.W. Effect of Electromechanical Stimulation on the Maturation of Myotubes on Aligned Electrospun Fibers. Cell Mol. Bioeng. 2008, 1, 133–145. [Google Scholar] [CrossRef]
- De Coppi, P.; Bellini, S.; Conconi, M.T.; Sabatti, M.; Simonato, E.; Gamba, P.G.; Nussdorfer, G.G.; Parnigotto, P.P. Myoblast-acellular skeletal muscle matrix constructs guarantee a long-term repair of experimental full-thickness abdominal wall defects. Tissue Eng. 2006, 12, 1929–1936. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.J.; Yates, A.J.; Weber, D.J.; Qureshi, I.R.; Stolz, D.B.; Gilbert, T.W.; Badylak, S.F., Jr. Xenogeneic extracellular matrix as an inductive scaffold for regeneration of a functioning musculotendinous junction. Tissue Eng. Part A 2010, 16, 3309–3317. [Google Scholar] [CrossRef] [PubMed]
- Rhim, C.; Lowell, D.A.; Reedy, M.C.; Slentz, D.H.; Zhang, S.J.; Kraus, W.E.; Truskey, G.A. Morphology and ultrastructure of differentiating three-dimensional mammalian skeletal muscle in a collagen gel. Muscle Nerve 2007, 36, 71–80. [Google Scholar] [CrossRef]
- Khodabukus, A.; Prabhu, N.; Wang, J.; Bursac, N. In Vitro Tissue-Engineered Skeletal Muscle Models for Studying Muscle Physiology and Disease. Adv. Healthc. Mater. 2018, 7, e1701498. [Google Scholar] [CrossRef] [PubMed]
- Vandenburgh, H.H.; Karlisch, P.; Farr, L. Maintenance of highly contractile tissue-cultured avian skeletal myotubes in collagen gel. In Vitro Cell Dev. Biol. 1988, 24, 166–174. [Google Scholar] [CrossRef]
- Shansky, J.; Del Tatto, M.; Chromiak, J.; Vandenburgh, H. A simplified method for tissue engineering skeletal muscle organoids in vitro. In Vitro Cell Dev. Biol. Anim. 1997, 33, 659–661. [Google Scholar] [CrossRef] [PubMed]
- Powell, C.A.; Smiley, B.L.; Mills, J.; Vandenburgh, H.H. Mechanical stimulation improves tissue-engineered human skeletal muscle. Am. J. Physiol. Cell Physiol. 2002, 283, C1557–C1565. [Google Scholar] [CrossRef]
- Kjaer, M. Role of extracellular matrix in adaptation of tendon and skeletal muscle to mechanical loading. Physiol. Rev. 2004, 84, 649–698. [Google Scholar] [CrossRef] [PubMed]
- Brady, M.A.; Lewis, M.P.; Mudera, V. Synergy between myogenic and non-myogenic cells in a 3D tissue-engineered craniofacial skeletal muscle construct. J. Tissue Eng. Regen. Med. 2008, 2, 408–417. [Google Scholar] [CrossRef]
- Gholobova, D.; Decroix, L.; Van Muylder, V.; Desender, L.; Gerard, M.; Carpentier, G.; VanDenburgh, H.; Thorrez, L. Endothelial Network Formation Within Human Tissue-Engineered Skeletal Muscle. Tissue Eng. Part A 2015, 21, 2548–2558. [Google Scholar] [CrossRef] [PubMed]
- Mann, C.J.; Perdiguero, E.; Kharraz, Y.; Aguilar, S.; Pessina, P.; Serrano, A.L.; Muñoz-Cánoves, P. Aberrant repair and fibrosis development in skeletal muscle. Skelet. Muscle 2011, 1, 21. [Google Scholar] [CrossRef]
- Stearns-Reider, K.M.; D’Amore, A.; Beezhold, K.; Rothrauff, B.; Cavalli, L.; Wagner, W.; Vorp, D.A.; Tsamis, A.; Shinde, S.; Zhang, C.; et al. Aging of the skeletal muscle extracellular matrix drives a stem cell fibrogenic conversion. Aging Cell 2017, 16, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Hinds, S.; Bian, W.; Dennis, R.G.; Bursac, N. The role of extracellular matrix composition in structure and function of bioengineered skeletal muscle. Biomaterials 2011, 32, 3575–3583. [Google Scholar] [CrossRef] [PubMed]
- Thorsteinsdóttir, S.; Deries, M.; Cachaço, A.S.; Bajanca, F. The extracellular matrix dimension of skeletal muscle development. Dev. Biol. 2011, 354, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Ito, A.; Kawabe, Y.; Nagamori, E.; Kamihira, M. Enhanced contractile force generation by artificial skeletal muscle tissues using IGF-I gene-engineered myoblast cells. J. Biosci. Bioeng. 2011, 112, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Mosesson, M.W. Fibrinogen and fibrin structure and functions. J. Thromb. Haemost. 2005, 3, 1894–1904. [Google Scholar] [CrossRef]
- Ahmed, T.A.E.; Dare, E.V.; Hincke, M. Fibrin: A versatile scaffold for tissue engineering applications. Tissue Eng. Part B Rev. 2008, 14, 199–215. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, F.; Lelkes, P.I. Fine-tuning of a three-dimensional microcarrier-based angiogenesis assay for the analysis of endothelial-mesenchymal cell co-cultures in fibrin and collagen gels. Angiogenesis 2006, 9, 111–125. [Google Scholar] [CrossRef]
- Chiron, S.; Tomczak, C.; Duperray, A.; Lainé, J.; Bonne, G.; Eder, A.; Hansen, A.; Eschenhagen, T.; Verdier, C.; Coirault, C. Complex interactions between human myoblasts and the surrounding 3D fibrin-based matrix. PLoS ONE 2012, 7, e36173. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef]
- Maffioletti, S.M.; Sarcar, S.; Henderson, A.B.H.; Mannhardt, I.; Pinton, L.; Moyle, L.; Steele-Stallard, H.; Cappellari, O.; Wells, K.E.; Ferrari, G.; et al. Three-Dimensional Human iPSC-Derived Artificial Skeletal Muscles Model Muscular Dystrophies and Enable Multilineage Tissue Engineering. Cell Rep. 2018, 23, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Madden, L.; Juhas, M.; Kraus, W.E.; Truskey, G.A.; Bursac, N. Bioengineered human myobundles mimic clinical responses of skeletal muscle to drugs. Elife 2015, 4, e04885. [Google Scholar] [CrossRef] [PubMed]
- Khodabukus, A.; Baar, K. Factors That Affect Tissue-Engineered Skeletal Muscle Function and Physiology. Cells Tissues Organs. 2016, 202, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Langelaan, M.L.P.; Boonen, K.J.M.; Rosaria-Chak, K.Y.; van der Schaft, D.W.J.; Post, M.J.; Baaijens, F.P.T. Advanced maturation by electrical stimulation: Differences in response between C2C12 and primary muscle progenitor cells. J. Tissue Eng. Regen. Med. 2011, 5, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Weist, M.R.; Wellington, M.S.; Bermudez, J.E.; Kostrominova, T.Y.; Mendias, C.L.; Arruda, E.M.; Larkin, L.M. TGF-β1 enhances contractility in engineered skeletal muscle. J. Tissue Eng. Regen. Med. 2013, 7, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Roche, S.M.; Gumucio, J.P.; Brooks, S.V.; Mendias, C.L.; Claflin, D.R. Measurement of Maximum Isometric Force Generated by Permeabilized Skeletal Muscle Fibers. J. Vis. Exp. 2015, 100, e52695. [Google Scholar] [CrossRef]
- Bach, A.D.; Beier, J.P.; Stern-Staeter, J.; Horch, R.E. Skeletal muscle tissue engineering. J. Cell Mol. Med. 2004, 8, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kim, I.; Seol, Y.J.; Ko, I.K.; Yoo, J.J.; Atala, A.; Lee, S.J. Neural cell integration into 3D bioprinted skeletal muscle constructs accelerates restoration of muscle function. Nat. Commun. 2020, 11, 1025. [Google Scholar] [CrossRef]
- Bersini, S.; Gilardi, M.; Ugolini, G.S.; Sansoni, V.; Talò, G.; Perego, S.; Zanotti, S.; Ostano, P.; Mora, M.; Soncini, M.; et al. Engineering an Environment for the Study of Fibrosis: A 3D Human Muscle Model with Endothelium Specificity and Endomysium. Cell Rep. 2018, 25, 3858–3868.e4. [Google Scholar] [CrossRef] [PubMed]
- Afshar Bakooshli, M.; Lippmann, E.S.; Mulcahy, B.; Iyer, N.; Nguyen, C.T.; Tung, K.; Stewart, B.A.; van den Dorpel, H.; Fuehrmann, T.; Shoichet, M.; et al. A 3D culture model of innervated human skeletal muscle enables studies of the adult neuromuscular junction. Elife 2019, 8, e44530. [Google Scholar] [CrossRef]
- Gholobova, D.; Terrie, L.; Gerard, M.; Declercq, H.; Thorrez, L. Vascularization of tissue-engineered skeletal muscle constructs. Biomaterials 2020, 235, 119708. [Google Scholar] [CrossRef] [PubMed]
- Perry, L.; Flugelman, M.Y.; Levenberg, S. Elderly Patient-Derived Endothelial Cells for Vascularization of Engineered Muscle. Mol. Ther. 2017, 25, 935–948. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.S.; Lee, S.H.; Park, W.J.; Lee, J.E.; Kim, B.; Han, D.W. Advanced Techniques for Skeletal Muscle Tissue Engineering and Regeneration. Bioengineering 2020, 7, 99. [Google Scholar] [CrossRef] [PubMed]
- Ostrovidov, S.; Salehi, S.; Costantini, M.; Suthiwanich, K.; Ebrahimi, M.; Sadeghian, R.B.; Fujie, T.; Shi, X.; Cannata, S.; Gargioli, C.; et al. 3D Bioprinting in Skeletal Muscle Tissue Engineering. Small 2019, 15, e1805530. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Jun, Y.J.; Kim, D.Y.; Yi, H.-G.; Chae, S.-H.; Kang, J.; Lee, J.; Gao, G.; Kong, J.-S.; Jang, J.; et al. A 3D cell printed muscle construct with tissue-derived bioink for the treatment of volumetric muscle loss. Biomaterials 2019, 206, 160–169. [Google Scholar] [CrossRef]
- Xu, C.; Tabebordbar, M.; Iovino, S.; Ciarlo, C.; Liu, J.; Castiglioni, A.; Price, E.; Liu, M.; Barton, E.R.; Kahn, C.R.; et al. A zebrafish embryo culture system defines factors that promote vertebrate myogenesis across species. Cell 2013, 155, 909–921. [Google Scholar] [CrossRef]
- Mueller, A.L.; Bloch, R.J. Skeletal muscle cell transplantation: Models and methods. J. Muscle Res. Cell Motil. 2020, 41, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Seol, Y.J.; Ko, I.K.; Kang, H.-W.; Lee, Y.K.; Yoo, J.J.; Atala, A.; Lee, S.J. 3D Bioprinted Human Skeletal Muscle Constructs for Muscle Function Restoration. Sci. Rep. 2018, 8, 12307. [Google Scholar] [CrossRef]
- Zhang, Y.; King, O.D.; Rahimov, F.; Jones, T.I.; Ward, C.W.; Kerr, J.P.; Liu, N.; Emerson, C.P.; Kunkel, L.M.; Partridge, T.A.; et al. Human skeletal muscle xenograft as a new preclinical model for muscle disorders. Hum. Mol. Genet. 2014, 23, 3180–3188. [Google Scholar] [CrossRef] [PubMed]
- Britson, K.A.; Black, A.D.; Wagner, K.R.; Lloyd, T.E. Performing Human Skeletal Muscle Xenografts in Immunodeficient Mice. J. Vis. Exp. 2019, 151, e59966. [Google Scholar] [CrossRef] [PubMed]
| Digestive Enzyme | Isolation Method | Plate Coating | Markers | Reference |
|---|---|---|---|---|
| Collagenase II | Enzymatic digestion | ----- | Desmin+ | [33] |
| Collagenase II | FACS | Collagen I | CD56+ | [34] |
| Collagenase II | Enzymatic digestion | Collagen I | Pax7+ MyoD1+ | [35] |
| Collagenase II | Enzymatic digestion | ----- | ----- | [36] |
| Collagenase II/ Dispase | Pre-plating to remove fibroblasts | Laminin | MyoD1+ | [37] |
| Collagenase II/ Dispase | MACS | ----- | CD56+ | [38] |
| Collagenase II/ Trypsin/EDTA | Pre-plating to remove fibroblasts | ----- | CD56+ CD29+ CD34- | [39] |
| Collagenase II/ Dispase | FACS | ECM proteins | CD34- CD45- CD31- | [40] |
| Collagenase II/Trypsin | MACS/FACS | Laminin | CD56+ CD29+ CXCR4+ | [41] |
| Trypsin | Serial Plating | Laminin/ Collagen I | Desmin+ | [43] |
| Trypsin/EDTA | Cell Cloning | ECM proteins | ----- | [45] |
| Collagenase II | Enzymatic digestion | Matrigel | Pax7+ | [46] |
| Trypsin/EDTA | Enzymatic digestion | ECM protein | ----- | [48] |
| Trypsin/EDTA | Cell Cloning | Collagen I | ----- | [53] |
| Trypsin/EDTA | FACS Cell Cloning | Collagen I | CD56+ | [54] |
| Trypsin/EDTA | FACS | Collagen I | CD56+ | [57] |
| Trypsin/EDTA | Pre-plating to remove fibroblasts | ECM proteins | ----- | [73] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romagnoli, C.; Iantomasi, T.; Brandi, M.L. Available In Vitro Models for Human Satellite Cells from Skeletal Muscle. Int. J. Mol. Sci. 2021, 22, 13221. https://doi.org/10.3390/ijms222413221
Romagnoli C, Iantomasi T, Brandi ML. Available In Vitro Models for Human Satellite Cells from Skeletal Muscle. International Journal of Molecular Sciences. 2021; 22(24):13221. https://doi.org/10.3390/ijms222413221
Chicago/Turabian StyleRomagnoli, Cecilia, Teresa Iantomasi, and Maria Luisa Brandi. 2021. "Available In Vitro Models for Human Satellite Cells from Skeletal Muscle" International Journal of Molecular Sciences 22, no. 24: 13221. https://doi.org/10.3390/ijms222413221
APA StyleRomagnoli, C., Iantomasi, T., & Brandi, M. L. (2021). Available In Vitro Models for Human Satellite Cells from Skeletal Muscle. International Journal of Molecular Sciences, 22(24), 13221. https://doi.org/10.3390/ijms222413221







