Serum Metabolomic and Lipidomic Profiling Reveals Novel Biomarkers of Efficacy for Benfotiamine in Alzheimer’s Disease
Abstract
:1. Introduction
2. Results
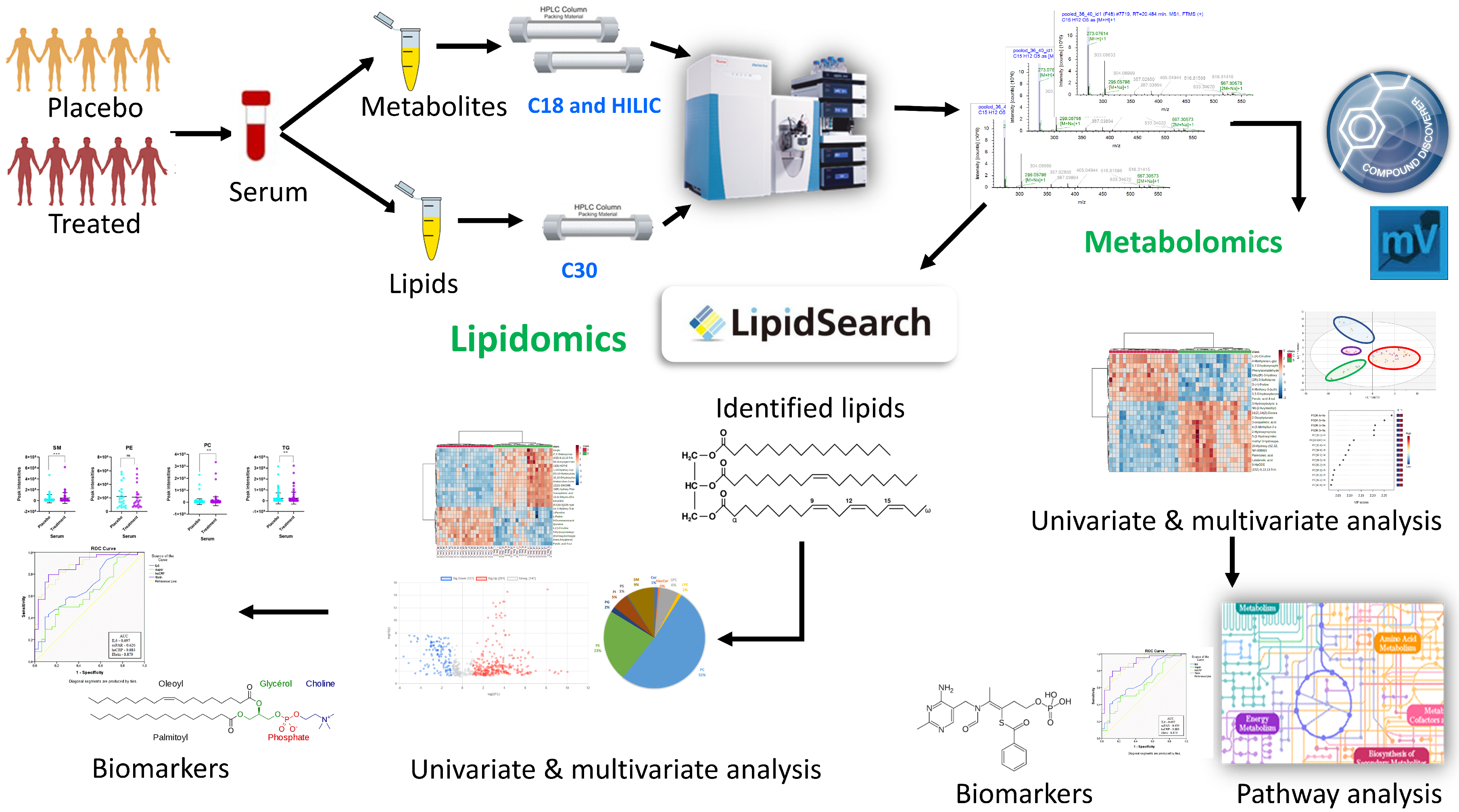
2.1. Study Characteristics and Parallel Metabolomics and Lipidomics Workflows
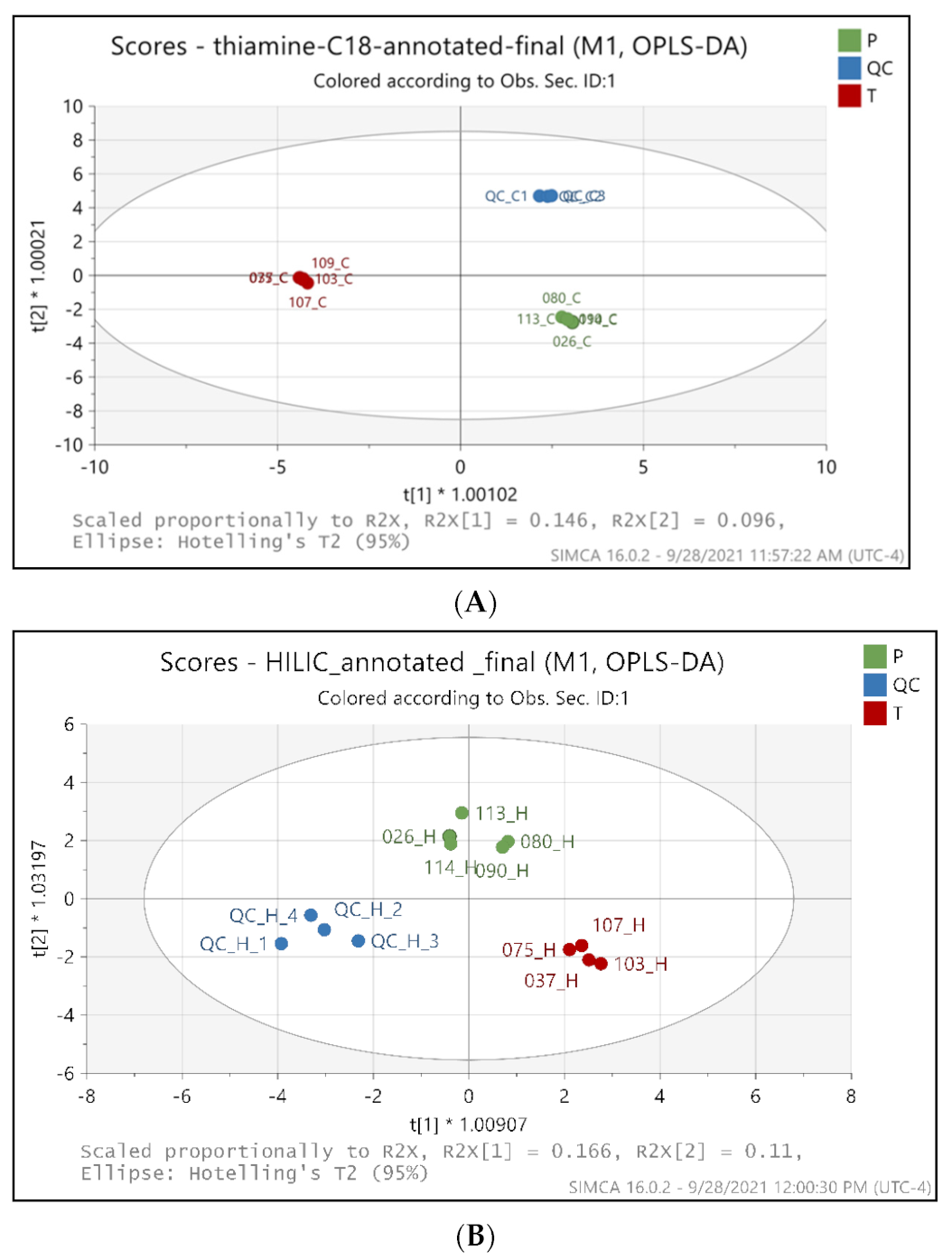
2.2. Metabolic Profiling Distinguishes the BFT Treatment from Placebo Groups
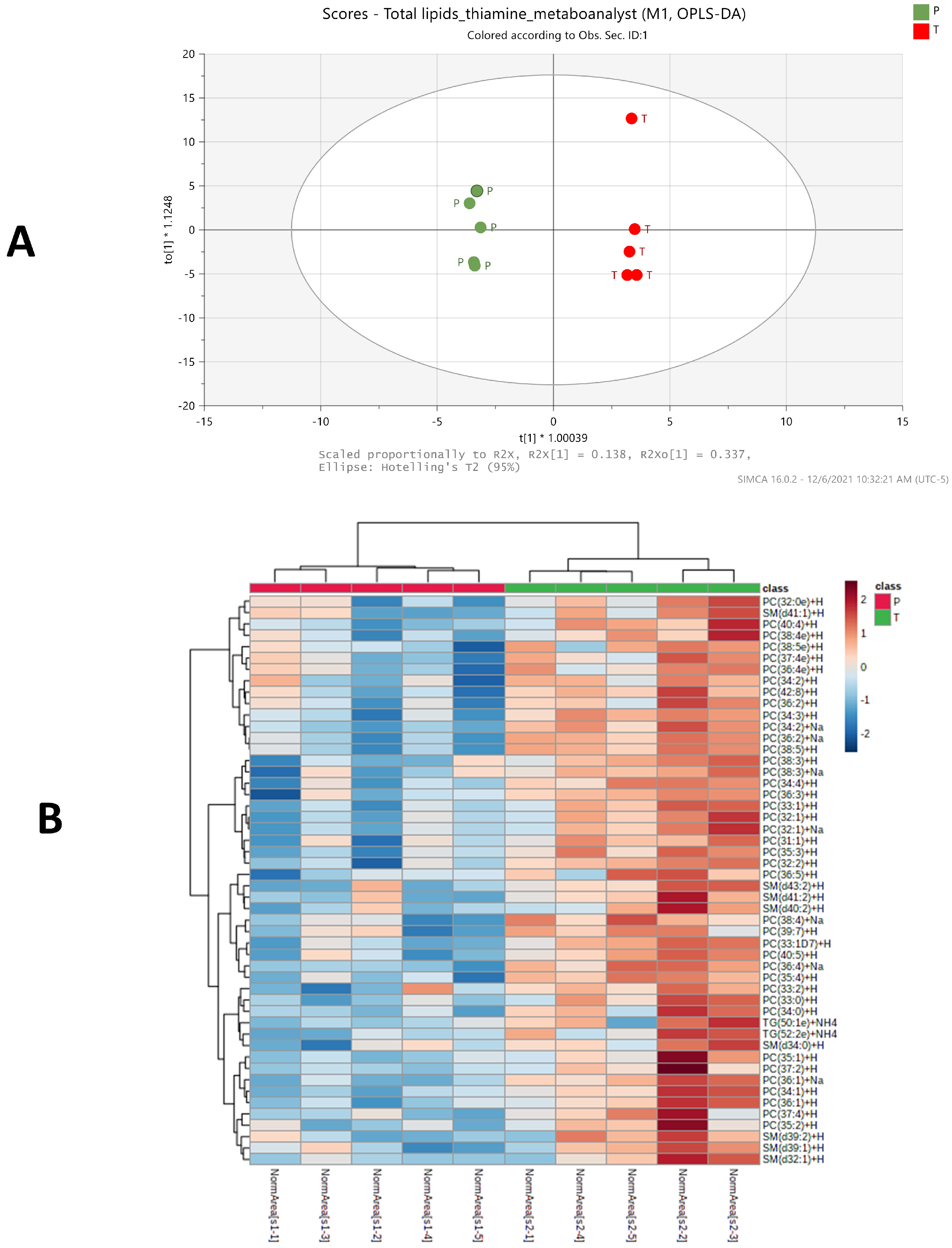
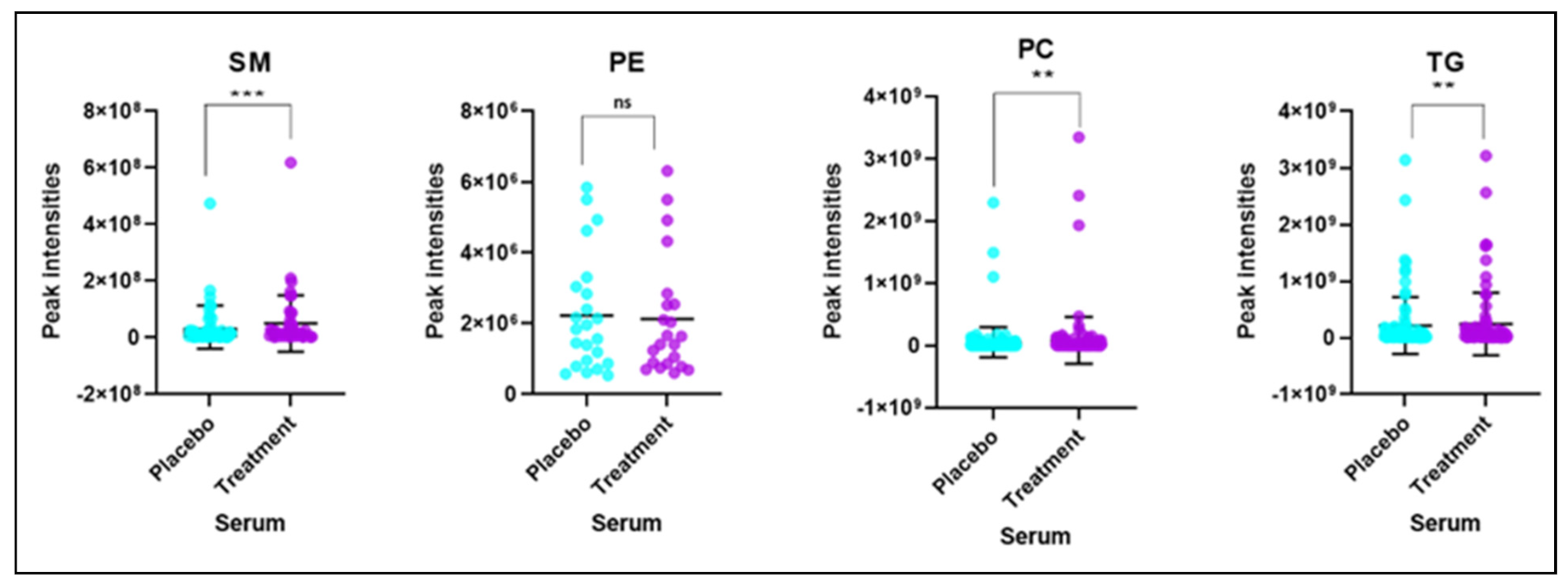
2.3. Significant Changes of Phosphatidylcholine, Sphingomyelin and Triglycerides Species Levels in BFT Treated Group
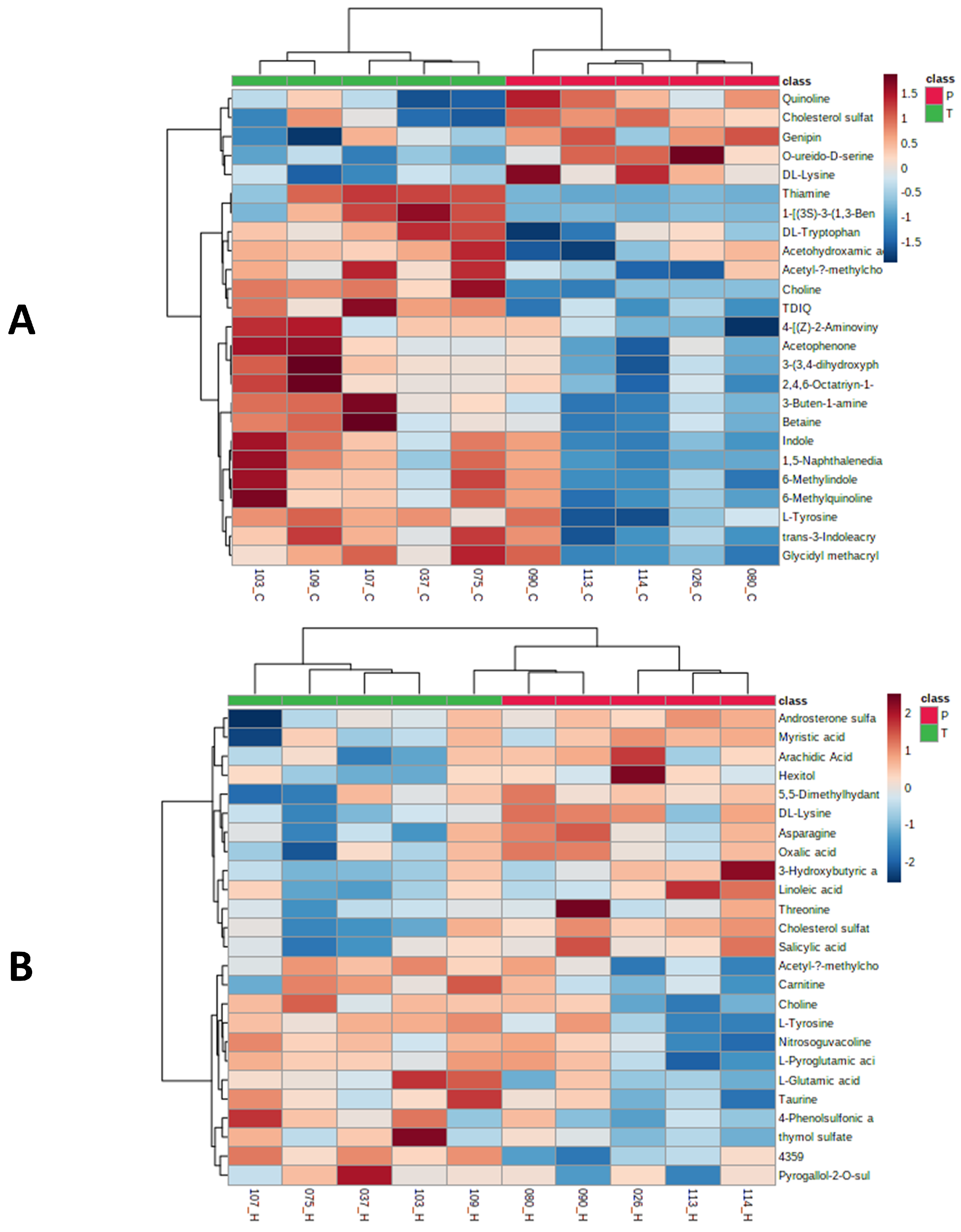
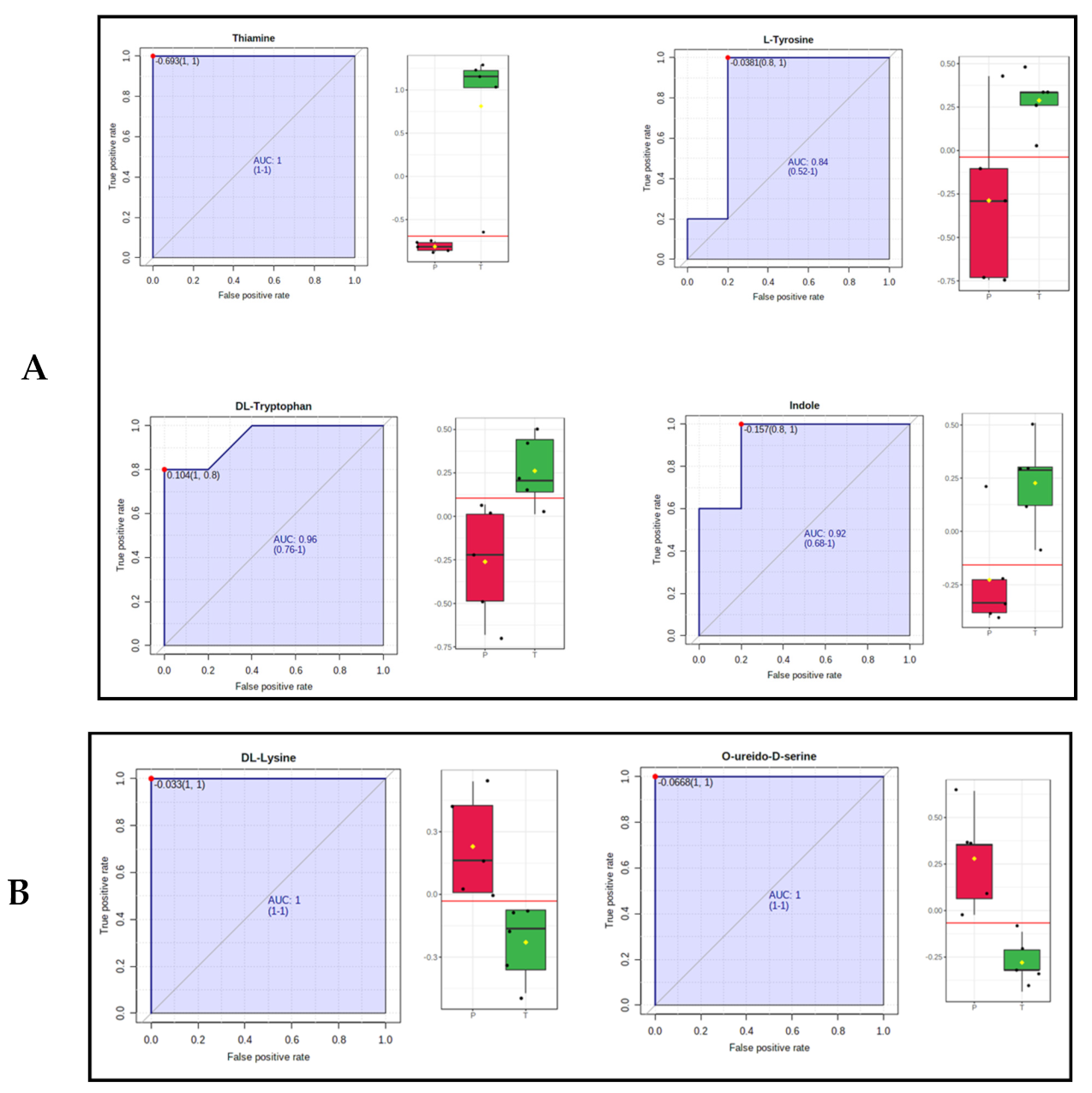
2.4. Identification of Metabolic, Lipid Signatures and Biomarker Candidates
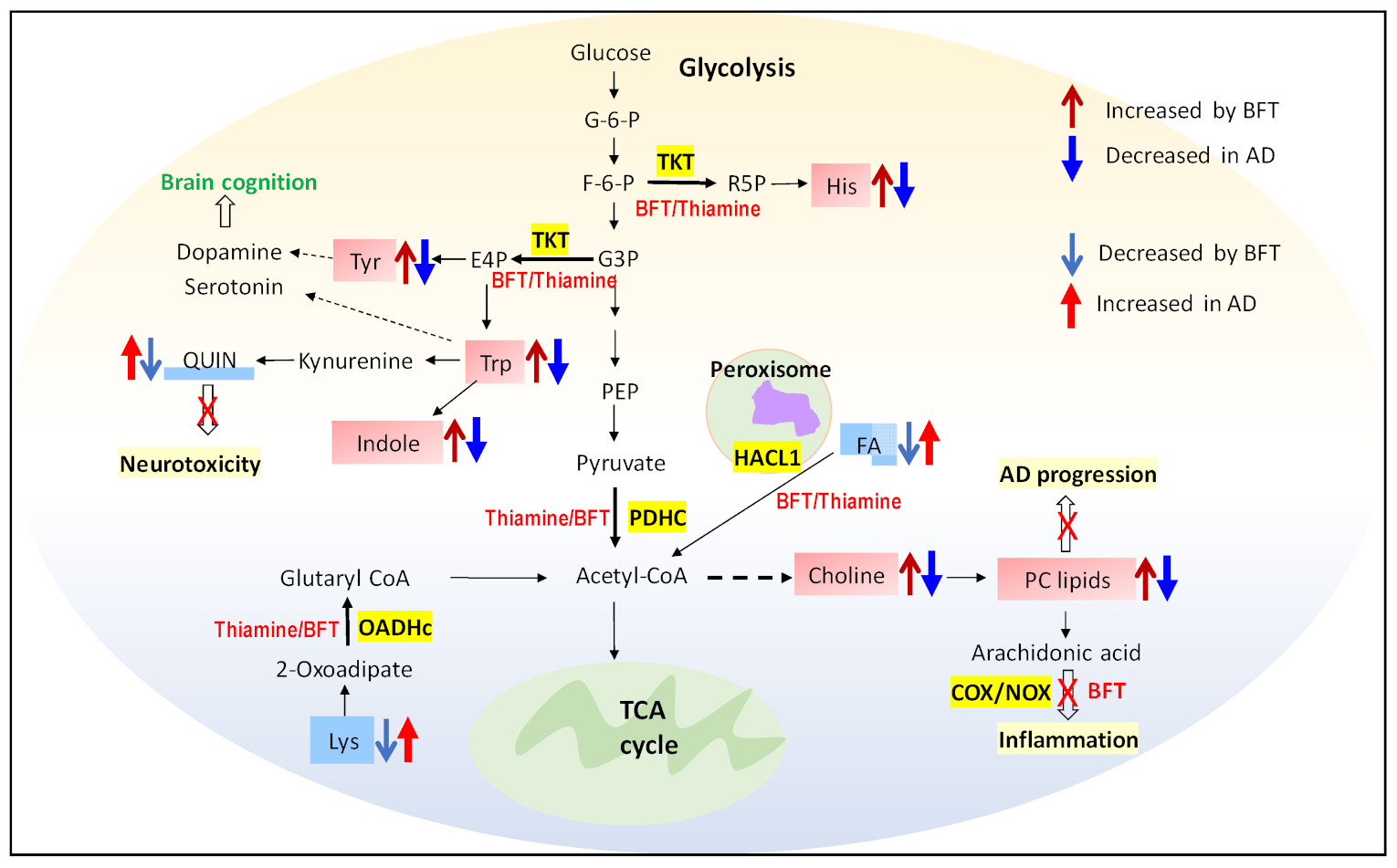
2.5. Pathway and Enrichment Analysis of Altered Metabolites
3. Discussion
4. Materials and Methods
4.1. Serum Sample Collection for Metabolomics and Lipidomics
4.2. Untargeted Metabolomics Analysis of Serum Samples
4.3. Metabolomics Data Processing and Statistical Analysis
4.4. Untargeted Lipidomics Analysis of Serum Samples
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beltramo, E.; Berrone, E.; Tarallo, S.; Porta, M. Effects of thiamine and benfotiamine on intracellular glucose metabolism and relevance in the prevention of diabetic complications. Acta Diabetol. 2008, 45, 131–141. [Google Scholar] [CrossRef]
- Kennedy, D.O. B Vitamins and the Brain: Mechanisms, Dose and Efficacy—A Review. Nutrients 2016, 8, 68. [Google Scholar] [CrossRef] [Green Version]
- Dhir, S.; Tarasenko, M.; Napoli, E.; Giulivi, C. Neurological, Psychiatric, and Biochemical Aspects of Thiamine Deficiency in Children and Adults. Front. Psychiatry 2019, 10, 207. [Google Scholar] [CrossRef] [Green Version]
- Yadav, U.C.; Kalariya, N.M.; Srivastava, S.K.; Ramana, K.V. Protective role of benfotiamine, a fat-soluble vitamin B1 analogue, in lipopolysaccharide-induced cytotoxic signals in murine macrophages. Free Radic. Biol. Med. 2010, 48, 1423–1434. [Google Scholar] [CrossRef] [Green Version]
- Gibson, G.E.; Hirsch, J.A.; Fonzetti, P.; Jordan, B.D.; Cirio, R.T.; Elder, J. Vitamin B1 (thiamine) and dementia. Ann. N. Y. Acad. Sci. 2016, 1367, 21–30. [Google Scholar] [CrossRef]
- Gibson, G.E.; Luchsinger, J.A.; Cirio, R.; Chen, H.; Franchino-Elder, J.; Hirsch, J.A.; Bettendorff, L.; Chen, Z.; Flowers, S.A.; Gerber, L.M.; et al. Benfotiamine and Cognitive Decline in Alzheimer’s Disease: Results of a Randomized Placebo-Controlled Phase IIa Clinical Trial. J. Alzheimers Dis. 2020, 78, 989–1010. [Google Scholar] [CrossRef]
- Volvert, M.L.; Seyen, S.; Piette, M.; Evrard, B.; Gangolf, M.; Plumier, J.C.; Bettendorff, L. Benfotiamine, a synthetic S-acyl thiamine derivative, has different mechanisms of action and a different pharmacological profile than lipid-soluble thiamine disulfide derivatives. BMC Pharmacol. 2008, 8, 10. [Google Scholar] [CrossRef] [Green Version]
- Moraes, R.C.M.; Singulani, M.P.; Goncalves, A.C.; Portari, G.V.; Torrao, A.D.S. Oral benfotiamine reverts cognitive deficit and increase thiamine diphosphate levels in the brain of a rat model of neurodegeneration. Exp. Gerontol. 2020, 141, 111097. [Google Scholar] [CrossRef]
- Murphy, M.P.; LeVine, H., 3rd. Alzheimer’s disease and the amyloid-beta peptide. J. Alzheimers Dis. 2010, 19, 311–323. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.K.; Srivastav, S.; Yadav, A.K.; Srikrishna, S.; Perry, G. Overview of Alzheimer’s Disease and Some Therapeutic Approaches Targeting Abeta by Using Several Synthetic and Herbal Compounds. Oxid. Med. Cell Longev. 2016, 2016, 7361613. [Google Scholar] [CrossRef] [Green Version]
- Jagust, W. Imaging the evolution and pathophysiology of Alzheimer disease. Nat. Rev. Neurosci. 2018, 19, 687–700. [Google Scholar] [CrossRef]
- DeTure, M.A.; Dickson, D.W. The neuropathological diagnosis of Alzheimer’s disease. Mol. Neurodegener. 2019, 14, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Bhawal, R.; Xu, H.; Chen, H.; Anderson, E.T.; Haroutunian, V.; Cross, A.C.; Zhang, S.; Gibson, G.E. The human brain acetylome reveals that decreased acetylation of mitochondrial proteins associates with Alzheimer’s disease. J. Neurochem. 2021, 158, 282–296. [Google Scholar] [CrossRef]
- Clish, C.B. Metabolomics: An emerging but powerful tool for precision medicine. Cold Spring Harb. Mol. Case Stud. 2015, 1, a000588. [Google Scholar] [CrossRef] [Green Version]
- Tada, I.; Tsugawa, H.; Meister, I.; Zhang, P.; Shu, R.; Katsumi, R.; Wheelock, C.E.; Arita, M.; Chaleckis, R. Creating a Reliable Mass Spectral-Retention Time Library for All Ion Fragmentation-Based Metabolomics. Metabolites 2019, 9, 251. [Google Scholar] [CrossRef] [Green Version]
- Lokhov, P.G.; Balashova, E.E.; Trifonova, O.P.; Maslov, D.L.; Ponomarenko, E.A.; Archakov, A.I. Mass Spectrometry-Based Metabolomics Analysis of Obese Patients’ Blood Plasma. Int. J. Mol. Sci. 2020, 21, 568. [Google Scholar] [CrossRef] [Green Version]
- Wilkins, J.M.; Trushina, E. Application of Metabolomics in Alzheimer’s Disease. Front. Neurol. 2017, 8, 719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niedzwiecki, M.M.; Walker, D.I.; Howell, J.C.; Watts, K.D.; Jones, D.P.; Miller, G.W.; Hu, W.T. High-resolution metabolomic profiling of Alzheimer’s disease in plasma. Ann. Clin. Transl. Neurol. 2020, 7, 36–45. [Google Scholar] [CrossRef] [Green Version]
- Yi, M.; Zhang, C.; Zhang, Z.; Yi, P.; Xu, P.; Huang, J.; Peng, W. Integrated Metabolomic and Lipidomic Analysis Reveals the Neuroprotective Mechanisms of Bushen Tiansui Formula in an Abeta1-42-Induced Rat Model of Alzheimer’s Disease. Oxid. Med. Cell Longev. 2020, 2020, 5243453. [Google Scholar] [CrossRef]
- Ledet, M.M.; Harman, R.M.; Fan, J.C.; Schmitt-Matzen, E.; Diaz-Rubio, M.E.; Zhang, S.; Van de Walle, G.R. Secreted sphingomyelins modulate low mammary cancer incidence observed in certain mammals. Sci. Rep. 2020, 10, 20580. [Google Scholar] [CrossRef]
- Hunsberger, H.C.; Greenwood, B.P.; Tolstikov, V.; Narain, N.R.; Kiebish, M.A.; Denny, C.A. Divergence in the metabolome between natural aging and Alzheimer’s disease. Sci. Rep. 2020, 10, 12171. [Google Scholar] [CrossRef]
- Trushina, E.; Mielke, M.M. Recent advances in the application of metabolomics to Alzheimer’s Disease. Biochim. Biophys. Acta 2014, 1842, 1232–1239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varma, V.R.; Oommen, A.M.; Varma, S.; Casanova, R.; An, Y.; Andrews, R.M.; O’Brien, R.; Pletnikova, O.; Troncoso, J.C.; Toledo, J.; et al. Brain and blood metabolite signatures of pathology and progression in Alzheimer disease: A targeted metabolomics study. PLoS Med. 2018, 15, e1002482. [Google Scholar] [CrossRef] [PubMed]
- Bhawal, R.; Oberg, A.L.; Zhang, S.; Kohli, M. Challenges and Opportunities in Clinical Applications of Blood-Based Proteomics in Cancer. Cancers 2020, 12, 2428. [Google Scholar] [CrossRef] [PubMed]
- Proitsi, P.; Kim, M.; Whiley, L.; Simmons, A.; Sattlecker, M.; Velayudhan, L.; Lupton, M.K.; Soininen, H.; Kloszewska, I.; Mecocci, P.; et al. Association of blood lipids with Alzheimer’s disease: A comprehensive lipidomics analysis. Alzheimers Dement. 2017, 13, 140–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metab. 2013, 1, 92–107. [Google Scholar]
- Monteleone, P.; Monteleone, A.M.; Troisi, J.; Dalle Grave, R.; Corrivetti, G.; Calugi, S.; Scala, G.; Patriciello, G.; Zanetti, A.; Maj, M. Metabolomics signatures of acutely ill and short-term weight recovered women with anorexia nervosa. Mol. Psychiatry 2021, 26, 3980–3991. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.W.; Bradshaw, P.C. Amino Acid Catabolism in Alzheimer’s Disease Brain: Friend or Foe? Oxid. Med. Cell Longev. 2017, 2017, 5472792. [Google Scholar] [CrossRef] [Green Version]
- Trushina, E.; Dutta, T.; Persson, X.M.; Mielke, M.M.; Petersen, R.C. Identification of altered metabolic pathways in plasma and CSF in mild cognitive impairment and Alzheimer’s disease using metabolomics. PLoS ONE 2013, 8, e63644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haglin, L.; Domellof, M.; Backman, L.; Forsgren, L. Low plasma thiamine and phosphate in male patients with Parkinson’s disease is associated with mild cognitive impairment. Clin. Nutr. ESPEN 2020, 37, 93–99. [Google Scholar] [CrossRef]
- Gonzalez-Dominguez, R.; Garcia-Barrera, T.; Gomez-Ariza, J.L. Metabolite profiling for the identification of altered metabolic pathways in Alzheimer’s disease. J. Pharm. Biomed. Anal. 2015, 107, 75–81. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, W.; Zan, J.; Wu, C.; Tan, W. Untargeted lipidomics reveals progression of early Alzheimer’s disease in APP/PS1 transgenic mice. Sci. Rep. 2020, 10, 14509. [Google Scholar] [CrossRef]
- Kao, Y.C.; Ho, P.C.; Tu, Y.K.; Jou, I.M.; Tsai, K.J. Lipids and Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 1505. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Sun, Y.P.; Luo, Y.M.; Peng, D.H.; Li, X.; Yang, B.Y.; Wang, Q.H.; Kuang, H.X. Biomarkers for the Clinical Diagnosis of Alzheimer’s Disease: Metabolomics Analysis of Brain Tissue and Blood. Front. Pharmacol. 2021, 12, 700587. [Google Scholar] [CrossRef]
- Altmaier, E.; Fobo, G.; Heier, M.; Thorand, B.; Meisinger, C.; Romisch-Margl, W.; Waldenberger, M.; Gieger, C.; Illig, T.; Adamski, J.; et al. Metabolomics approach reveals effects of antihypertensives and lipid-lowering drugs on the human metabolism. Eur. J. Epidemiol. 2014, 29, 325–336. [Google Scholar] [CrossRef] [Green Version]
- Xuan, J.; Pan, G.; Qiu, Y.; Yang, L.; Su, M.; Liu, Y.; Chen, J.; Feng, G.; Fang, Y.; Jia, W.; et al. Metabolomic profiling to identify potential serum biomarkers for schizophrenia and risperidone action. J. Proteome Res. 2011, 10, 5433–5443. [Google Scholar] [CrossRef]
- Nie, Q.; Xing, M.; Chen, H.; Hu, J.; Nie, S. Metabolomics and Lipidomics Profiling Reveals Hypocholesterolemic and Hypolipidemic Effects of Arabinoxylan on Type 2 Diabetic Rats. J. Agric. Food Chem. 2019, 67, 10614–10623. [Google Scholar] [CrossRef]
- Nuzzo, T.; Mancini, A.; Miroballo, M.; Casamassa, A.; Di Maio, A.; Donati, G.; Sansone, G.; Gaetani, L.; Paoletti, F.P.; Isidori, A.; et al. High performance liquid chromatography determination of L-glutamate, L-glutamine and glycine content in brain, cerebrospinal fluid and blood serum of patients affected by Alzheimer’s disease. Amino Acids 2021, 53, 435–449. [Google Scholar] [CrossRef]
- Vignisse, J.; Sambon, M.; Gorlova, A.; Pavlov, D.; Caron, N.; Malgrange, B.; Shevtsova, E.; Svistunov, A.; Anthony, D.C.; Markova, N.; et al. Thiamine and benfotiamine prevent stress-induced suppression of hippocampal neurogenesis in mice exposed to predation without affecting brain thiamine diphosphate levels. Mol. Cell. Neurosci. 2017, 82, 126–136. [Google Scholar] [CrossRef]
- Richard, D.M.; Dawes, M.A.; Mathias, C.W.; Acheson, A.; Hill-Kapturczak, N.; Dougherty, D.M. L-Tryptophan: Basic Metabolic Functions, Behavioral Research and Therapeutic Indications. Int. J. Tryptophan Res. 2009, 2, 45–60. [Google Scholar] [CrossRef] [Green Version]
- Gostner, J.M.; Geisler, S.; Stonig, M.; Mair, L.; Sperner-Unterweger, B.; Fuchs, D. Tryptophan Metabolism and Related Pathways in Psychoneuroimmunology: The Impact of Nutrition and Lifestyle. Neuropsychobiology 2020, 79, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Whiley, L.; Chappell, K.E.; D’Hondt, E.; Lewis, M.R.; Jimenez, B.; Snowden, S.G.; Soininen, H.; Kloszewska, I.; Mecocci, P.; Tsolaki, M.; et al. Metabolic phenotyping reveals a reduction in the bioavailability of serotonin and kynurenine pathway metabolites in both the urine and serum of individuals living with Alzheimer’s disease. Alzheimers Res. Ther. 2021, 13, 20. [Google Scholar] [CrossRef]
- Ramos-Chavez, L.A.; Roldan-Roldan, G.; Garcia-Juarez, B.; Gonzalez-Esquivel, D.; Perez de la Cruz, G.; Pineda, B.; Ramirez-Ortega, D.; Garcia Munoz, I.; Jimenez Herrera, B.; Rios, C.; et al. Low Serum Tryptophan Levels as an Indicator of Global Cognitive Performance in Nondemented Women over 50 Years of Age. Oxid. Med. Cell Longev. 2018, 2018, 8604718. [Google Scholar] [CrossRef]
- Attipoe, S.; Zeno, S.A.; Lee, C.; Crawford, C.; Khorsan, R.; Walter, A.R.; Deuster, P.A. Tyrosine for Mitigating Stress and Enhancing Performance in Healthy Adult Humans, a Rapid Evidence Assessment of the Literature. Mil. Med. 2015, 180, 754–765. [Google Scholar] [CrossRef] [Green Version]
- van de Rest, O.; Bloemendaal, M.; de Heus, R.; Aarts, E. Dose-Dependent Effects of Oral Tyrosine Administration on Plasma Tyrosine Levels and Cognition in Aging. Nutrients 2017, 9, 1279. [Google Scholar] [CrossRef] [Green Version]
- Young, S.N. L-tyrosine to alleviate the effects of stress? J. Psychiatry Neurosci. 2007, 32, 224. [Google Scholar] [PubMed]
- Agarwal, M.; Khan, S. Plasma Lipids as Biomarkers for Alzheimer’s Disease: A Systematic Review. Cureus 2020, 12, e12008. [Google Scholar] [CrossRef]
- Smriga, M.; Kameishi, M.; Uneyama, H.; Torii, K. Dietary L-lysine deficiency increases stress-induced anxiety and fecal excretion in rats. J. Nutr. 2002, 132, 3744–3746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jordan, F.; Nemeria, N.; Gerfen, G. Human 2-Oxoglutarate Dehydrogenase and 2-Oxoadipate Dehydrogenase Both Generate Superoxide/H2O2 in a Side Reaction and Each Could Contribute to Oxidative Stress in Mitochondria. Neurochem. Res. 2019, 44, 2325–2335. [Google Scholar] [CrossRef]
- Bodner, O.; Radzishevsky, I.; Foltyn, V.N.; Touitou, A.; Valenta, A.C.; Rangel, I.F.; Panizzutti, R.; Kennedy, R.T.; Billard, J.M.; Wolosker, H. D-Serine Signaling and NMDAR-Mediated Synaptic Plasticity Are Regulated by System A-Type of Glutamine/D-Serine Dual Transporters. J. Neurosci. 2020, 40, 6489–6502. [Google Scholar] [CrossRef]
- Madeira, C.; Lourenco, M.V.; Vargas-Lopes, C.; Suemoto, C.K.; Brandao, C.O.; Reis, T.; Leite, R.E.; Laks, J.; Jacob-Filho, W.; Pasqualucci, C.A.; et al. d-serine levels in Alzheimer’s disease: Implications for novel biomarker development. Transl. Psychiatry 2015, 5, e561. [Google Scholar] [CrossRef]
- Di Cara, F.; Andreoletti, P.; Trompier, D.; Vejux, A.; Bulow, M.H.; Sellin, J.; Lizard, G.; Cherkaoui-Malki, M.; Savary, S. Peroxisomes in Immune Response and Inflammation. Int. J. Mol. Sci. 2019, 20, 3877. [Google Scholar] [CrossRef] [Green Version]
- Raj, V.; Ojha, S.; Howarth, F.C.; Belur, P.D.; Subramanya, S.B. Therapeutic potential of benfotiamine and its molecular targets. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3261–3273. [Google Scholar]
- Romas, S.N.; Tang, M.X.; Berglund, L.; Mayeux, R. APOE genotype, plasma lipids, lipoproteins, and AD in community elderly. Neurology 1999, 53, 517–521. [Google Scholar] [CrossRef]
- Dorninger, F.; Moser, A.B.; Kou, J.; Wiesinger, C.; Forss-Petter, S.; Gleiss, A.; Hinterberger, M.; Jungwirth, S.; Fischer, P.; Berger, J. Alterations in the Plasma Levels of Specific Choline Phospholipids in Alzheimer’s Disease Mimic Accelerated Aging. J. Alzheimers Dis. 2018, 62, 841–854. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Dominguez, R.; Gonzalez-Dominguez, A.; Sayago, A.; Fernandez-Recamales, A. Recommendations and Best Practices for Standardizing the Pre-Analytical Processing of Blood and Urine Samples in Metabolomics. Metabolites 2020, 10, 229. [Google Scholar] [CrossRef]
- Noordwijk, K.J.; Qin, R.; Diaz-Rubio, M.E.; Zhang, S.; Su, J.; Mahal, L.K.; Reesink, H.L. Metabolism and global protein glycosylation are differentially expressed in healthy and osteoarthritic equine carpal synovial fluid. Equine Vet. J. 2021. [Google Scholar] [CrossRef]
- Hao, L.; Wang, J.; Page, D.; Asthana, S.; Zetterberg, H.; Carlsson, C.; Okonkwo, O.C.; Li, L. Comparative Evaluation of MS-based Metabolomics Software and Its Application to Preclinical Alzheimer’s Disease. Sci. Rep. 2018, 8, 9291. [Google Scholar] [CrossRef] [Green Version]

| Primary ID | VIP Score | FC of T/P | log2(FC) of T/P | p Value | BFT Treatment | Literature FC for AD/C [28,29,30,31] |
|---|---|---|---|---|---|---|
| Thiamine | 2.974 | 101.07 | 6.66 | 0.0110 | ↑ | 0.18↓ |
| 4-[(Z)-2-Aminovinyl]phenol * | 1.273 | 2.16 | 1.11 | 0.0306 | ↑ | NA |
| 3-Buten-1-amine * | 1.121 | 1.77 | 0.82 | 0.0036 | ↑ | NA |
| Tyrosine | 1.329 | 1.66 | 0.73 | 0.0496 | ↑ | 0.86↓ |
| Betaine* | 1.128 | 1.641 | 0.71 | 0.0154 | ↑ | NA |
| Indole | 1.244 | 1.59 | 0.67 | 0.0182 | ↑ | 0.86↓ |
| Tryptophan | 1.518 | 1.55 | 0.63 | 0.0323 | ↑ | 0.75↓ |
| Choline | 1.466 | 1.77 | 0.83 | 0.0632 | ↑ | NA |
| Histidine | 1.174 | 1.29 | 0.37 | 0.0626 | ↑ | 0.83↓ |
| Platelet-activating factor | 1.035 | 1.10 | 0.14 | 0.4183 | ↑ | NA |
| 1-Palmitoyl-2-hydroxy-sn-glycero-3-PE | 1.235 | 0.69 | −0.54 | 0.3572 | ↓ | NA |
| O-Ureido-D-serine | 1.222 | 0.78 | −0.36 | 0.0019 | ↓ | NA |
| Carboxymethyl-L-lysine | 1.653 | 0.73 | −0.45 | 0.5331 | ↓ | 1.37↑ |
| 9-HpODE | 1.027 | 0.71 | −0.49 | 0.1830 | ↓ | NA |
| 10-Undecenoic acid | 1.274 | 0.50 | −1.01 | 0.1024 | ↓ | NA |
| Palmitic acid | 1.150 | 0.50 | −1.01 | 0.1033 | ↓ | 0.65↓ |
| N-Undecanoylglycine | 1.054 | 0.50 | −1.01 | 0.1024 | ↓ | NA |
| Androsterone sulfate | 1.219 | 0.48 | −1.06 | 0.0343 | ↓ | 1.93↑ |
| Uric acid | 1.117 | 0.43 | −1.22 | 0.1340 | ↓ | 1.20↑ |
| 9(S)-HpOTrE | 1.249 | 0.41 | −1.28 | 0.1733 | ↓ | NA |
| Linoleic acid | 1.351 | 0.37 | −1.43 | 0.0817 | ↓ | 1.28↑ |
| 5α-Dihydrotestosterone glucuronide | 1.461 | 0.37 | −1.45 | 0.1190 | ↓ | NA |
| 16-Hydroxyhexadecanoic acid | 1.220 | 0.25 | −2.01 | 0.0241 | ↓ | NA |
| 2-Isopropylmalic acid | 1.605 | 0.15 | −2.73 | 0.2055 | ↓ | NA |
| Lysine | 1.777 | 0.08 | −3.56 | 0.0272 | ↓ | 1.28↑ |
| Primary ID | VIP | FC of T/P | log2(FC) of T/P | p Value | BFT Treatment | Literature FC for AD/C [32,33] |
|---|---|---|---|---|---|---|
| PC(32:2) + H | 2.08683 | 3.65 | 1.87 | 0.0176 | ↑ | NA |
| PC(32:1) + H | 1.72063 | 2.78 | 1.47 | 0.0079 | ↑ | --↓ |
| PC(40:6) + H | 1.69282 | 2.67 | 1.42 | 0.0003 | ↑ | --↓ |
| PC(38:5) + H | 1.71809 | 2.55 | 1.35 | 0.0032 | ↑ | NA |
| PC(40:4) + H | 1.61852 | 2.48 | 1.31 | 0.0055 | ↑ | --↓ |
| PC(34:4) + H | 1.8971 | 3.00 | 1.58 | 0.0035 | ↑ | NA |
| PC(36:3) + H | 2.13179 | 3.58 | 1.84 | 0.0023 | ↑ | 0.90↓ |
| PC(36:5) + H | 1.07928 | 2.27 | 1.18 | 0.0414 | ↑ | 0.59↓ |
| PC(34:3) + H | 1.60953 | 2.20 | 1.14 | 0.0003 | ↑ | --↓ |
| TG(48:3) + NH4 | 1.4194 | 1.78 | 0.83 | 0.0808 | ↑ | --↓ |
| PC(38:6) + H | 1.51857 | 1.75 | 0.80 | 0.3515 | ↑ | 0.78↓ |
| SM(d39:2) + H | 1.19304 | 1.73 | 0.79 | 0.0064 | ↑ | --↑ |
| SM(d41:1) + H | 1.05188 | 1.49 | 0.57 | 0.0113 | ↑ | --↓ |
| TG(48:1) + NH4 | 1.52317 | 1.39 | 0.48 | 0.2737 | ↑ | --↓ |
| SM(d40:2) + H | 1.09519 | 1.30 | 0.38 | 0.0271 | ↑ | NA |
| TG(58:2) + NH4 | 1.6588 | 2.83 | 1.50 | 0.405 | ↑ | NA |
| TG(53:6) + NH4 | 1.16343 | 0.77 | −0.37 | 0.098 | ↓ | >4.0↑ |
| TG(54:0) + NH4 | 1.06078 | 0.84 | −0.26 | 0.0454 | ↓ | NA |
| TG(58:7) + NH4 | 1.24209 | 0.68 | −0.55 | 0.0736 | ↓ | >4.0↑ |
| TG(54:6) + NH4 | 1.31772 | 0.67 | −0.57 | 0.0907 | ↓ | NA |
| TG(60:10) + NH4 | 1.12928 | 0.61 | −0.71 | 0.0517 | ↓ | >4.0↑ |
| TG(54:6) + H | 1.04225 | 0.73 | −0.438 | 0.00958 | ↓ | --↑ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhawal, R.; Fu, Q.; Anderson, E.T.; Gibson, G.E.; Zhang, S. Serum Metabolomic and Lipidomic Profiling Reveals Novel Biomarkers of Efficacy for Benfotiamine in Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 13188. https://doi.org/10.3390/ijms222413188
Bhawal R, Fu Q, Anderson ET, Gibson GE, Zhang S. Serum Metabolomic and Lipidomic Profiling Reveals Novel Biomarkers of Efficacy for Benfotiamine in Alzheimer’s Disease. International Journal of Molecular Sciences. 2021; 22(24):13188. https://doi.org/10.3390/ijms222413188
Chicago/Turabian StyleBhawal, Ruchika, Qin Fu, Elizabeth T. Anderson, Gary E. Gibson, and Sheng Zhang. 2021. "Serum Metabolomic and Lipidomic Profiling Reveals Novel Biomarkers of Efficacy for Benfotiamine in Alzheimer’s Disease" International Journal of Molecular Sciences 22, no. 24: 13188. https://doi.org/10.3390/ijms222413188
APA StyleBhawal, R., Fu, Q., Anderson, E. T., Gibson, G. E., & Zhang, S. (2021). Serum Metabolomic and Lipidomic Profiling Reveals Novel Biomarkers of Efficacy for Benfotiamine in Alzheimer’s Disease. International Journal of Molecular Sciences, 22(24), 13188. https://doi.org/10.3390/ijms222413188







