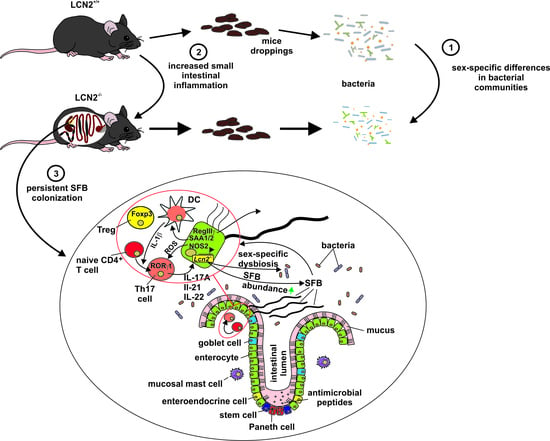
Depletion of Lipocalin 2 (LCN2) in Mice Leads to Dysbiosis and Persistent Colonization with Segmented Filamentous Bacteria
Abstract
1. Introduction
2. Results
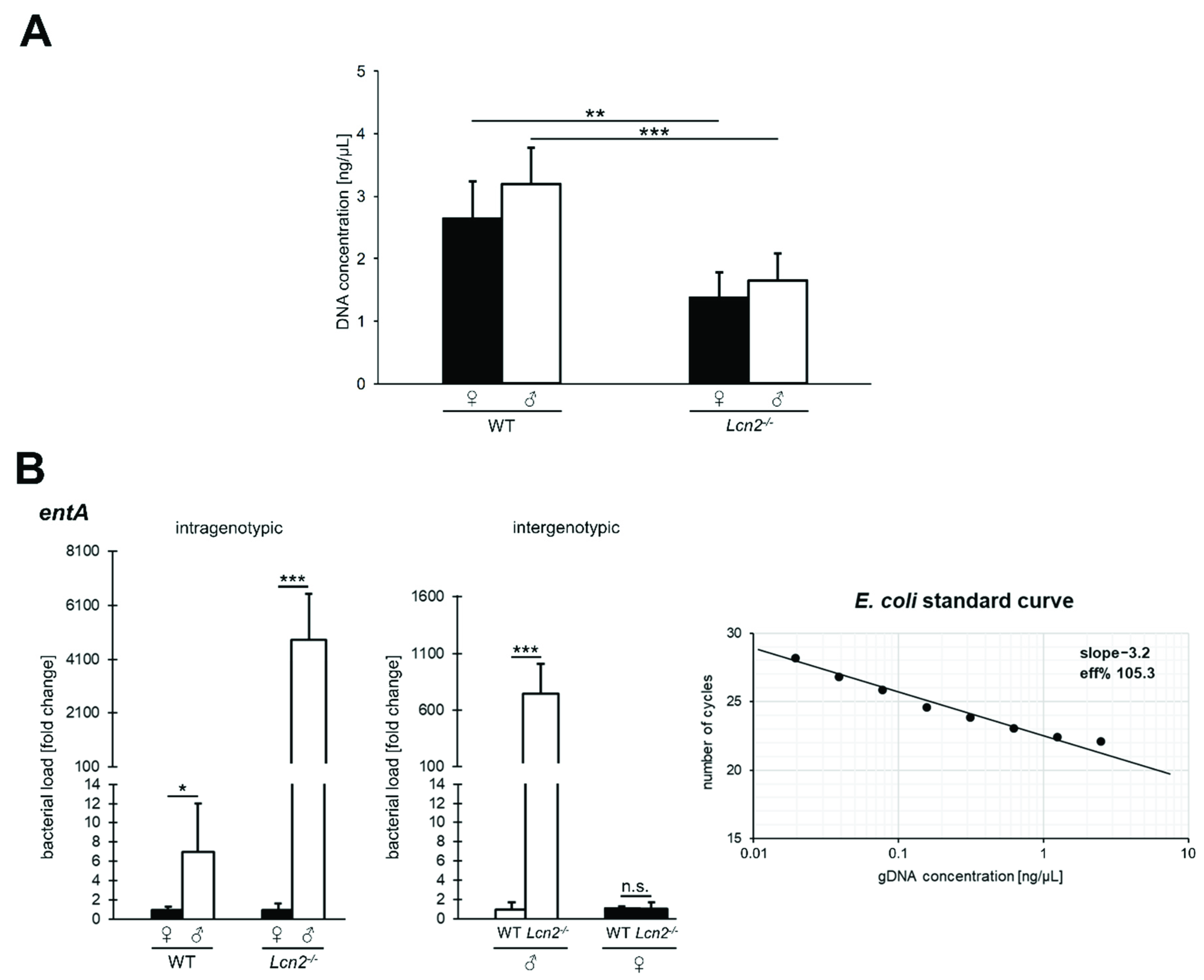
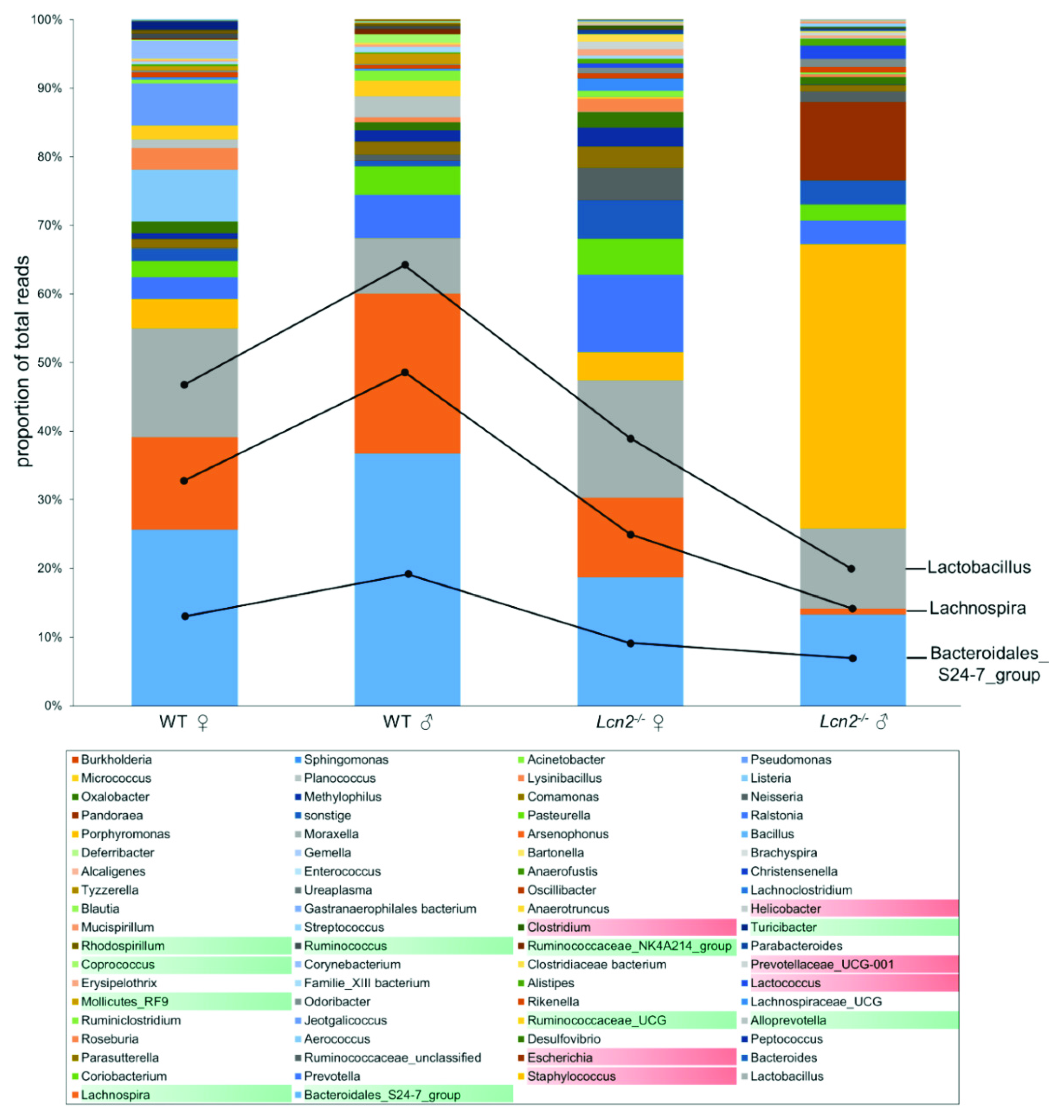
2.1. Gender-Specific Alteration in the Quantity and Quality of Intestinal Microbiota in Lcn2 Null Mice
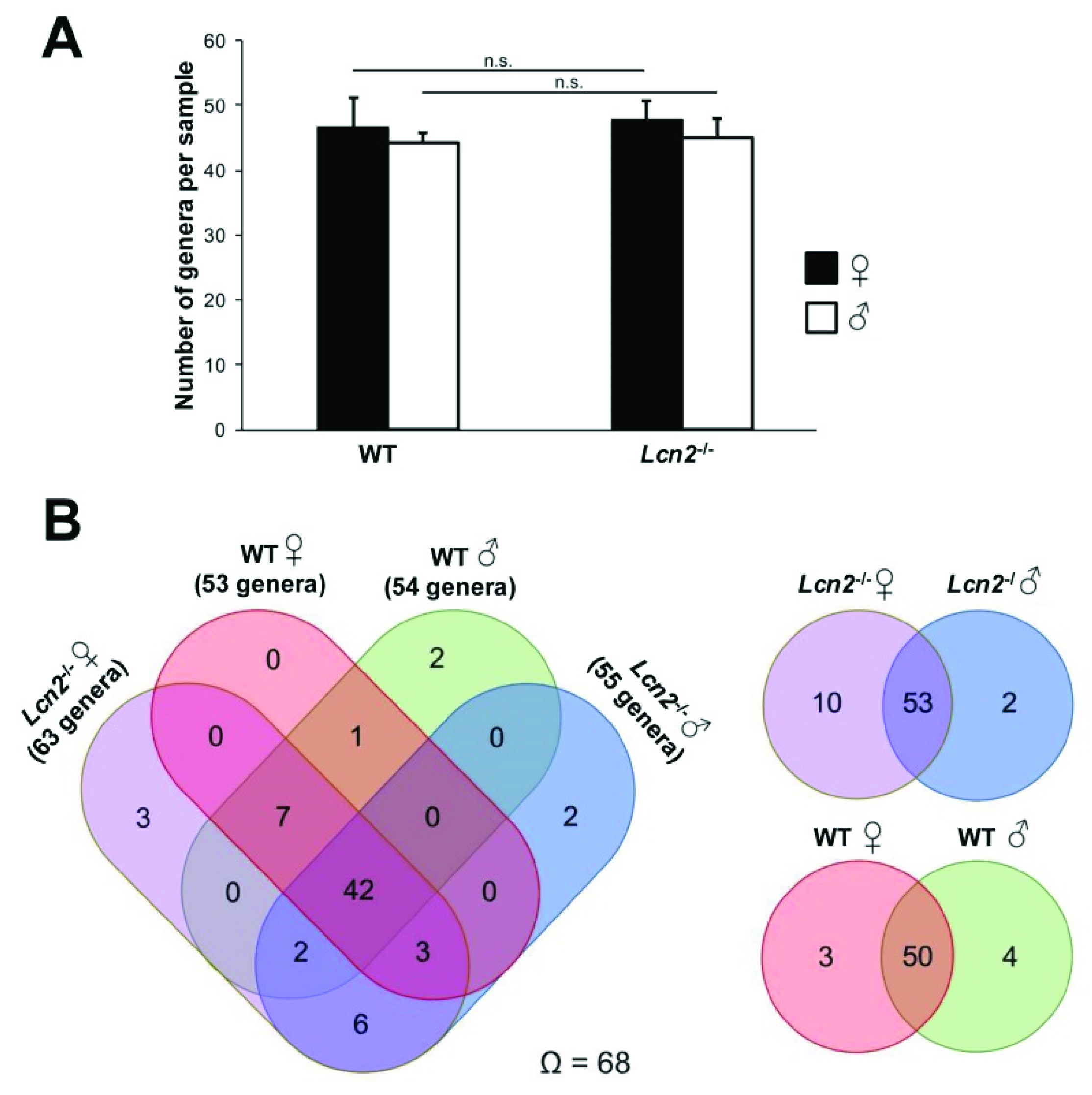
2.2. The Observed Intestinal Microbiota Diversity between Wild Type and Lcn2 Null Mice Is Homogenous
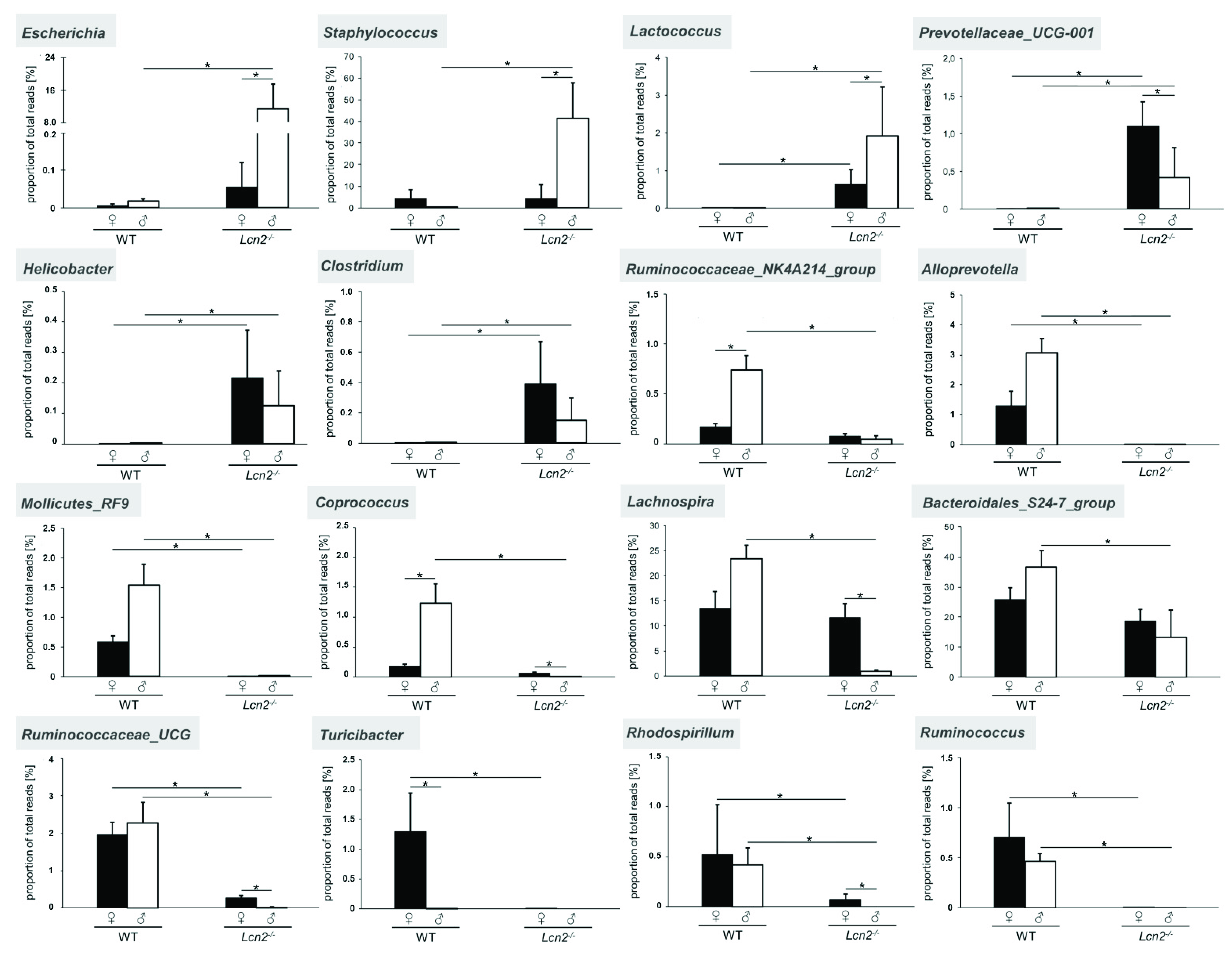
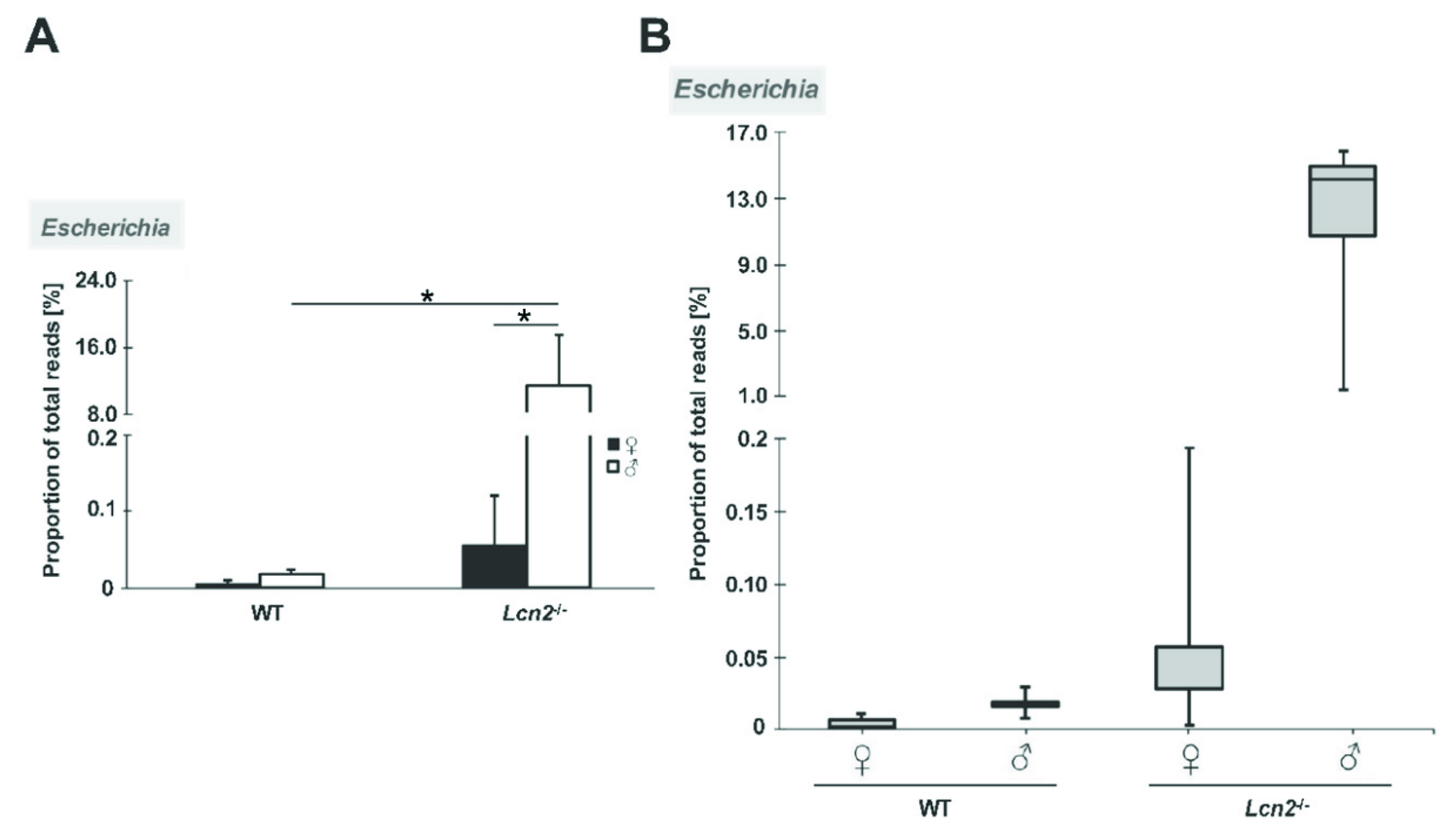
2.3. Distribution and Abundance of Identified Genera
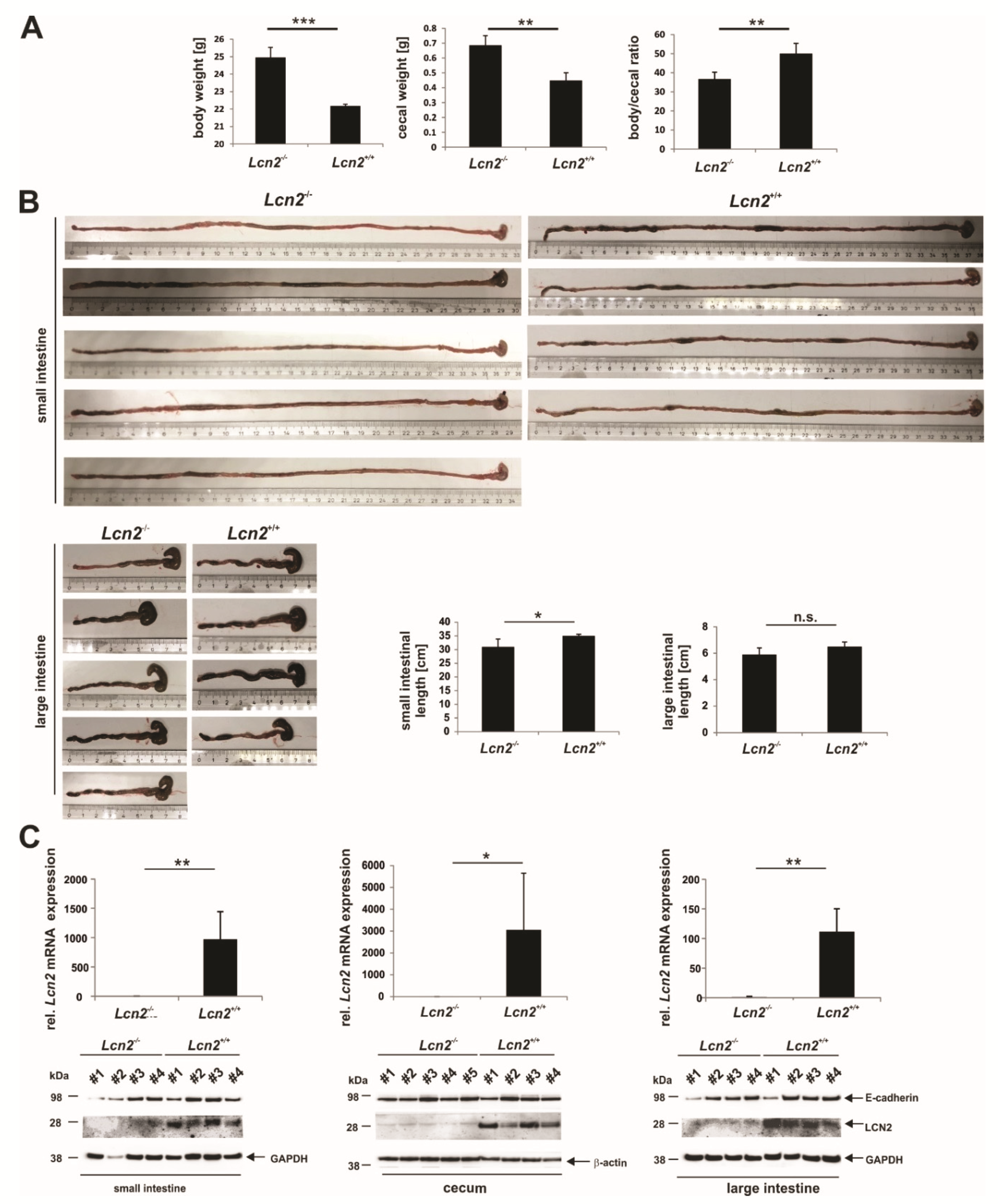
2.4. LCN2 Deficiency Leads to an Increase in Cecal Weight
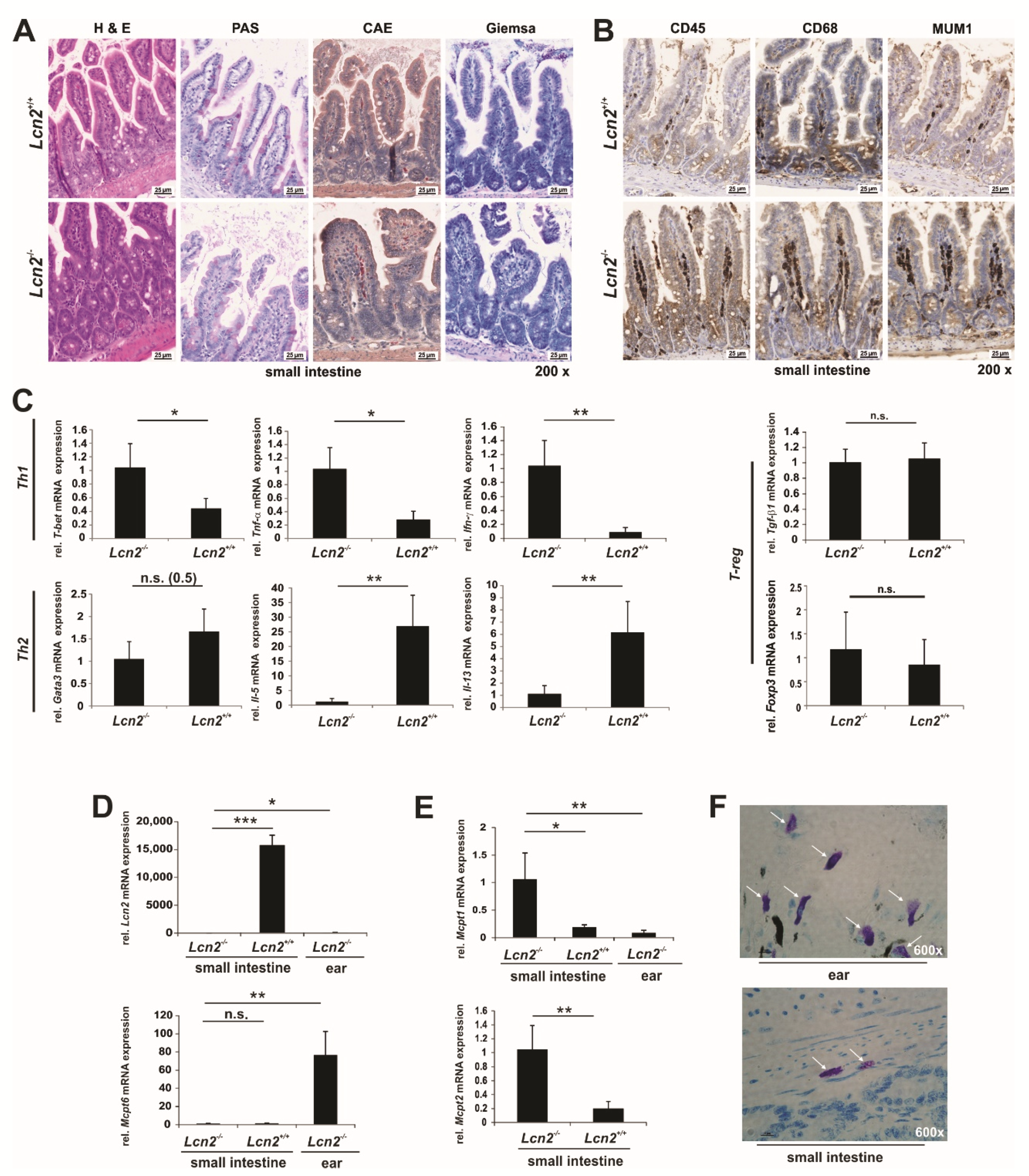
2.5. LCN2 Deficiency Triggers the Adaptive and Innate Immune Response in the Small Intestine
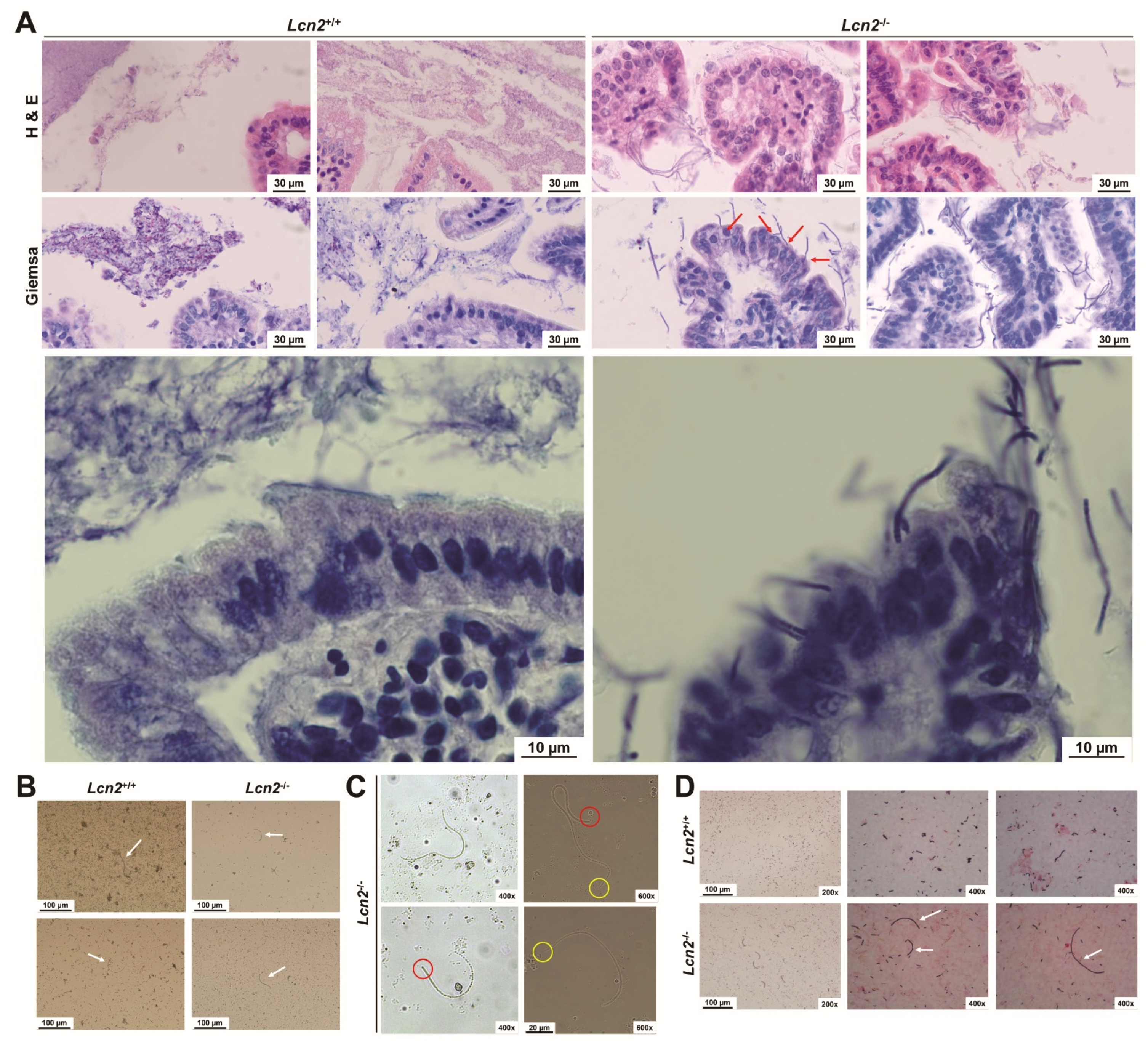
2.6. Loss of LCN2 Leads to Elevated and Persistant Colonization with Segmented Filamentous Bacteria
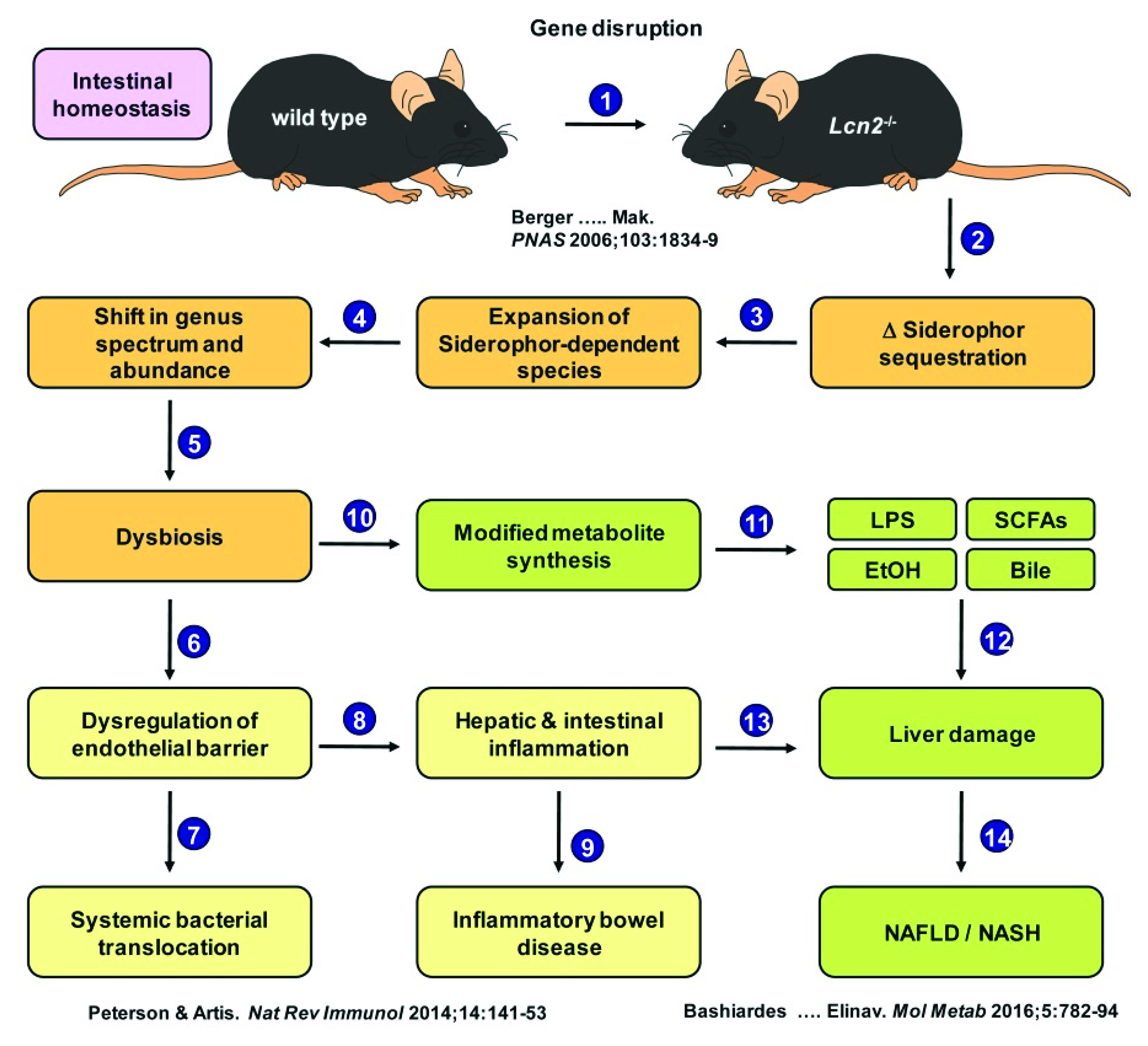
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Collection of Fecal Samples
4.3. Cultivation of Microorganisms from Murine Feces Samples
4.4. Liquid Culture of Unknown Isolates
4.5. Identification of Cultured Microorganisms Using MALDI-TOF MS
4.6. Preparation of Small Intestinal Fecal Smears and Gram-Stains
4.7. DNA Extraction from Murine Feces Samples and Quantification of the Total Bacterial Load
4.8. Extraction of Chromosomal DNA from Bacterial Isolates
4.9. 16S rRNA Amplicon and Index PCR
4.10. PCR Amplification of SFB 16S rDNA
4.11. Bioinformatic and Statistical Data Analysis
4.12. RNA Expression Analysis
4.13. Immunohistochemistry and Tissue Staining
4.14. Sodium Dodecylsulfate Polyacrylamide Gel Electrophoresis and Western Blot Analysis of Proteins from the Distal Small Intestine
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CAE | Chloroacetate esterase |
| CD | Crohn disease |
| ctMC | connective tissue mast cell(s) |
| EMT | epithelial-to-mesenchymal transition |
| Ent | Enterobactin |
| H & E | Hematoxylin & eosin |
| IECs | intestinal epithelial cells |
| LCN2 | Lipocalin 2 |
| MALDI-TOF | matrix-assisted laser desorption/ionization-time of flight |
| MC | Mast cell(s) |
| mMC | mucosal mast cell(s) |
| MUM-1 | Multiple myeloma oncogene 1 |
| MyD88 | Myeloid differentiation factor 88 |
| NGAL | Neutrophil gelatinase associated lipocalin |
| NGS | Next-Generation Sequencing |
| NOS2 | epithelial derived inducible NO synthase |
| ORFs | open reading frames |
| OTUs | operational taxonomic units |
| PAS | Periodic acid-Schiff |
| PCoA | Principal coordinates analysis |
| RegIIIγ | regeneration islet-derived protein 3 γ |
| Rorγt | receptor-related orphan receptor γt |
| ROS | reactive oxygen species |
| SAA1/2 | Serum amyloid A1 or A2 |
| SFB | segmented filamentous bacteria |
| Th17 | T helper 17 |
| TLR | Toll like receptor |
| WT | wild type |
References
- Kjeldsen, L.; Johnsen, A.H.; Sengeløv, H.; Borregaard, N. Isolation and primary structure of NGAL, a novel protein associated with human neutrophil gelatinase. J. Biol. Chem. 1993, 268, 10425–10432. [Google Scholar] [CrossRef] [PubMed]
- Borkham-Kamphorst, E.; Drews, F.; Weiskirchen, R. Induction of lipocalin-2 expression in acute and chronic experimental liver injury moderated by pro-inflammatory cytokines interleukin-1β through nuclear factor-κB activation. Liver Int. 2011, 31, 656–665. [Google Scholar] [CrossRef]
- Asimakopoulou, A.; Weiskirchen, S.; Weiskirchen, R. Lipocalin 2 (LCN2) Expression in hepatic malfunction and therapy. Front. Physiol. 2016, 7, 430. [Google Scholar] [CrossRef]
- Schröder, S.K.; Asimakopoulou, A.; Weiskirchen, A. Lipocalin 2 as a potential diagnostic and/or prognostic biomarker in prostate, lung and liver cancer. Clin. Oncol. 2018, 1, 1–16. [Google Scholar]
- Borkham-Kamphorst, E.; van de Leur, E.; Zimmermann, H.W.; Karlmark, K.R.; Tihaa, L.; Haas, U.; Tacke, F.; Berger, T.; Mak, T.W.; Weiskirchen, R. Protective effects of lipocalin-2 (LCN2) in acute liver injury suggest a novel function in liver homeostasis. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2013, 1832, 660–673. [Google Scholar] [CrossRef]
- Singh, V.; Yeoh, B.S.; Chassaing, B.; Zhang, B.; Saha, P.; Xiao, X.; Awasthi, D.; Shashidharamurthy, R.; Dikshit, M.; Gewirtz, A.; et al. Microbiota-inducible innate immune siderophore binding protein Lipocalin 2 is critical for intestinal homeostasis. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 482–498.e6. [Google Scholar] [CrossRef]
- Stallhofer, J.; Friedrich, M.; Konrad-Zerna, A.; Wetzke, M.; Lohse, P.; Glas, J.; Tillack-Schreiber, C.; Schnitzler, F.; Beigel, F.; Brand, S. Lipocalin-2 is a disease activity marker in inflammatory bowel disease regulated by IL-17A, IL-22, and TNF-α and modulated by IL23R genotype status. Inflamm. Bowel Dis. 2015, 21, 2327–2340. [Google Scholar] [CrossRef] [PubMed]
- Yvon, S.; Olier, M.; Leveque, M.; Jard, G.; Tormo, H.; Haimoud-Lekhal, D.A.; Peter, M.; Eutamène, H. Donkey milk consumption exerts anti-inflammatory properties by normalizing antimicrobial peptides levels in Paneth’s cells in a model of ileitis in mice. Eur. J. Nutr. 2016, 57, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Thorsvik, S.; Bakke, I.; Granlund, A.V.B.; Røyset, E.S.; Damås, J.K.; Østvik, A.E.; Sandvik, A.K. Expression of neutrophil gelatinase-associated lipocalin (NGAL) in the gut in Crohn’s disease. Cell Tissue Res. 2018, 374, 339–348. [Google Scholar] [CrossRef]
- Mori, K.; Suzuki, T.; Minamishima, S.; Igarashi, T.; Inoue, K.; Nishimura, D.; Seki, H.; Yamada, T.; Kosugi, S.; Katori, N.; et al. Neutrophil gelatinase-associated lipocalin regulates gut microbiota of mice. J. Gastroenterol. Hepatol. 2015, 31, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Craven, M.; Egan, C.E.; Dowd, S.; McDonough, S.P.; Dogan, B.; Denkers, E.Y.; Bowman, D.; Scherl, E.J.; Simpson, K.W. Inflammation drives dysbiosis and bacterial invasion in murine models of ileal Crohn’s disease. PLoS ONE 2012, 7, e41594. [Google Scholar] [CrossRef] [PubMed]
- Scanlan, P.D.; Shanahan, F.; O’Mahony, C.; Marchesi, J.R. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn’s disease. J. Clin. Microbiol. 2006, 44, 3980–3988. [Google Scholar] [CrossRef] [PubMed]
- Manichanh, C.; Rigottier-Gois, L.; Bonnaud, E.; Gloux, K.; Pelletier, E.; Frangeul, L.; Nalin, R.; Jarrin, C.; Chardon, P.; Marteau, P.; et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006, 55, 205–211. [Google Scholar] [CrossRef]
- Schaubeck, M.; Clavel, T.; Calasan, J.; Lagkouvardos, I.; Haange, S.B.; Jehmlich, N.; Basic, M.; Dupont, A.; Hornef, M.; von Bergen, M.; et al. Dysbiotic gut microbiota causes transmissible Crohn’s disease-like ileitis independent of failure in antimicrobial defence. Gut 2015, 65, 225–237. [Google Scholar] [CrossRef]
- Hold, G.; Smith, M.; Grange, C.; Watt, E.R.; El-Omar, E.M.; Mukhopadhy, I. Role of the gut microbiota in inflammatory bowel disease pathogenesis: What have we learnt in the past 10 years? World J. Gastroenterol. 2014, 20, 1192–1210. [Google Scholar] [CrossRef] [PubMed]
- Kleessen, B.; Kroesen, A.J.; Buhr, H.J.; Blaut, M. Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand. J. Gastroenterol. 2002, 37, 1034–1041. [Google Scholar] [CrossRef]
- González-Castro, A.M.; Martínez, C.; Salvo-Romero, E.; Fortea, M.; Pardo-Camacho, C.; Perez, M.V.; Alonso-Cotoner, C.; Santos, J.; Vicario, M. Mucosal pathobiology and molecular signature of epithelial barrier dysfunction in the small intestine in irritable bowel syndrome. J. Gastroenterol. Hepatol. 2017, 32, 53–63. [Google Scholar] [CrossRef]
- Menendez, A.; Willing, B.; Montero, M.; Wlodarska, M.; So, C.; Bhinder, G.; Vallance, B.; Finlay, B. Bacterial stimulation of the TLR-MyD88 pathway modulates the homeostatic expression of ileal Paneth cell α-defensins. J. Innate Immun. 2012, 5, 39–49. [Google Scholar] [CrossRef]
- Salzman, N.H. Paneth cell defensins and the regulation of the microbiome. Gut Microbes 2010, 1, 401–406. [Google Scholar] [CrossRef]
- Guaní-Guerra, E.; Santos-Mendoza, T.; Reyes, S.O.L.; Teran, L.M. Antimicrobial peptides: General overview and clinical implications in human health and disease. Clin. Immunol. 2010, 135, 1–11. [Google Scholar] [CrossRef]
- Salzman, N.H.; Bevins, C.L. Dysbiosis—A consequence of Paneth cell dysfunction. Semin. Immunol. 2013, 25, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Jäger, S.; Stange, E.F.; Wehkamp, J. Antimicrobial peptides in gastrointestinal inflammation. Int. J. Inflamm. 2010, 2010, 1–11. [Google Scholar] [CrossRef]
- Wehkamp, J.; Salzman, N.; Porter, E.; Nuding, S.; Weichenthal, M.; Petras, R.E.; Shen, B.; Schaeffeler, E.; Schwab, M.; Linzmeier, R.; et al. Reduced Paneth cell-defensins in ileal Crohn’s disease. Proc. Natl. Acad. Sci. USA 2005, 102, 18129–18134. [Google Scholar] [CrossRef]
- Wehkamp, J.; Wang, G.; Kübler, I.; Nuding, S.; Gregorieff, A.; Schnabel, A.; Kays, R.J.; Fellermann, K.; Burk, O.; Schwab, M.; et al. The Paneth cell α-defensin deficiency of ileal Crohn’s disease is linked to Wnt/Tcf-4. J. Immunol. 2007, 179, 3109–3118. [Google Scholar] [CrossRef] [PubMed]
- Cadwell, K.; Patel, K.K.; Komatsu, M.; Virgin, I.H.W.; Stappenbeck, T.S. A common role for Atg16L1, Atg5, and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy 2009, 5, 250–252. [Google Scholar] [CrossRef] [PubMed]
- Berger, T.; Togawa, A.; Duncan, G.S.; Elia, A.J.; You-Ten, A.; Wakeham, A.; Fong, H.E.H.; Cheung, C.C.; Mak, T.W. Lipocalin 2-deficient mice exhibit increased sensitivity to Escherichia coli infection but not to ischemia-reperfusion injury. Proc. Natl. Acad. Sci. USA 2006, 103, 1834–1839. [Google Scholar] [CrossRef]
- Goetz, D.H.; Holmes, M.A.; Borregaard, N.; Bluhm, M.E.; Raymond, K.N.; Strong, R.K. The neutrophil Lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell 2002, 10, 1033–1043. [Google Scholar] [CrossRef]
- Holmes, M.A.; Paulsene, W.; Jide, X.; Ratledge, C.; Strong, R.K. Siderocalin (Lcn 2) also binds carboxymycobactins, potentially defending against mycobacterial infections through iron sequestration. Structure 2005, 13, 29–41. [Google Scholar] [CrossRef]
- Xu, M.-J.; Feng, D.; Wu, H.; Wang, H.; Chan, Y.; Kolls, J.; Borregaard, N.; Porse, B.; Berger, T.; Mak, T.W.; et al. Liver is the major source of elevated serum lipocalin-2 levels after bacterial infection or partial hepatectomy: A critical role for IL-6/STAT3. Hepatology 2014, 61, 692–702. [Google Scholar] [CrossRef]
- Moschen, A.; Gerner, R.R.; Wang, J.; Klepsch, V.; Adolph, T.E.; Reider, S.; Hackl, H.; Pfister, A.; Schilling, J.; Moser, P.L.; et al. Lipocalin 2 protects from inflammation and tumorigenesis associated with gut microbiota alterations. Cell Host Microbe 2016, 19, 455–469. [Google Scholar] [CrossRef]
- Qi, B.; Han, M. Microbial Siderophore enterobactin promotes mitochondrial iron uptake and development of the host via interaction with ATP synthase. Cell 2018, 175, 571–582.e11. [Google Scholar] [CrossRef] [PubMed]
- Moschen, A.R.; Adolph, T.E.; Gerner, R.R.; Wieser, V.; Tilg, H. Lipocalin-2: A master mediator of intestinal and metabolic inflammation. Trends Endocrinol. Metab. 2017, 28, 388–397. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, Y.; Zhu, L.; Liu, W.; Liao, N.; Jiang, M.; Zhu, B.; Yu, H.; Xiang, C.; Wang, X. Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J. 2012, 7, 615–621. [Google Scholar] [CrossRef]
- Golonka, R.; Yeoh, B.S.; Vijay-Kumar, M. The iron tug-of-war between bacterial siderophores and innate immunity. J. Innate Immun. 2019, 11, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Clifton, M.C.; Rupert, P.B.; Hoette, T.M.; Raymond, K.N.; Abergel, R.J.; Strong, R.K. Parsing the functional specificity of Siderocalin/Lipocalin 2/NGAL for siderophores and related small-molecule ligands. J. Struct. Biol. X 2019, 2, 100008. [Google Scholar] [CrossRef]
- Peterson, L.W.; Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Bashiardes, S.; Shapiro, H.; Rozin, S.; Shibolet, O.; Elinav, E. Non-alcoholic fatty liver and the gut microbiota. Mol. Metab. 2016, 5, 782–794. [Google Scholar] [CrossRef] [PubMed]
- Hedblom, G.A.; Reiland, H.A.; Sylte, M.; Johnson, T.J.; Baumler, D.J. Segmented filamentous bacteria—metabolism meets immunity. Front. Microbiol. 2018, 9, 1991. [Google Scholar] [CrossRef]
- Ericsson, A.C.; Hagan, C.E.; Davis, D.J.; Franklin, C.L. Segmented filamentous bacteria: Commensal microbes with potential effects on research. Comp. Med. 2014, 64, 90–98. [Google Scholar]
- Snel, J.; Hermsen, C.C.; Smits, H.J.; Bos, N.A.; Eling, W.; Cebra, J.J.; Heidt, P.J. Interactions between gut-associated lymphoid tissue and colonization levels of indigenous, segmented, filamentous bacteria in the small intestine of mice. Can. J. Microbiol. 1998, 44, 1177–1182. [Google Scholar] [CrossRef]
- Macpherson, A.J.; Uhr, T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 2004, 303, 1662–1665. [Google Scholar] [CrossRef] [PubMed]
- Talham, G.L.; Jiang, H.-Q.; Bos, N.A.; Cebra, J.J. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect. Immun. 1999, 67, 1992–2000. [Google Scholar] [CrossRef]
- Flannigan, K.L.; Denning, T.L. Segmented filamentous bacteria-induced immune responses: A balancing act between host protection and autoimmunity. Immunology 2018, 154, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Meek, B.; Doi, Y.; Muramatsu, M.; Chiba, T.; Honjo, T.; Fagarasan, S. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl. Acad. Sci. USA 2004, 101, 1981–1986. [Google Scholar] [CrossRef]
- Nakajima, A.; Vogelzang, A.; Maruya, M.; Miyajima, M.; Murata, M.; Son, A.; Kuwahara, T.; Tsuruyama, T.; Yamada, S.; Matsuura, M.; et al. IgA regulates the composition and metabolic function of gut microbiota by promoting symbiosis between bacteria. J. Exp. Med. 2018, 215, 2019–2034. [Google Scholar] [CrossRef]
- Luu, M.; Binder, K.; Hartmann, S.; Kespohl, M.; Bazant, J.; Romero, R.; Schütz, B.; Steinhoff, U.; Visekruna, A. Transcription factor c-Rel mediates communication between commensal bacteria and mucosal lymphocytes. J. Leukoc. Biol. 2021. [Google Scholar] [CrossRef]
- Schnupf, P.; Gaboriau-Routhiau, V.; Cerf-Bensussan, N. Host interactions with Segmented filamentous bacteria: An unusual trade-off that drives the post-natal maturation of the gut immune system. Semin. Immunol. 2013, 25, 342–351. [Google Scholar] [CrossRef]
- Sanford, S.E. Light and electron microscopic observations of a segmented filamentous bacterium Attached to the mucosa of the terminal ileum of pigs. J. Veter-Diagn. Investig. 1991, 3, 328–333. [Google Scholar] [CrossRef]
- Caselli, M.; Holton, J.; Boldrini, P.; Vaira, D.; Calò, G. Morphology of segmented filamentous bacteria and their patterns of contact with the follicle-associated epithelium of the mouse terminal ileum. Gut Microbes 2010, 1, 367–372. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Meyerholz, D.K.; Stabel, T.J.; Cheville, N.F. Segmented filamentous bacteria interact with intraepithelial mononuclear cells. Infect. Immun. 2002, 70, 3277–3280. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sczesnak, A.; Segata, N.; Qin, X.; Gevers, D.; Petrosino, J.F.; Huttenhower, C.; Littman, D.R.; Ivanov, I.I. The genome of Th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe 2011, 10, 260–272. [Google Scholar] [CrossRef]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef]
- Schnupf, P.; Gaboriau-Routhiau, V.; Sansonetti, P.J.; Cerf-Bensussan, N. Segmented filamentous bacteria, Th17 inducers and helpers in a hostile world. Curr. Opin. Microbiol. 2017, 35, 100–109. [Google Scholar] [CrossRef]
- Strobel, S.; Miller, H.R.; Ferguson, A. Human intestinal mucosal mast cells: Evaluation of fixation and staining techniques. J. Clin. Pathol. 1981, 34, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Flannigan, K.L.; Ngo, V.L.; Geem, D.; Harusato, A.; Hirota, S.A.; Parkos, C.A.; Lukacs, N.W.; Nusrat, A.; Gaboriau-Routhiau, V.; Cerf-Bensussan, N.; et al. IL-17A-mediated neutrophil recruitment limits expansion of segmented filamentous bacteria. Mucosal Immunol. 2016, 10, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Schroll, A.; Eller, K.; Feistritzer, C.; Nairz, M.; Sonnweber, T.; Moser, P.A.; Rosenkranz, A.R.; Theurl, I.; Weiss, G. Lipocalin-2 ameliorates granulocyte functionality. Eur. J. Immunol. 2012, 42, 3346–3357. [Google Scholar] [CrossRef] [PubMed]
- Panea, C.; Farkas, A.M.; Goto, Y.; Abdollahi-Roodsaz, S.; Lee, C.; Koscso, B.; Gowda, K.; Hohl, T.M.; Bogunovic, M.; Ivanov, I.I. Intestinal monocyte-derived macrophages control commensal-specific Th17 responses. Cell Rep. 2015, 12, 1314–1324. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; Panea, C.; Nakato, G.; Cebula, A.; Lee, C.; Diez, M.G.; Laufer, T.M.; Ignatowicz, L.; Ivanov, I.I. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 2014, 40, 594–607. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Ando, M.; Kamada, N.; Nagano, Y.; Narushima, S.; Suda, W.; Imaoka, A.; Setoyama, H.; Nagamori, T.; et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 2015, 163, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Cash, H.L.; Whitham, C.V.; Hooper, L.V. Refolding, purification, and characterization of human and murine RegIII proteins expressed in Escherichia coli. Protein Expr. Purif. 2006, 48, 151–159. [Google Scholar] [CrossRef]
- Vaishnava, S.; Yamamoto, M.; Severson, K.M.; Ruhn, K.A.; Yu, X.; Koren, O.; Ley, R.; Wakeland, E.K.; Hooper, L.V. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science 2011, 334, 255–258. [Google Scholar] [CrossRef]
- Wang, Y.; Yin, Y.; Chen, X.; Zhao, Y.; Wu, Y.; Li, Y.; Wang, X.; Chen, H.; Xiang, C. Induction of intestinal Th17 cells by flagellins from segmented filamentous bacteria. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Shaw, M.H.; Kamada, N.; Kim, Y.-G.; Núñez, G. Microbiota-induced IL-1β, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J. Exp. Med. 2012, 209, 251–258. [Google Scholar] [CrossRef]
- Meurer, S.K.; Tezcan, O.; Lammers, T.; Weiskirchen, R. Differential regulation of Lipocalin 2 (LCN2) in doxorubicin-resistant 4T1 triple negative breast cancer cells. Cell. Signal. 2020, 74, 109731. [Google Scholar] [CrossRef] [PubMed]
- Klaasen, H.L.; Van der Heijden, P.J.; Stok, W.; Poelma, F.G.; Koopman, J.P.; Brink, M.E.V.D.; Bakker, M.H.; Eling, W.M.; Beynen, A.C. Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune system of mice. Infect. Immun. 1993, 61, 303–306. [Google Scholar] [CrossRef]
- Umesaki, Y.; Okada, Y.; Matsumoto, S.; Imaoka, A.; Setoyama, H. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol. Immunol. 1995, 39, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Snel, J.; Bakker, M.; Heidt, P. Quantification of antigen-specific immunoglobulin A after oral booster immunization with ovalbumin in mice mono-associated with segmented filamentous bacteria or Clostridium innocuum. Immunol. Lett. 1997, 58, 25–28. [Google Scholar] [CrossRef]
- Liao, N.; Yin, Y.; Sun, G.; Xiang, C.; Liu, D.; Yu, H.D.; Wang, X. Colonization and distribution of segmented filamentous bacteria (SFB) in chicken gastrointestinal tract and their relationship with host immunity. FEMS Microbiol. Ecol. 2012, 81, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Bielecki, M.; Antonyuk, S.; Strange, R.W.; Siemińska, K.; Smalley, J.W.; Mackiewicz, P.; Śmiga, M.; Cowan, M.; Capper, M.J.; Ślęzak, P.; et al. Prevotella intermedia produces two proteins homologous to Porphyromonas gingivalis HmuY but with different heme coordination mode. Biochem. J. 2020, 477, 381–405. [Google Scholar] [CrossRef] [PubMed]
- Dauner, M.; Skerra, A. Scavenging bacterial siderophores with engineered lipocalin proteins as an alternative antimicrobial strategy. ChemBioChem 2019, 21, 601–606. [Google Scholar] [CrossRef]
- Hastie, J.L.; Hanna, P.C.; Carlson, P.E. Transcriptional response of Clostridium difficile to low iron conditions. Pathog. Dis. 2018, 76. [Google Scholar] [CrossRef]
- Marchetti, M.; De Bei, O.; Bettati, S.; Campanini, B.; Kovachka, S.; Gianquinto, E.; Spyrakis, F.; Ronda, L. Iron metabolism at the interface between host and pathogen: From nutritional immunity to antibacterial development. Int. J. Mol. Sci. 2020, 21, 2145. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, N.; Sapra, A. Gram Staining. StatPerarls Publishing. Available online: https://www.ncbi.nlm.nih.gov/books/NBK562156/ (accessed on 29 October 2021).
- The Greengenes Database. Available online: https://greengenes.secondgenome.com/ (accessed on 29 October 2021).
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.G.; Neufeld, J.D. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinform. 2012, 13, 31. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Silva High Quality Ribosomal RNA Databases. Available online: https://www.arb-silva.de (accessed on 29 October 2021).
- NCBI Taxonomy: A Comprehensive Update on Curation, Resources and Tools. Available online: https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi (accessed on 29 October 2021).
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A tool for visualizing high-throughput microbial community data. GigaScience 2013, 2, 16. [Google Scholar] [CrossRef]
- Navas-Molina, J.A.; Peralta-Sánchez, J.M.; González, A.; McMurdie, P.J.; Vázquez-Baeza, Y.; Xu, Z.; Ursell, L.K.; Lauber, C.; Zhou, H.; Song, S.J.; et al. Advancing our understanding of the human microbiome using QIIME. Methods Enzymol. 2013, 531, 371–444. [Google Scholar] [CrossRef]
- Jiang, L.; Amir, A.; Morton, J.T.; Heller, R.; Arias-Castro, E.; Knight, R. Discrete false-discovery rate improves identification of differentially abundant microbes. mSystems 2017, 2, e00092-17. [Google Scholar] [CrossRef]
- Biocore/dsFDR. Available online: https://github.com/biocore/dsfdr (accessed on 26 November 2021).
- Asimakopoulou, A.; Fülöp, A.; Borkham-Kamphorst, E.; Van de Leur, E.; Gassler, N.; Berger, T.; Weiskirchen, R. Altered mitochondrial and peroxisomal integrity in lipocalin-2-deficient mice with hepatic steatosis. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2017, 1863, 2093–2110. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Thompson, S.W.; Hunt, R.D. Selected Histochemical and Histopathological Methods; Charles, C., Ed.; Thomas: Springfield, IL, USA, 1966; ISBN-10: 0398031320. [Google Scholar]
- Sheehan, D.C.; Hrapchak, B.B. Theory and Practice of Histotechnology, 2nd ed.; Battelle Memorial Institute: Columbus, OH, USA, 1987; ISBN-10: 0935470395. [Google Scholar]
- Kashima, T.G.; Inagaki, Y.; Grammatopoulos, G.; Athanasou, N.A. Use of chloroacetate esterase staining for the histological diagnosis of prosthetic joint infection. Virchows Arch. 2015, 466, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Grigorev, I.; Korzhevskii, D. Modern imaging technologies of mast cells for biology and medicine (Review). Sovrem. Tech. Med. 2021, 13, 93. [Google Scholar] [CrossRef] [PubMed]
- Meurer, S.; Wimmer, A.E.; Van De Leur, E.; Weiskirchen, R. Leur Endoglin trafficking/exosomal targeting in liver cells depends on N-glycosylation. Cells 2019, 8, 997. [Google Scholar] [CrossRef] [PubMed]
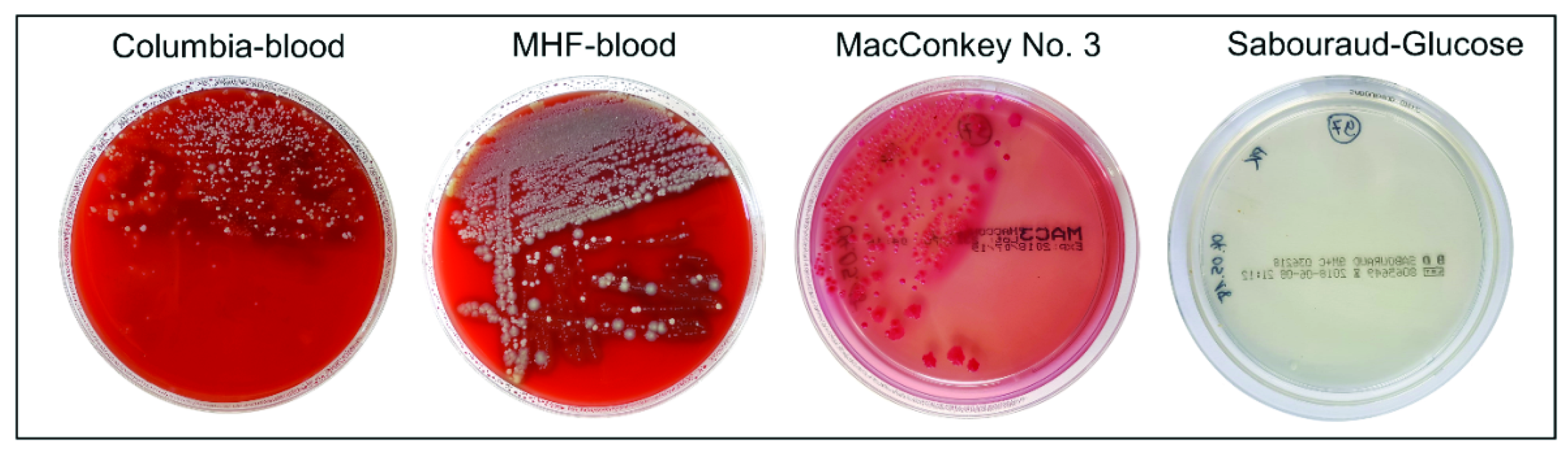


| Species | Wild Type (♀) | Wild Type (♂) | Lcn2-/- (♀) | Lcn2-/- (♂) |
|---|---|---|---|---|
| Escherichia coli | M (99.9) | M (99.9) | M (99.9) | M (99.9) |
| Staphylococcus xylosus | M (99.9) | M (99.9) | M (99.9) | M (99.9) |
| Aerococcus viridans | M (99.9) | M (99.9) | M (99.9) | M (99.9) |
| Enterococcus faecalis | M (99.9) | |||
| Lactobacillus murinus/ Lactobacillus animalis | S (99.0) | |||
| Lactobacillus johnsonii | S (98.0) | |||
| Lactococcus formosensis | S (99.0) | |||
| Streptococcus danieliae | S (99.0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klüber, P.; Meurer, S.K.; Lambertz, J.; Schwarz, R.; Zechel-Gran, S.; Braunschweig, T.; Hurka, S.; Domann, E.; Weiskirchen, R. Depletion of Lipocalin 2 (LCN2) in Mice Leads to Dysbiosis and Persistent Colonization with Segmented Filamentous Bacteria. Int. J. Mol. Sci. 2021, 22, 13156. https://doi.org/10.3390/ijms222313156
Klüber P, Meurer SK, Lambertz J, Schwarz R, Zechel-Gran S, Braunschweig T, Hurka S, Domann E, Weiskirchen R. Depletion of Lipocalin 2 (LCN2) in Mice Leads to Dysbiosis and Persistent Colonization with Segmented Filamentous Bacteria. International Journal of Molecular Sciences. 2021; 22(23):13156. https://doi.org/10.3390/ijms222313156
Chicago/Turabian StyleKlüber, Patrick, Steffen K. Meurer, Jessica Lambertz, Roman Schwarz, Silke Zechel-Gran, Till Braunschweig, Sabine Hurka, Eugen Domann, and Ralf Weiskirchen. 2021. "Depletion of Lipocalin 2 (LCN2) in Mice Leads to Dysbiosis and Persistent Colonization with Segmented Filamentous Bacteria" International Journal of Molecular Sciences 22, no. 23: 13156. https://doi.org/10.3390/ijms222313156
APA StyleKlüber, P., Meurer, S. K., Lambertz, J., Schwarz, R., Zechel-Gran, S., Braunschweig, T., Hurka, S., Domann, E., & Weiskirchen, R. (2021). Depletion of Lipocalin 2 (LCN2) in Mice Leads to Dysbiosis and Persistent Colonization with Segmented Filamentous Bacteria. International Journal of Molecular Sciences, 22(23), 13156. https://doi.org/10.3390/ijms222313156