Recombinant Bovine Growth Hormone-Induced Metabolic Remodelling Enhances Growth of Gilthead Sea-Bream (Sparus aurata): Insights from Stable Isotopes Composition and Proteomics
Abstract
1. Introduction
2. Results
2.1. Growth Performance, Proximal and Isotopic Composition
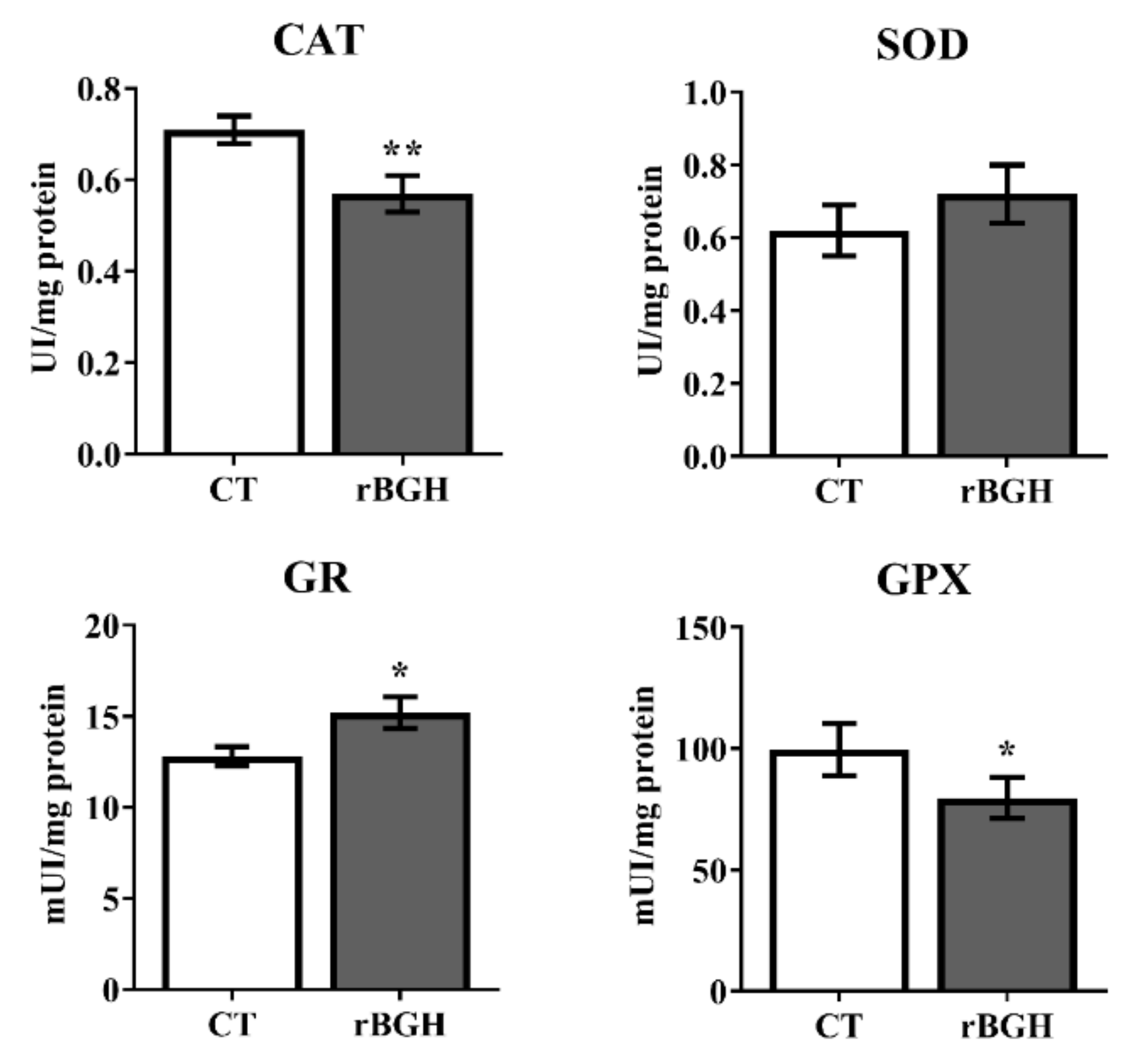
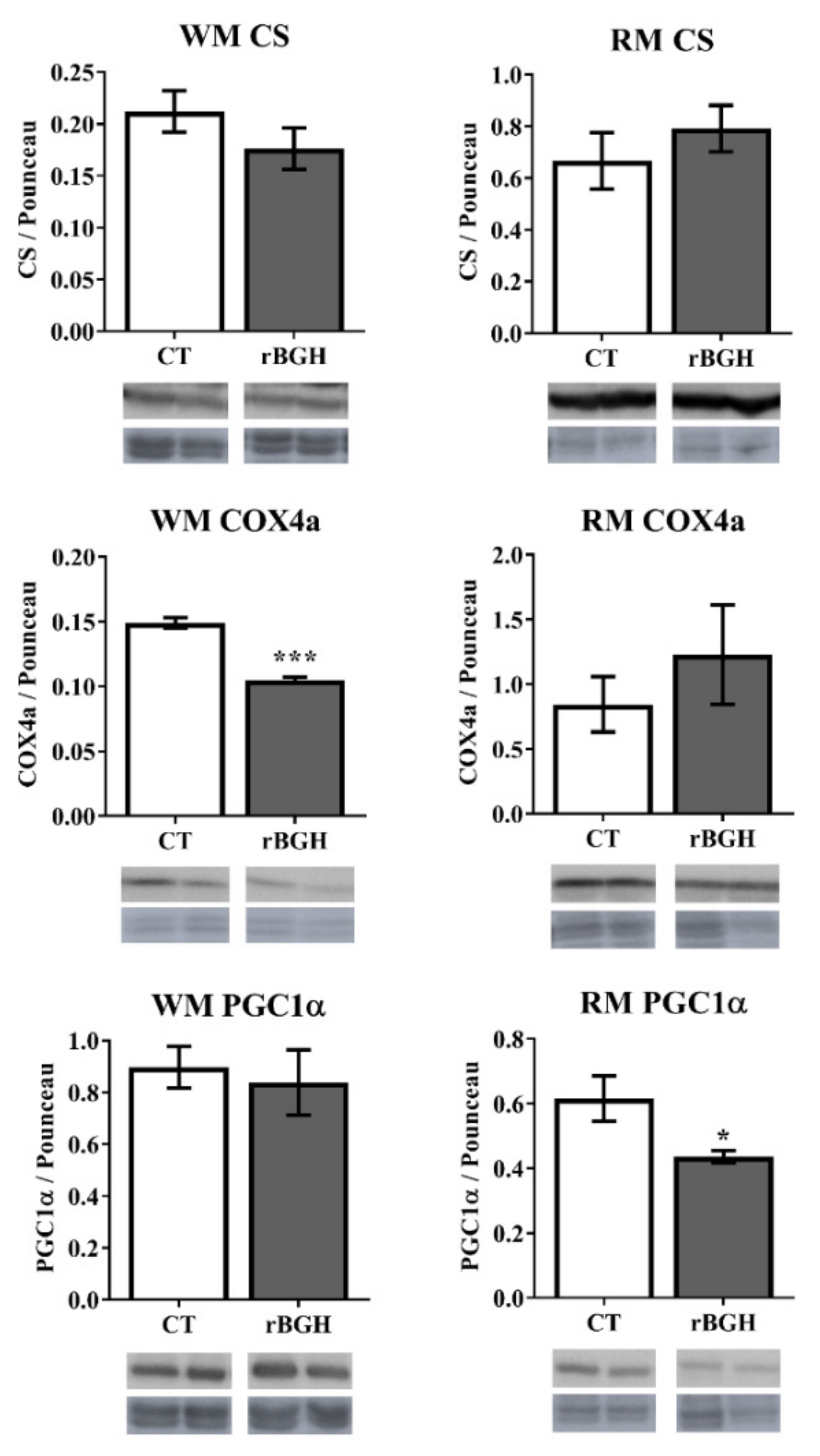
2.2. Enzyme Activities and Protein Expression
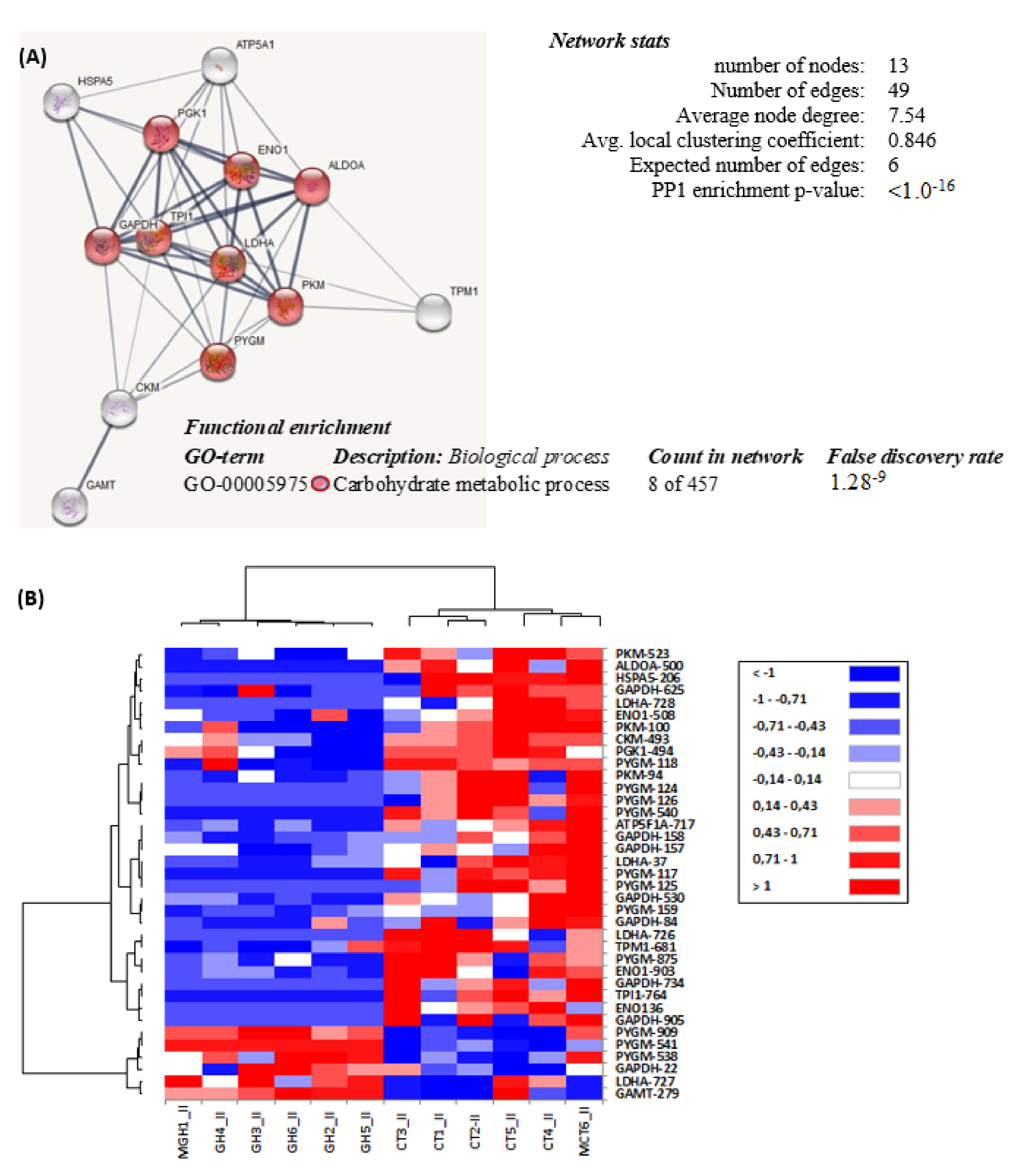
2.3. White Muscle Proteome
3. Discussion
4. Materials and Methods
4.1. Fish and Experimental Design
4.1.1. Gilthead Sea Bream Fingerlings
4.1.2. Gilthead Sea Bream Juveniles
4.2. Proximal and Isotopic Composition (δ15N and δ13C) of Tissues
4.3. Nucleic Acid Quantification and Enzyme Activity
4.4. Western Blot
4.5. Proteome Analysis
4.5.1. Protein Extraction and 2-Dimesional Electrophoresis Separation
4.5.2. Gel Image Analysis
4.5.3. LC-MS/MS Analysis and Database Search
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Árnason, T.; Gunnarsson, A.; Steinarsson, A.; Danñielsdóttir, A.K.; Björnsson, B.T. Impact of temperature and growth hormone on growth physiology of junenile Atlantic wolffish (Anarhichas lupus). Aquaculture 2019, 504, 404–413. [Google Scholar] [CrossRef]
- Devlin, R.H.; Biagi, C.A.; Yesaki, T.Y.; Smailus, D.E.; Byatt, J.C. Growth of domesticated transgenic fish. Nature 2001, 409, 781–782. [Google Scholar] [CrossRef]
- Duan, C. The insulin-like growth factor system and its biological actions in fish. Integr. Comp. Biol. 1997, 37, 491–503. [Google Scholar] [CrossRef]
- McLean, E.; Devlin, R.H. Seaweeds and invertebrates. In Recent Advances in Marine Biotechnology, Aquaculture: Part A; Fingerman, M., Nagabhushanam, R., Eds.; Science Publishers, Inc.: Endfield, CT, USA, 2000; Volume 4. [Google Scholar]
- Leedom, T.A.; Uchida, K.; Yada, T.; Richman, N.H.; Byatt, J.C.; Collier, R.J.; Hirano, T.; Grau, E.G. Recombinant bovine growth hormone treatment of tilapia: Growth response, metabolic clearance, receptor binding and immunoglobulin production. Aquaculture 2002, 207, 359–380. [Google Scholar] [CrossRef]
- Peterson, B.C.; Small, B.C.; Bosworth, B.G. Effects of bovine growth hormone (Posilac®) on growth performance, body composition, and IGFBPs in two strains of channel catfish. Aquaculture 2004, 232, 651–663. [Google Scholar] [CrossRef]
- Bower, N.I.; Johnston, I.A. Transcriptional regulation of the IGF signaling pathway by amino acids and insulin-like growth factors during myogenesis in Atlantic salmon. PLoS ONE 2010, 5, e11100. [Google Scholar] [CrossRef] [PubMed]
- Raven, P.A.; Sakhrani, D.; Beckman, B.; Neregård, L.; Sundström, L.F.; Björnsson, B.T.; Devlin, R.H. Growth and endocrine effects of recombinant bovine growth hormone treatment in non-transgenic and growth hormone transgenic coho salmon. Gen. Comp. Endocrinol. 2012, 177, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, E.N.; Valdés, J.A.; Molina, A.; Björnsson, B.T. Regulation of skeletal muscle growth in fish by the growth hormone--insulin-like growth factor system. Gen. Comp. Endocrinol. 2013, 192, 136–148. [Google Scholar] [CrossRef]
- Vélez, E.J.; Lutfi, E.; Azizi, S.; Perelló, M.; Salmerón, C.; Riera-Codina, M.; Ibarz, A.; Fernández-Borràs, J.; Blasco, J.; Capilla, E.; et al. Understanding fish muscle growth regulation to optimize aquaculture production. Aquaculture 2017, 467, 28–40. [Google Scholar] [CrossRef]
- Vélez, E.J.; Perelló, M.; Azizi, S.; Moya, A.; Lutfi, E.; Pérez-Sánchez, J.; Calduch-Giner, J.A.; Navarro, I.; Blasco, J.; Fernández-Borràs, J.; et al. Recombinant bovine growth hormone (rBGH) enhances somatic growth by regulating the GH-IGF axis in fingerlings of gilthead sea bream (Sparus aurata). Gen. Comp. Endocrinol. 2018, 257, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Vélez, E.J.; Perelló-Amorós, M.; Lutfi, E.; Azizi, S.; Capilla, E.; Navarro, I.; Pérez-Sánchez, J.; Calduch-Giner, J.A.; Blasco, J.; Fernández-Borràs, J.; et al. A long–term growth hormone treatment stimulates growth and lipolysis in gilthead sea bream juveniles. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2019, 232, 67–78. [Google Scholar] [CrossRef]
- Le Roith, D.; Bondy, C.; Yakar, S.; Liu, J.L.; Butler, A. The somatomedin hypothesis. Endocr. Rev. 2001, 22, 53–74. [Google Scholar] [CrossRef] [PubMed]
- Reindl, K.M.; Sheridan, M.A. Peripheral regulation of the growth hormone-insulin-like growth factor system in fish and other vertebrates. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 2012, 163, 231–245. [Google Scholar] [CrossRef]
- Vélez, E.J.; Unniappan, S.A. Comparative update on the neuroendocrine regulation of growth hormone in vertebrates. Front. Endocrinol. 2021, 11, 614981. [Google Scholar] [CrossRef]
- Sweeney, G. Leptin signalling. Cell Signal. 2002, 14, 655–663. [Google Scholar] [CrossRef]
- Reinecke, M.; Björnsson, B.T.; Dickhoff, W.W.; McCormick, S.D.; Navarro, I.; Power, D.M.; Gutierrez, J. Growth hormone and insulin-like growth factors in fish: Where we are and where to go. Gen. Comp. Endocrinol. 2005, 142, 20–24. [Google Scholar] [CrossRef]
- Møller, N.; Jørgensen, J.O.L. Effects of growth hormone on glucose, lipid, and protein metabolism in human subjects. Endocr. Rev. 2009, 30, 152–177. [Google Scholar] [CrossRef]
- Mommsen, T.P. Growth and metabolism. In The Physiology of Fishes; Evans, D.H., Ed.; CRC Press: Boca Raton, FL, USA, 1998; pp. 65–97. [Google Scholar]
- Sangiao-Alvarellos, S.; Míguez, J.M.; Soengas, J.L. Actions of growth hormone on carbohydrate metabolism and osmoregulation of rainbow trout (Oncorhynchus mykiss). Gen. Comp. Endocrinol. 2005, 141, 214–225. [Google Scholar] [CrossRef]
- O’Connor, P.K.; Reich, B.; Sheridan, M.A. Growth hormone stimulates hepatic lipid mobilization in rainbow trout, Oncorhynchus mykiss. J. Comp. Physiol. 1993, 163B, 427–431. [Google Scholar] [CrossRef]
- Leena, S.; Oommen, O.V. Growth hormone and prolactin action in a teleost Anabas testudineus (Bloch): Effect of the time of administration. Biol. Rhythm Res. 2001, 32, 501–510. [Google Scholar] [CrossRef]
- Leung, T.C.; Ng, T.B.; Woo, N.Y.S. Metabolic effects of bovine growth hormone in the tilapia Oreochromis mossambicus. Comp. Biochem. Physiol. A 1991, 99, 633–636. [Google Scholar] [CrossRef]
- Matty, A.J. Effects of mammalian growth hormone on Cottus scorpius blood. Nature 1962, 195, 506–507. [Google Scholar] [CrossRef]
- McKeown, B.A.; Leatherland, J.F.; John, T.M. The effect of growth hormone and prolactin on the mobilization of free fatty acids and glucose in the kokanee salmon (Oncorhynchus nerka). Comp. Biochem. Physiol. 1975, 50B, 425–430. [Google Scholar] [CrossRef]
- Sweeting, R.M.; Wagner, G.F.; McKeown, B.A. Changes in plasma glucose, amino acid nitrogen and growth hormone during smoltification and seawater adaptation in coho salmon, Oncorhynchus kisutch. Aquaculture 1985, 45, 185–197. [Google Scholar] [CrossRef]
- Aas-Hansen, Ø.; Jørgensen, E.H.; Vijayan, M.M. Fasting modulates metabolic responses to cortisol, GH and IGF-I in Arctic charr hepatocytes. J. Fish Biol. 2005, 67, 1631–1645. [Google Scholar] [CrossRef]
- Foster, A.R.; Houlihan, D.F.; Gray, C.; Medale, F.; Fauconneau, B.; Kaushik, S.; le Bail, P.Y. The effects of ovine growth hormone on protein turnover in rainbow trout. Gen. Comp. Endocrinol. 1991, 82, 111–120. [Google Scholar] [CrossRef]
- Fauconneau, B.; Mady, M.; Le Bail, P.Y. Effect of growth hormone on muscle protein synthesis in rainbow trout Oncorhynchus mykiss and Atlantic salmon Salmo salar. Fish Physiol. Biochem. 1996, 15, 49–56. [Google Scholar] [CrossRef]
- Herbert, N.A.; Armstrong, J.D.; Björnsson, B.T. Evidence that growth hormone-induced elevation in routine metabolism of juvenile Atlantic salmon is a result of increased spontaneous activity. J. Fish Biol. 2001, 59, 754–757. [Google Scholar] [CrossRef]
- Causey, D.R.; Kim, J.H.; Stead, D.A.; Martin, S.A.M.; Devlin, R.H.; Macqueen, D.J. Proteomic comparison of selective breeding and growth hormone transgenesis in fish: Unique pathways to enhanced growth. J. Proteom. 2019, 192, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Martínez Del Rio, C.; Wolf, N.; Carleton, S.A.; Gannes, L.Z. Isotopic ecology ten years after a call for more laboratory experiments. Biol. Rev. 2009, 84, 91–111. [Google Scholar] [CrossRef]
- Gannes, L.Z.; Marti, C. Natural abundance variations in stable isotopes and their potential uses in animal physiological ecology. Comp. Biochem.Physiol. Part A. 1998, 119, 725–737. [Google Scholar] [CrossRef]
- Martin-Pérez, M.; Fernández-Borràs, J.; Ibarz, A.; Felip, O.; Gutierrez, J.; Blasco, J. Stable isotope analysis combined with metabolic indices discriminates between gilthead sea bream (Sparus aurata) fingerlings produced in various hatcheries. J. Agric. Food Chem. 2011, 59, 10261–10270. [Google Scholar] [CrossRef]
- Beltrán, M.; Fernández-Borrás, J.; Médale, F.; Pérez-Sánchez, J.; Kaushik, S.; Blasco, J. Natural abundance of 15N and 13C in fish tissues and the use of stable isotopes as dietary protein tracers in rainbow trout and gilthead sea bream. Aquac. Nutr. 2009, 15, 9–18. [Google Scholar] [CrossRef]
- Martin-Pérez, M.; Fernández-Borràs, J.; Ibarz, A.; Millan-Cubillo, A.; Felip, O.; de Oliveira, E.; Blasco, J. New insights into fish swimming: A proteomic and isotopic approach in gilthead sea bream. J. Proteome Res. 2012, 11, 3533–3547. [Google Scholar] [CrossRef] [PubMed]
- Martin-Pérez, M.; Fernández-Borràs, J.; Ibarz, A.; Felip, O.; Fontanillas, R.; Gutiérrez, J. Naturally occurring stable isotopes reflect changes in protein turnover and growth in gilthead sea bream (Sparus aurata) juveniles under different dietary protein levels. J. Agric. Food Chem. 2013, 61, 8924–8933. [Google Scholar] [CrossRef] [PubMed]
- Biga, P.R.; Meyer, J. Growth hormone differentially regulates growth and growth-related gene expression in closely related fish species. Comp. Biochem. Physiol. A 2009, 154, 465–473. [Google Scholar] [CrossRef]
- Fenn, C.M.; Small, B.C. Exogenous recombinant bovine growth hormone stimulates growth and hepatic IGF expression in shovelnose sturgeon Scaphirhynchus platorhynchus. Comp. Biochem. Physiol. A 2015, 180, 18–22. [Google Scholar] [CrossRef]
- McLean, E.; Devlin, R.H.; Byatt, J.C.; Clarke, W.C.; Donaldson, E.M. Impact of a controlled release formulation of recombinant bovine growth hormone upon growth and seawater adaptation in coho (Oncorhynchus kisutch) and chinook (Oncorhynchus tshawytscha) and salmon. Aquaculture 1997, 156, 113–128. [Google Scholar] [CrossRef]
- Garber, M.J.; DeYonge, K.G.; Byatt, J.C.; Lellis, W.A.; Honeyfield, D.C.; Bull, R.C.; Schelling, G.T.; Roeder, R.A. Dose-response effects of recombinant bovine somatotropin (Posilac) on growth performance and body composition of two-year-old rainbow trout (Oncorhynchus mykiss). J. Anim. Sci. 1995, 73, 3216–3222. [Google Scholar] [CrossRef]
- Johnsson, J.I.; Björnsson, B.T. Growth hormone increases growth rate, appetite and dominance in juvenile rainbow trout, Oncorhynchus mykiss. Anim. Behav. 1994, 47, 177–186. [Google Scholar] [CrossRef]
- Kling, P.; Jönsson, E.; Nilsen, T.O.; Einarsdottir, I.E.; Rønnestad, I.; Stefansson, S.O.; Björnsson, B.T. The role of growth hormone in growth, lipid homeostasis, energy utilization and partitioning in rainbow trout: Interactions with leptin, ghrelin and insulin-like growth factor I. Gen. Comp. Endocrinol. 2012, 175, 153–162. [Google Scholar] [CrossRef]
- Fauconneau, B.; Andre, S.; Chmaitilly, J.; Bail, P.Y.; Krieg, F.; Kaushik, S.J. Control of skeletal muscle fibres and adipose cells size in the flesh of rainbow trout. J. Fish Biol. 1997, 50, 296–314. [Google Scholar] [CrossRef]
- Leatherland, J.F.; Nuti, R.N. Effects of bovine growth hormone on plasma FFA concentrations and liver, muscle and carcass lipid content in rainbow trout, Salmo gairdneri Richardson. J. Fish Biol. 1981, 19, 487–498. [Google Scholar] [CrossRef]
- Albalat, A.; Gómez-Requeni, P.; Rojas, P.; Médale, F.; Kaushik, S.; Vianen, G.J.; van den Thillart, G.; Gutiérrez, J.; Pérez-Sánchez, J.; Navarro, I. Nutritional and hormonal control of lipolysis in isolated gilthead seabream (Sparus aurata) adipocytes. Am. J. Physiol. Reg. Integr. Comp. Physiol. 2005, 289, R259–R265. [Google Scholar] [CrossRef]
- Bergan, H.E.; Kittilson, J.D.; Sheridan, M.A. PKC and ERK mediate Gh stimulated lipolysis. J. Mol. Endocrinol. 2013, 51, 213–224. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bergan, H.E.; Kittilson, J.D.; Sheridan, M.A. Nutritional state modulates growth hormone-stimulated lipolysis. Gen. Comp. Endocrinol. 2015, 217, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Blasco, J.; Moya, A.; Millán-Cubillo, A.; Vélez, E.J.; Capilla, E.; Pérez-Sánchez, J.; Gutiérrez, J.; Fernández-Borràs, J. Growth-promoting effects of sustained swimming in fingerlings of gilthead sea bream (Sparus aurata L.). J. Comp. Physiol. B. 2015, 185, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, R.S.; Rønsholdt, B.; Ostenfeld, T.H.; McLean, E.; Byatt, J.C. Growth, feed utilisation, carcass composition and sensory characteristics of rainbow trout treated with recombinant bovine placental lactogen and growth hormone. Aquaculture 2001, 195, 367–384. [Google Scholar] [CrossRef]
- McKenzie, D.J.; Martínez, R.; Morales, A.; Acosta, J.; Morales, R.; Taylor, E.W.; Steffensen, J.F.; Estrada, M.P. Effects of growth hormone transgenesis on metabolic rate, exercise performance and hypoxia tolerance in tilapia hybrids. J. Fish. Biol. 2003, 63, 398–409. [Google Scholar] [CrossRef]
- Short, K.R.; Møller, N.; Bigelow, M.L.; Coenen-Schimke, J.; Nair, K.S. Enhancement of muscle mitochondrial function by growth hormone. J. Clin. Endocrinol. Metab. 2008, 93, 597–604. [Google Scholar] [CrossRef]
- Lange, K.H.; Isaksson, F.; Juul, A.; Rasmussen, M.H.; Bülow, J.; Kjaer, M. Growth hormone enhances effects of endurance training on oxidative muscle metabolism in elderly women. Am. J. Physiol. Endocrinol. Metab. 2000, 279, E989–E996. [Google Scholar] [CrossRef]
- Leggatt, ·R.A.; Brauner, C.J.; Iwama, G.K.; Devlin, R.H. The glutathione antioxidant system is enhanced in growth hormone transgenic coho salmon (Oncorhynchus kisutch). J. Comp. Physiol. B 2007, 177, 413–422. [Google Scholar] [CrossRef]
- Tolomeo, A.M.; Carraro, A.; · Bakiu, R.; Toppo, S.; Garofalo, F.; Pellegrino, D.; Gerdol, M.; Ferro, D.; Place, S.P.; Santovito, G. Molecular characterization of novel mitochondrial peroxiredoxins from the Antarctic emerald rockcod and their gene expression in response to environmental warming. Comp. Biochem. Physiol. C 2019, 225, 108580. [Google Scholar]
- Scarpulla, R.C. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim. Biophys. Acta Mol. Cell Res. 2011, 1813, 1269–1278. [Google Scholar] [CrossRef]
- Wenz, T. Regulation of mitochondrial biogenesis and PGC-1α under cellular stress. Mitochondrion 2013, 13, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Mueller, I.A.; Grim, J.M.; Beers, J.M.; Crockett, E.L.; O’Brien, K.M. Interrelationship between mitochondrial function and susceptibility to oxidative stress in red- and white-blooded Antarctic notothenioid fishes. J. Exp. Biol. 2011, 214, 3732–3741. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.M. Mitochondrial biogenesis in cold-bodied fishes. J. Exp. Biol. 2011, 214, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Bermejo-Nogales, A.; Nederlof, M.; Benedito-Palos, L.; Ballester-Lozano, G.F.; Folkedal, O.; Olsen, R.E.; Sitjà-Bobadilla, A.; Pérez-Sánchez, J. Metabolic and transcriptional responses of gilthead sea bream (Sparus aurata L.) to environmental stress: New insights in fish mitochondrial phenotyping. Gen. Comp. Endocrinol. 2014, 205, 305–315. [Google Scholar] [CrossRef]
- Panserat, S.; Kamalam, B.S.; Fournier, J.; Plagnes-Juan, E.; Woodward, K.; Devlin, R.H. Glucose metabolic gene expression in growth hormone transgenic coho salmon. Comp. Biochem. Physiol. 2014, 170, 38–45. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Good, C.A.; Kramer, H.; Somogyi, M. The determination of glycogen. J. Biol. Chem. 1933, 100, 485–491. [Google Scholar] [CrossRef]
- Fraga, F. Determinación de glucógeno en moluscos con el reactivo de antrona. Inv. Pesq. 1956, 3, 69–74. [Google Scholar]
- Buckley, L.J.; Bulow, F.J. Techniques for the Estimation of RNA, DNA and Protein in Fish; Iowa State University Press: Ames, IA, USA, 1987; p. 357. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Srere, P.A. Citrate synthase: EC 4.1.3.7. Citrate oxaloacetate-lyase (CoA-acetylating). In Methods in Enzymology; Lowenstein, J.M., Ed.; Academic Press: New York, NY, USA, 1969; Volume 13, pp. 3–11. [Google Scholar]
- Sánchez-Nuño, S.; Sanahuja, I.; Ferández-Alacid, L.; Ordóñez-Grande, B.; Fontanillas, R.; Fernández-Borràs, J.; Blasco, J.; Carbonell, T.; Ibarz, A. Redox challenge in a cultured temperate marine species during low temperature and temperature recovery. Front. Physiol. 2018, 9, 923. [Google Scholar] [CrossRef] [PubMed]
- McCord, J.M.; Fridovich, I. The utility of superoxide dismutase in studying free radical reactions. J. Biol. Chem. 1969, 244, 6056–6063. [Google Scholar] [CrossRef]
- Furné, M.; García-Gallego, M.; Hidalgo, M.C.; Morales, A.E.; Domezain, A.; Domezain, J.; Sanz, A. Oxidative stress parameters during starvation and refeeding periods in Adriatic sturgeon (Acipenser naccarii) and rainbow trout (Oncorhynchus mykiss). Aquac. Nutr. 2009, 15, 587–595. [Google Scholar] [CrossRef]
- Aebi, H. Catalase in vitro. Meth. Enzymol. 1984, 105, 121–126. [Google Scholar]
- Bell, J.G.; Cowey, C.B.; Adron, J.W.; Shanks, A.M. Some effects of vitamin E and selenium deprivation on tissue enzyme levels and indices of tissue peroxidation in rainbow trout (Salmo gairdneri). Br. J. Nutr. 1985, 53, 149–157. [Google Scholar] [CrossRef]
- Carlberg, I.; Mannervik, B. Glutathione reductase. Methods Enzymol. 1985, 113, 484–490. [Google Scholar] [PubMed]
| Fingerlings | Juveniles | |||||
|---|---|---|---|---|---|---|
| Control | rbGH | Control | rbGH | |||
| Initial B.W. | 1.04 ± 0.02 | 1.01 ± 0.05 | 16.3 ± 0.06 | 16.3 ± 0.34 | ||
| Final B.W. | 8.07 ± 0.25 | 8.84 ± 0.18 | * | 57.2 ± 1.49 | 70.6 ± 2.20 | ** |
| SGR | 4.9 ± 0.08 | 5.2 ± 0.07 | * | 1.98 ± 0.02 | 2.33 ± 0.02 | *** |
| CF | 1.5 ± 0.01 | 1.5 ± 0.01 | 2.53 ± 0.03 | 2.39 ± 0.04 | ||
| HSI | 1.6 ± 0.04 | 1.4 ± 0.04 | ** | 1.5 ± 0.06 | 1.2 ± 0.05 | ** |
| MSI | - | - | 34.0 ± 0.63 | 34.8 ± 0.34 | ||
| MFI | 1.5 ± 0.07 | 1.3 ± 0.07 | 1.5 ± 0.06 | 1.2 ± 0.06 | ||
| FCR | 1.7 ± 0.13 | 1.6 ± 0.12 | 1.7± 0.14 | 1.3 ± 0.05 | * | |
| Fingerlings | Juveniles | |||||
|---|---|---|---|---|---|---|
| Control | rbGH | Control | rbGH | |||
| Composition | ||||||
| Protein (% w.w.) | 19.2 ± 0.17 | 19.2 ± 0.18 | 19.8 ± 0.28 | 19.7 ± 0.22 | ||
| Lipids (% w.w.) | 2.2 ± 0.12 | 1.5 ± 0.08 | *** | 2.4 ± 0.12 | 1.5 ± 0.10 | *** |
| Glycogen (% w.w.) | 0.15 ± 0.01 | 0.17 ± 0.02 | 0.25 ± 0.04 | 0.46 ± 0.05 | ** | |
| Wet weight (%) | 78.5 ± 0.14 | 78.5 ± 0.14 | 75.1 ± 0.26 | 76.5 ± 0.22 | * | |
| RNA (µg/mg prot) | 6.7 ± 0.19 | 7.3 ± 0.56 | 4.1 ± 0.09 | 5.4 ± 0.20 | *** | |
| DNA (µg/mg prot) | 1.46 ± 0.07 | 1.42 ± 0.13 | 0.9 ± 0.03 | 1.0 ± 0.05 | ||
| RNA/DNA | 4.7 ± 0.27 | 4.8 ± 0.28 | 4.7 ± 0.18 | 5.7 ± 0.32 | ** | |
| Enzyme activities | ||||||
| CS 1 | 77.7 ± 3.50 | 81.0 ± 5.07 | 42.3 ± 1.98 | 44.7 ± 1.81 | ||
| COX 1 | 27.5 ± 1.25 | 26.9 ± 2.19 | 15.1 ± 0.56 | 14.5 ± 0.83 | ||
| COX/CS | 0.35 ± 0.02 | 0.34 ± 0.03 | 0.37 ± 0.03 | 0.33 ± 0.02 | ||
| Juveniles | ||||||
|---|---|---|---|---|---|---|
| Red Muscle | Liver | |||||
| Control | rbGH | Control | rbGH | |||
| Composition | ||||||
| Protein (% w.w.) | 15.7 ± 1.29 | 16.9 ± 1.49 | 12.5 ± 0.62 | 12.5 ± 0.79 | ||
| Glycogen (% w.w.) | 0.45 ± 0.06 | 0.75 ± 0.02 | ** | 12.5 ± 0.85 | 9.9 ± 0.51 | * |
| Lipids (% w.w.) | 19.2 ± 0.87 | 14.4 ± 1.70 | 16.8 ± 0.77 | 13.6 ± 0.97 | * | |
| Enzyme activities | ||||||
| CS 1 | 467 ± 23.1 | 515 ± 51.2 | 34.3 ± 1.76 | 40.1 ± 1.37 | * | |
| COX 1 | 131 ± 10.3 | 125 ± 9.92 | 121.8 ± 3.8 | 114.4 ± 5.20 | ||
| HOAD 2 | 5.09 ± 0.35 | 5.14 ± 0.20 | ||||
| LDH 1 | 8.01 ± 0.68 | 8.07 ± 0.41 | ||||
| ALAT 1 | 45.6 ± 1.22 | 39.4 ± 1.01 | ** | |||
| ASAT 1 | 77.6 ± 4.53 | 81.3 ± 3.16 | ||||
| Fingerlings | Juveniles | |||||
|---|---|---|---|---|---|---|
| Control | rbGH | Control | rbGH | |||
| δ13 C-muscle | −20.60 ± 0.03 | −20.45 ± 0.03 | ** | −20.49 ± 0.12 | −19.77 ± 0.06 | *** |
| δ13 C-lipid | −26.15 ± 0.05 | −25.97 ± 0.06 | * | −25.61 ± 0.02 | −25.46 ± 0.03 | *** |
| δ13 C-glycogen | −20.91 ± 0.14 | −20.60 ± 0.14 | −19.93 ± 0.19 | −19.41 ± 0.17 | * | |
| δ13 C-protein | −21.51 ± 0.05 | −21.89 ± 0.36 | −20.49 ± 0.02 | −20.56 ± 0.05 | ||
| δ15 N-muscle | 12.19 ± 0.04 | 12.13 ± 0.06 | 9.64 ± 0.11 | 9.24 ± 0.06 | ** | |
| δ15 N-protein | 13.47 ± 0.06 | 13.19 ± 0.09 | * | 10.59 ± 0.10 | 10.22 ± 0.07 | ** |
| ∆15 N-muscle 1 | 2.57 ± 0.04 | 2.51 ± 0.07 | 0.02 ± 0.11 | -0.38 ± 0.06 | ** | |
| ∆15 N-protein 1 | 3.73 ± 0.05 | 3.45 ± 0.09 | * | 0.85 ± 0.10 | 0.48 ± 0.07 | * |
| a SPOT | Accession No. | b Protein Name | Species | c Symbol | Theorical KDa/pI | Observed Kda/pI | d score | e Peptides (Unique) | f SC (%) | g FC | h p-Value | UniprotKB | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cellular Metabolic Process: GO: 0044237 | |||||||||||||
| Carbohydrate Metabolic Process: GO: 0005975 (p-Value: 2.3−47) | |||||||||||||
| 125 | I3JBN0 | Alpha-1,4 glucan phosphorylase | Oreochromis niloticus | PYGM | 97.1/7.1 | 95.0/8.5 | 233.68 | 28 (2) | 34.68 | 0.24 | 0.0102 | P11217 | |
| 538 | I3JBN0 | Alpha-1,4 glucan phosphorylase | Oreochromis niloticus | PYGM | 97.1/7.1 | 96.0/6.9 | 991.63 | 35 (6) | 40.50 | 1.39 | 0.0422 | P11217 | |
| 540 | I3JBN0 | Alpha-1,4 glucan phosphorylase | Oreochromis niloticus | PYGM | 97.1/7.1 | 96.0/6.9 | 991.11 | 33 (5) | 39.07 | 0.74 | 0.0026 | P11217 | |
| 117 | Q4SFP9 | Alpha-1,4 glucan phosphorylase (Fragment) | Tetraodon nigroviridis | PYGM | 97.2/6.9 | 95.0/7.3 | 1085.21 | 36 (1) | 41.74 | 0.40 | 0.0003 | P11217 | |
| 541 | G3QBP8 | Alpha-1,4 glucan phosphorylase | Gasterosteus aculeatus | PYGM | 83.5/6.9 | 97.0/6.8 | 100.04 | 7 (2) | 13.36 | 1.51 | 0.0001 | P11217 | |
| 909 | Q4SFP9 | Alpha-1,4 glucan phosphorylase (Fragment) | Tetraodon nigroviridis | PYGM | 97.2/6.9 | 95.0/7.2 | 244.66 | 30 (2) | 37.57 | 3.21 | 0.0018 | P11217 | |
| 124 | I3JBN0 | Alpha-1,4 glucan phosphorylase | Oreochromis niloticus | PYGM | 97.1/7.1 | 96.0/8.3 | 524.38 | 36 (3) | 40.26 | 0.32 | 0.0117 | P11217 | |
| 118 | A0A147AQX3 | Alpha-1,4 glucan phosphorylase | Fundulus heteroclitus | PYGM | 102.7/7.3 | 97.0/7.4 | 907.09 | 33 (3) | 37.95 | 0.34 | 0.0285 | P11217 | |
| 126 | A0A147AQX3 | Alpha-1,4 glucan phosphorylase | Fundulus heteroclitus | PYGM | 102.7/7.3 | 96.0/8.6 | 304.63 | 31 (4) | 33.22 | 0.45 | 0.0275 | P11217 | |
| 875 | I3JBN0 | Alpha-1,4 glucan phosphorylase | Oreochromis niloticus | PYGM | 97.1/7.1 | 109.0/6.8 | 309.30 | 32 (3) | 38.36 | 0.23 | 0.0211 | P11217 | |
| 494 | I3KL67 | Phosphoglycerate kinase | Oreochromis niloticus | PGK1 | 44.5/6.9 | 50.0/6.9 | 905.26 | 20 (3) | 48.68 | 0.70 | 0.0244 | P00558 | |
| 157 | Q155W8 | Glyceraldehyde-3-phosphate dehydrogenase | Sparus aurata | GAPDH | 36.0/8.4 | 20.2/8.2 | 705.47 | 19 (6) | 72.67 | 0.46 | 0.0341 | P04406 | |
| 158 | Q155W8 | Glyceraldehyde-3-phosphate dehydrogenase | Sparus aurata | GAPDH | 36.0/8.4 | 24.9/7.8 | 463.46 | 15 (7) | 69.07 | 0.29 | 0.0548 | P04406 | |
| 84 | Q155W8 | Glyceraldehyde-3-phosphate dehydrogenase | Sparus aurata | GAPDH | 36.0/8.4 | 67.0/8.2 | 1083.31 | 15 (7) | 67.27 | 0.34 | 0.0289 | P04406 | |
| 625 | Q155W8 | Glyceraldehyde-3-phosphate dehydrogenase | Sparus aurata | GAPDH | 36.0/8.4 | 25.3/8.0 | 401.04 | 11 (11) | 60.96 | 0.53 | 0.0227 | P04406 | |
| 905 | Q155W8 | Glyceraldehyde-3-phosphate dehydrogenase | Sparus aurata | GAPDH | 36.0/8.4 | 70.0/7.9 | 33.25 | 6 (6) | 32.73 | 0.18 | 0.0368 | P04406 | |
| 22 | Q155W8 | Glyceraldehyde-3-phosphate dehydrogenase | Sparus aurata | GAPDH | 36.0/8.4 | 44.0/6.8 | 126.74 | 15 (6) | 57.06 | 2.40 | 0.0285 | P04406 | |
| 530 | Q155W8 | Glyceraldehyde-3-phosphate dehydrogenase | Sparus aurata | GAPDH | 36.0/8.4 | 87.0/8.1 | 100.59 | 10 (5) | 49.25 | 0.14 | 0.0293 | P04406 | |
| 734 | Q155W8 | Glyceraldehyde-3-phosphate dehydrogenase | Sparus aurata | GAPDH | 36.0/8.4 | 108/8.2 | 83.44 | 12 (7) | 62.46 | 0.65 | 0.0081 | P04406 | |
| 37 | O13276 | L-lactate dehydrogenase A chain | Sphyraena argentea | LDHA | 36.4/8.0 | 69.0/6.9 | 942.71 | 17 (2) | 37.95 | 0.37 | 0.0176 | P00338 | |
| 728 | O13276 | L-lactate dehydrogenase A chain | Sphyraena argentea | LDHA | 36.4/8.0 | 44.0/6.9 | 557.60 | 12 (3) | 32.23 | 0.50 | 0.0348 | P00338 | |
| 726 | O13276 | L-lactate dehydrogenase A chain | Sphyraena argentea | LDHA | 36.4/8.0 | 41.0/6.9 | 868.73 | 14 (1) | 34.34 | 0.40 | 0.0094 | P00338 | |
| 727 | O13276 | L-lactate dehydrogenase A chain | Sphyraena argentea | LDHA | 36.4/8.0 | 48.0/6.9 | 559.10 | 14 (4) | 34.34 | 1.92 | 0.0279 | P00338 | |
| 508 | A0A147AE36 | Alpha-enolase | Fundulus heteroclitus | ENO1 | 47.5/6.8 | 64.0/7.3 | 1028.05 | 20 (1) | 58.29 | 0.55 | 0.0220 | P06733 | |
| 903 | A0A147AE36 | Alpha-enolase | Fundulus heteroclitus | ENO1 | 47.5/6.8 | 65.0/7.7 | 865.01 | 15 (1) | 47.24 | 0.40 | 0.0322 | P06733 | |
| 36 | A0A146XBN2 | Alpha-enolase | Fundulus heteroclitus | ENO1 | 46.4/6.7 | 65.0/6.6 | 901.83 | 15 (1) | 50.70 | 0.53 | 0.0086 | P06733 | |
| 94 | A0A0F8AK35 | Pyruvate kinase | Larimichthys crocea | PKM | 58.2/7.7 | 73.0/8.5 | 819.93 | 15 (4) | 36.04 | 0.39 | 0.0216 | P14618 | |
| 523 | Q8QGU8 | Pyruvate kinase | Takifugu rubripes | PKM | 58.0/7.9 | 74.0/7.6 | 657.85 | 18 (2) | 34.53 | 0.44 | 0.0037 | P14618 | |
| 100 | A0A0F8AK35 | Pyruvate kinase | Larimichthys crocea | PKM | 58.2/7.7 | 74.0/8.0 | 195.81 | 8 (1) | 21.51 | 0.22 | 0.0120 | P14618 | |
| 500 | H2TGY6 | Fructose-bisphosphate aldolase | Takifugu rubripes | ALDOA | 39.6/8.3 | 54.0/7.7 | 283.00 | 12 (1) | 40.22 | 0.56 | 0.0051 | P04075 | |
| 764 | A0A1A8A8E2 | Triosephosphate isomerase | Nothobranchius furzeri | TPI1 | 26.5/7.3 | 29.0/7.3 | 69.63 | 7 (7) | 36.84 | 0.29 | 0.0010 | P60174 | |
| Others | |||||||||||||
| 279 | Q71N41 | Guanidinoacetate N-methyltransferase | Danio rerio | GAMT | 26.7/6.3 | 33.0/6.0 | 513.76 | 5 (2) | 28.21 | 1.50 | 0.0138 | Q14353 | |
| 493 | A0A146WHL4 | Creatine kinase M-type | Fundulus heteroclitus | CKM | 42.8/6.8 | 53.0/6.9 | 1117.72 | 15 (1) | 41.10 | 0.46 | 0.0123 | P06732 | |
| 206 | A0A0F8AHC2 | Glucose-regulated protein | Larimichthys crocea | HSPA5 | 82.3/5.6 | 95.0/5.6 | 152.41 | 19 (3) | 29.88 | 0.64 | 0.0088 | P11021 | |
| 717 | A0A0F6MX10 | ATP synthase subunit alpha | Sparus aurata | ATP5F1A | 59.6/9.1 | 68.0/6.9 | 94.37 | 15 (15) | 38.66 | 0.15 | 0.0185 | P25705 | |
| 681 | D6PVP3 | Tropomyosin | Epinephelus coioides | TPM1 | 32.7/4.7 | 45.0/4.7 | 183.97 | 17 (2) | 46.13 | 0.34 | 0.0080 | P09493 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blasco, J.; Vélez, E.J.; Perelló-Amorós, M.; Azizi, S.; Capilla, E.; Fernández-Borràs, J.; Gutiérrez, J. Recombinant Bovine Growth Hormone-Induced Metabolic Remodelling Enhances Growth of Gilthead Sea-Bream (Sparus aurata): Insights from Stable Isotopes Composition and Proteomics. Int. J. Mol. Sci. 2021, 22, 13107. https://doi.org/10.3390/ijms222313107
Blasco J, Vélez EJ, Perelló-Amorós M, Azizi S, Capilla E, Fernández-Borràs J, Gutiérrez J. Recombinant Bovine Growth Hormone-Induced Metabolic Remodelling Enhances Growth of Gilthead Sea-Bream (Sparus aurata): Insights from Stable Isotopes Composition and Proteomics. International Journal of Molecular Sciences. 2021; 22(23):13107. https://doi.org/10.3390/ijms222313107
Chicago/Turabian StyleBlasco, Josefina, Emilio J. Vélez, Miquel Perelló-Amorós, Sheida Azizi, Encarnación Capilla, Jaume Fernández-Borràs, and Joaquim Gutiérrez. 2021. "Recombinant Bovine Growth Hormone-Induced Metabolic Remodelling Enhances Growth of Gilthead Sea-Bream (Sparus aurata): Insights from Stable Isotopes Composition and Proteomics" International Journal of Molecular Sciences 22, no. 23: 13107. https://doi.org/10.3390/ijms222313107
APA StyleBlasco, J., Vélez, E. J., Perelló-Amorós, M., Azizi, S., Capilla, E., Fernández-Borràs, J., & Gutiérrez, J. (2021). Recombinant Bovine Growth Hormone-Induced Metabolic Remodelling Enhances Growth of Gilthead Sea-Bream (Sparus aurata): Insights from Stable Isotopes Composition and Proteomics. International Journal of Molecular Sciences, 22(23), 13107. https://doi.org/10.3390/ijms222313107








