The Expression Pattern of Genes Related to Melanogenesis and Endogenous Opioids in Psoriasis
Abstract
1. Introduction
2. Results
2.1. mRNA Expression in Lesional, Non-Lesional, and Control Skin
2.2. Expression Level Interactions
3. Discussion
4. Materials and Methods
4.1. Patients and Healthy Controls
4.2. Skin Samples and Quantitative Real-Time-PCR
4.3. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kurd, S.K.; Gelfand, J.M. The prevalence of previously diagnosed and undiagnosed psoriasis in US adults: Results from NHANES 2003–2004. J. Am. Acad. Dermatol. 2009, 60, 218–224. [Google Scholar] [CrossRef]
- Parisi, R.; Symmons, D.P.; Griffiths, C.E.; Ashcroft, D.M. Global epidemiology of psoriasis: A systematic review of incidence and prevalence. J. Investig. Dermatol. 2013, 133, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Dowlatshahi, E.A.; van der Voort, E.A.; Arends, L.R.; Nijsten, T. Markers of systemic inflammation in psoriasis: A systematic review and meta-analysis. Br. J. Dermatol. 2013, 169, 266–282. [Google Scholar] [CrossRef]
- Ogawa, E.; Sato, Y.; Minagawa, A.; Okuyama, R. Pathogenesis of psoriasis and development of treatment. J. Dermatol. 2018, 45, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Dand, N.; Mahil, S.K.; Capon, F.; Smith, C.H.; Simpson, M.A.; Barker, J.N. Psoriasis and Genetics. Acta Derm. Venereol. 2020, 100, adv00030. [Google Scholar] [CrossRef]
- Slominski, A.T.; Kleszczynski, K.; Semak, I.; Janjetovic, Z.; Zmijewski, M.A.; Kim, T.K.; Slominski, R.M.; Reiter, R.J.; Fischer, T.W. Local melatoninergic system as the protector of skin integrity. Int. J. Mol. Sci. 2014, 15, 17705–17732. [Google Scholar] [CrossRef] [PubMed]
- Slominski, A.T.; Zmijewski, M.A.; Skobowiat, C.; Zbytek, B.; Slominski, R.M.; Steketee, J.D. Sensing the environment: Regulation of local and global homeostasis by the skin’s neuroendocrine system. Adv. Anat. Embryol. Cell Biol. 2012, 212, 1–115. [Google Scholar]
- Pasparakis, M.; Haase, I.; Nestle, F.O. Mechanisms regulating skin immunity and inflammation. Nat. Rev. Immunol. 2014, 14, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Slominski, A. Neuroendocrine system of the skin. Dermatology 2005, 211, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.J.; Montero-Melendez, T.; Perretti, M.; Pitzalis, C. Curbing Inflammation through Endogenous Pathways: Focus on Melanocortin Peptides. Int. J. Inflam. 2013, 2013, 985815. [Google Scholar] [CrossRef]
- Wang, W.; Guo, D.Y.; Lin, Y.J.; Tao, Y.X. Melanocortin Regulation of Inflammation. Front. Endocrinol. 2019, 10, 683. [Google Scholar] [CrossRef]
- Fu, C.; Chen, J.; Lu, J.; Yi, L.; Tong, X.; Kang, L.; Pei, S.; Ouyang, Y.; Jiang, L.; Ding, Y.; et al. Roles of inflammation factors in melanogenesis (Review). Mol. Med. Rep. 2020, 21, 1421–1430. [Google Scholar] [CrossRef]
- Abdel-Naser, M.B.; Liakou, A.I.; Elewa, R.; Hippe, S.; Knolle, J.; Zouboulis, C.C. Increased Activity and Number of Epidermal Melanocytes in Lesional Psoriatic Skin. Dermatology 2016, 232, 425–430. [Google Scholar] [CrossRef]
- Wang, C.Q.F.; Akalu, Y.T.; Suarez-Farinas, M.; Gonzalez, J.; Mitsui, H.; Lowes, M.A.; Orlow, S.J.; Manga, P.; Krueger, J.G. IL-17 and TNF synergistically modulate cytokine expression while suppressing melanogenesis: Potential relevance to psoriasis. J. Investig. Dermatol. 2013, 133, 2741–2752. [Google Scholar] [CrossRef]
- Vachiramon, V.; Thadanipon, K. Postinflammatory hypopigmentation. Clin. Exp. Dermatol. 2011, 36, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Loite, U.; Kingo, K.; Reimann, E.; Reemann, P.; Vasar, E.; Silm, H.; Koks, S. Gene expression analysis of the corticotrophin-releasing hormone-proopiomelanocortin system in psoriasis skin biopsies. Acta Derm. Venereol. 2013, 93, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Traks, T.; Keermann, M.; Karelson, M.; Ratsep, R.; Reimann, E.; Silm, H.; Vasar, E.; Koks, S.; Kingo, K. Polymorphisms in Corticotrophin-releasing Hormone-proopiomelanocortin (CRH-POMC) System Genes are Associated with Plaque Psoriasis. Acta Derm. Venereol. 2019, 99, 444–445. [Google Scholar] [CrossRef]
- Bigliardi, P.L.; Tobin, D.J.; Gaveriaux-Ruff, C.; Bigliardi-Qi, M. Opioids and the skin—Where do we stand? Exp. Dermatol. 2009, 18, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Slominski, A.; Wortsman, J.; Luger, T.; Paus, R.; Solomon, S. Corticotropin releasing hormone and proopiomelanocortin involvement in the cutaneous response to stress. Physiol. Rev. 2000, 80, 979–1020. [Google Scholar] [CrossRef]
- Veening, J.G.; Barendregt, H.P. The effects of beta-endorphin: State change modification. Fluids Barriers CNS 2015, 12, 3. [Google Scholar] [CrossRef]
- He, X.; Huang, L.; Qiu, S.; Yin, X.; Shen, Y.; Wu, Y.; Jiang, Y.; Fang, J. beta-Endorphin attenuates collagen-induced arthritis partially by inhibiting peripheral pro-inflammatory mediators. Exp. Ther. Med. 2018, 15, 4014–4018. [Google Scholar]
- Bigliardi, P.L.; Dancik, Y.; Neumann, C.; Bigliardi-Qi, M. Opioids and skin homeostasis, regeneration and ageing—What’s the evidence? Exp. Dermatol. 2016, 25, 586–591. [Google Scholar] [CrossRef]
- Rittner, H.L.; Brack, A. Leukocytes as mediators of pain and analgesia. Curr. Rheumatol. Rep. 2007, 9, 503–510. [Google Scholar] [CrossRef]
- Reimann, E.; Kingo, K.; Karelson, M.; Salum, T.; Aunin, E.; Reemann, P.; Abram, K.; Vasar, E.; Silm, H.; Koks, S. Analysis of the expression profile of CRH-POMC system genes in vitiligo skin biopsies. J. Dermatol. Sci. 2010, 60, 125–128. [Google Scholar] [CrossRef] [PubMed]
- D’Mello, S.A.; Finlay, G.J.; Baguley, B.C.; Askarian-Amiri, M.E. Signaling Pathways in Melanogenesis. Int. J. Mol. Sci. 2016, 17, 1144. [Google Scholar] [CrossRef]
- Busca, R.; Ballotti, R. Cyclic AMP a key messenger in the regulation of skin pigmentation. Pigment. Cell Res. 2000, 13, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Yasumoto, K.; Takeda, K.; Saito, H.; Watanabe, K.; Takahashi, K.; Shibahara, S. Microphthalmia-associated transcription factor interacts with LEF-1, a mediator of Wnt signaling. EMBO J. 2002, 21, 2703–2714. [Google Scholar] [CrossRef] [PubMed]
- Saha, B.; Singh, S.K.; Sarkar, C.; Bera, R.; Ratha, J.; Tobin, D.J.; Bhadra, R. Activation of the Mitf promoter by lipid-stimulated activation of p38-stress signalling to CREB. Pigment Cell Res. 2006, 19, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Doerschuk, C.M. The p38 mitogen-activated protein kinase mediates cytoskeletal remodeling in pulmonary microvascular endothelial cells upon intracellular adhesion molecule-1 ligation. J. Immunol. 2001, 166, 6877–6884. [Google Scholar] [CrossRef] [PubMed]
- Swope, V.B.; Abdel-Malek, Z.; Kassem, L.M.; Nordlund, J.J. Interleukins 1 alpha and 6 and tumor necrosis factor-alpha are paracrine inhibitors of human melanocyte proliferation and melanogenesis. J. Investig. Dermatol. 1991, 96, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Paik, P.K.; Chen, J.; Yarilina, A.; Kockeritz, L.; Lu, T.T.; Woodgett, J.R.; Ivashkiv, L.B. IFN-gamma suppresses IL-10 production and synergizes with TLR2 by regulating GSK3 and CREB/AP-1 proteins. Immunity 2006, 24, 563–574. [Google Scholar] [CrossRef]
- Srivastava, A.; Luo, L.; Lohcharoenkal, W.; Meisgen, F.; Pasquali, L.; Pivarcsi, A.; Sonkoly, E. Cross-talk between IFN-γ and TWEAK through miR-149 amplifies skin inflammation in psoriasis. J. Allergy Clin. Immunol. 2021, 147, 2225–2235. [Google Scholar] [CrossRef]
- Son, J.; Kim, M.; Jou, I.; Park, K.C.; Kang, H.Y. IFN-γ inhibits basal and α-MSH-induced melanogenesis. Pigment Cell Melanoma Res. 2014, 27, 201–208. [Google Scholar] [CrossRef]
- Kingo, K.; Aunin, E.; Karelson, M.; Rätsep, R.; Silm, H.; Vasar, E.; Kõks, S. Expressional changes in the intracellular melanogenesis pathways and their possible role in the pathogenesis of vitiligo. J. Dermatol. Sci. 2008, 52, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Johansen, C.; Kragballe, K.; Westergaard, M.; Henningsen, J.; Kristiansen, K.; Iversen, L. The mitogen-activated protein kinases p38 and ERK1/2 are increased in lesional psoriatic skin. Br. J. Dermatol. 2005, 152, 37–42. [Google Scholar] [CrossRef]
- Sakurai, K.; Dainichi, T.; Garcet, S.; Tsuchiya, S.; Yamamoto, Y.; Kitoh, A.; Honda, T.; Nomura, T.; Egawa, G.; Otsuka, A.; et al. Cutaneous p38 mitogen-activated protein kinase activation triggers psoriatic dermatitis. J. Allergy Clin. Immunol. 2019, 144, 1036–1049. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.; Kiger, A.A. Classes of phosphoinositide 3-kinases at a glance. J. Cell Sci. 2014, 127, 923–928. [Google Scholar] [CrossRef] [PubMed]
- Martelli, A.M.; Nyakern, M.; Tabellini, G.; Bortul, R.; Tazzari, P.L.; Evangelisti, C.; Cocco, L. Phosphoinositide 3-kinase/Akt signaling pathway and its therapeutical implications for human acute myeloid leukemia. Leukemia 2006, 20, 911–928. [Google Scholar] [CrossRef]
- Wu, P.Y.; You, Y.J.; Liu, Y.J.; Hou, C.W.; Wu, C.S.; Wen, K.C.; Lin, C.Y.; Chiang, H.M. Sesamol Inhibited Melanogenesis by Regulating Melanin-Related Signal Transduction in B16F10 Cells. Int. J. Mol. Sci. 2018, 19, 1108. [Google Scholar] [CrossRef]
- Berven, L.A.; Crouch, M.F. Cellular function of p70S6K: A role in regulating cell motility. Immunol. Cell Biol. 2000, 78, 447–451. [Google Scholar] [CrossRef]
- Busca, R.; Bertolotto, C.; Ortonne, J.P.; Ballotti, R. Inhibition of the phosphatidylinositol 3-kinase/p70(S6)-kinase pathway induces B16 melanoma cell differentiation. J. Biol. Chem. 1996, 271, 31824–31830. [Google Scholar] [CrossRef]
- Pimiento, J.M.; Larkin, E.M.; Smalley, K.S.; Wiersma, G.L.; Monks, N.R.; Fedorenko, I.V.; Peterson, C.A.; Nickoloff, B.J. Melanoma genotypes and phenotypes get personal. Lab. Investig. 2013, 93, 858–867. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, R.; Tsukamoto, K.; Harada, K.; Shimizu, A.; Shimada, S.; Kobayashi, T.; Imokawa, G. Mechanisms underlying the dysfunction of melanocytes in vitiligo epidermis: Role of SCF/KIT protein interactions and the downstream effector, MITF-M. J. Pathol. 2004, 202, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Serre, C.; Busuttil, V.; Botto, J.M. Intrinsic and extrinsic regulation of human skin melanogenesis and pigmentation. Int. J. Cosmet. Sci. 2018, 40, 328–347. [Google Scholar] [CrossRef] [PubMed]
- Watanuki, Y.; Takayasu, S.; Kageyama, K.; Iwasaki, Y.; Sakihara, S.; Terui, K.; Nigawara, T.; Suda, T. Involvement of Nurr-1/Nur77 in corticotropin-releasing factor/urocortin1-induced tyrosinase-related protein 1 gene transcription in human melanoma HMV-II cells. Mol. Cell. Endocrinol. 2013, 370, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.P.; McEvoy, A.; Conneely, O.M.; Bresnihan, B.; FitzGerald, O. Involvement of the nuclear orphan receptor NURR1 in the regulation of corticotropin-releasing hormone expression and actions in human inflammatory arthritis. Arthritis Rheum. 2001, 44, 782–793. [Google Scholar] [CrossRef]
- O’Kane, M.; Markham, T.; McEvoy, A.N.; Fearon, U.; Veale, D.J.; FitzGerald, O.; Kirby, B.; Murphy, E.P. Increased expression of the orphan nuclear receptor NURR1 in psoriasis and modulation following TNF-alpha inhibition. J. Investig. Dermatol. 2008, 128, 300–310. [Google Scholar] [CrossRef]
- Gunn, T.M.; Miller, K.A.; He, L.; Hyman, R.W.; Davis, R.W.; Azarani, A.; Schlossman, S.F.; Duke-Cohan, J.S.; Barsh, G.S. The mouse mahogany locus encodes a transmembrane form of human attractin. Nature 1999, 398, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Hida, T.; Wakamatsu, K.; Sviderskaya, E.V.; Donkin, A.J.; Montoliu, L.; Lynn Lamoreux, M.; Yu, B.; Millhauser, G.L.; Ito, S.; Barsh, G.S.; et al. Agouti protein, mahogunin, and attractin in pheomelanogenesis and melanoblast-like alteration of melanocytes: A cAMP-independent pathway. Pigment Cell Melanoma Res. 2009, 22, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Slominski, A.T.; Zmijewski, M.A.; Zbytek, B.; Brozyna, A.A.; Granese, J.; Pisarchik, A.; Szczesniewski, A.; Tobin, D.J. Regulated proenkephalin expression in human skin and cultured skin cells. J. Investig. Dermatol. 2011, 131, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, C.M.; Tobin, A.M.; Kirby, B. Innate immunity in the pathogenesis of psoriasis. Arch. Dermatol. Res. 2011, 303, 691–705. [Google Scholar] [CrossRef]
- Taneda, K.; Tominaga, M.; Negi, O.; Tengara, S.; Kamo, A.; Ogawa, H.; Takamori, K. Evaluation of epidermal nerve density and opioid receptor levels in psoriatic itch. Br. J. Dermatol. 2011, 165, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Stuber, F.; Stamer, U.M. Inflammatory mediators influence the expression of nociceptin and its receptor in human whole blood cultures. PLoS ONE 2013, 8, e74138. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gavioli, E.C.; Romao, P.R. NOP Receptor Ligands as Potential Agents for Inflammatory and Autoimmune Diseases. J. Amino Acids 2011, 2011, 836569. [Google Scholar] [CrossRef]
- Swindell, W.R.; Sarkar, M.K.; Liang, Y.; Xing, X.; Baliwag, J.; Elder, J.T.; Johnston, A.; Ward, N.L.; Gudjonsson, J.E. RNA-seq identifies a diminished differentiation gene signature in primary monolayer keratinocytes grown from lesional and uninvolved psoriatic skin. Sci. Rep. 2017, 7, 18045. [Google Scholar] [CrossRef] [PubMed]
- Pasquali, L.; Srivastava, A.; Meisgen, F.; Das Mahapatra, K.; Xia, P.; Xu Landén, N.; Pivarcsi, A.; Sonkoly, E. Keratinocyte Transcriptome in Psoriasis: Pathways Related to Immune Responses, Cell Cycle and Keratinization. Acta Derm. Venereol. 2019, 99, 196–205. [Google Scholar] [CrossRef]
- Mrowietz, U.; Kragballe, K.; Reich, K.; Spuls, P.; Griffiths, C.E.M.; Nast, A.; Franke, J.; Antoniou, C.; Arenberger, P.; Balieva, F.; et al. Definition of treatment goals for moderate to severe psoriasis: A European consensus. Arch. Dermatol. Res. 2011, 303, 1–10. [Google Scholar] [CrossRef]
 Receptor agonist
Receptor agonist  Receptor antagonist
Receptor antagonist  Positive regulation
Positive regulation  Negative regulation.
Negative regulation.
 Receptor agonist
Receptor agonist  Receptor antagonist
Receptor antagonist  Positive regulation
Positive regulation  Negative regulation.
Negative regulation.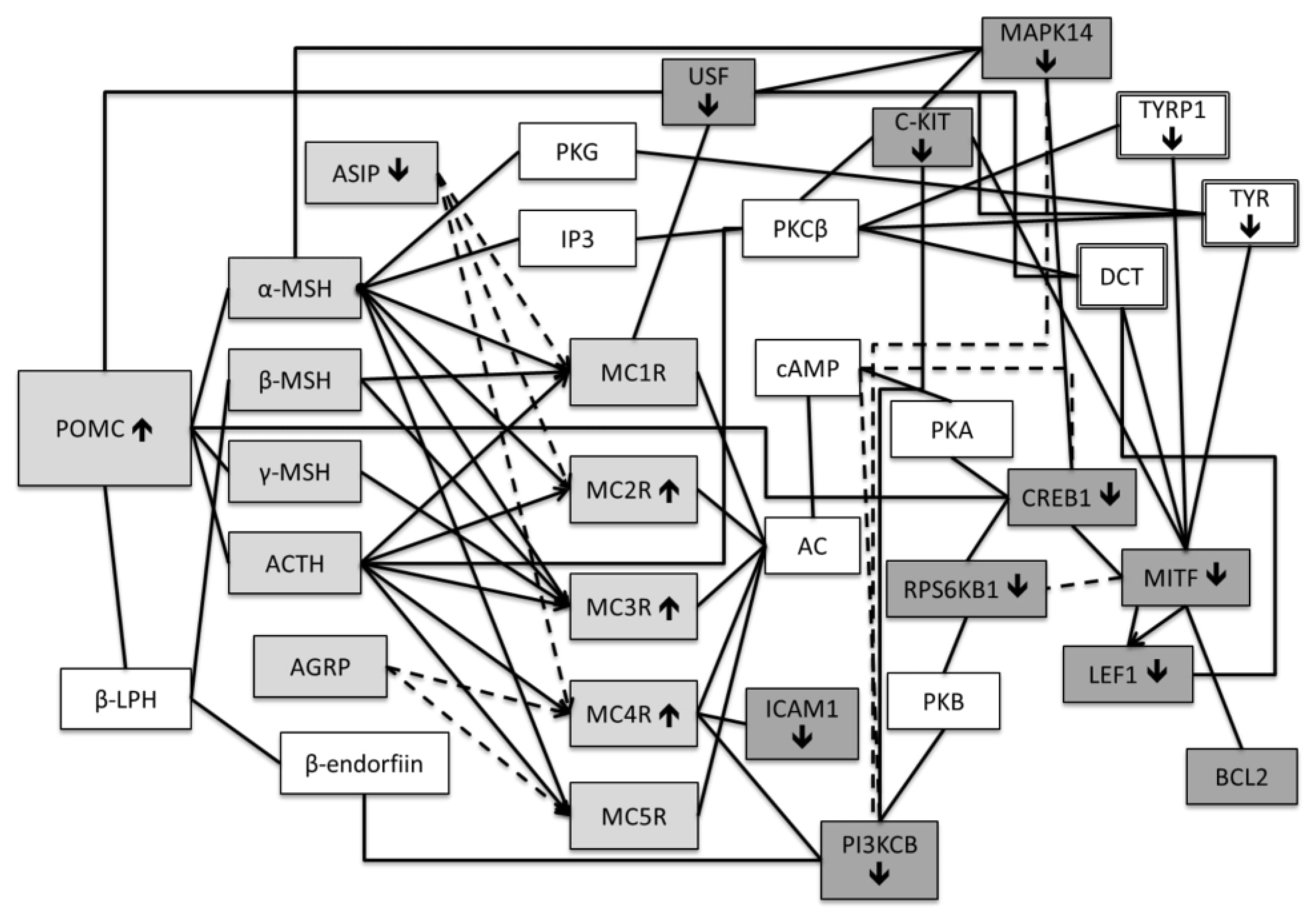


| BLC2 | CREB1 | LEF1 | PNOC | USF1 | PIK3CB | RPS6KB1 | MITF | MAPK14 | NURR1 | ATRN | KIT | ICAM1 | PDYN | PENK | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BLC2 | 1.00 | ||||||||||||||
| CREB1 | −0.16 | 1.00 | |||||||||||||
| LEF1 | −0.13 | −0.03 | 1.00 | ||||||||||||
| PNOC | −0.26 | −0.22 | 0.22 | 1.00 | |||||||||||
| USF1 | −0.11 | −0.17 | −0.20 | 0.58 * | 1.00 | ||||||||||
| PIK3CB | −0.21 | −0.14 | 0.05 | −0.08 | 0.33 | 1.00 | |||||||||
| RPS6KB1 | 0.00 | 0.27 | −0.03 | 0.06 | 0.11 | −0.11 | 1.00 | ||||||||
| MITF | 0.47 | −0.13 | −0.08 | −0.38 | −0.24 | −0.31 | −0.52 * | 1.00 | |||||||
| MAPK14 | −0.11 | 0.46 | 0.31 | −0.22 | −0.16 | −0.16 | 0.26 | 0.04 | 1.00 | ||||||
| NURR1 | 0.11 | −0.02 | −0.01 | 0.21 | 0.05 | −0.14 | 0.16 | 0.01 | 0.06 | 1.00 | |||||
| ATRN | −0.02 | −0.09 | −0.32 | 0.34 | 0.02 | −0.42 | 0.10 | 0.03 | −0.22 | −0.24 | 1.00 | ||||
| KIT | −0.10 | 0.22 | −0.09 | −0.07 | −0.06 | −0.15 | −0.17 | 0.09 | −0.28 | 0.44 | −0.27 | 1.00 | |||
| ICAM1 | 0.02 | −0.20 | −0.20 | 0.59 * | 0.51 * | −0.06 | 0.33 | −0.21 | 0.03 | 0.41 | 0.05 | −0.09 | 1.00 | ||
| PDYN | 0.25 | 0.24 | −0.23 | 0.14 | 0.10 | −0.34 | −0.10 | 0.12 | 0.03 | 0.08 | −0.15 | 0.06 | 0.18 | 1.00 | |
| PENK | 0.12 | −0.47 * | −0.42 | 0.22 | 0.23 | 0.16 | −0.29 | 0.12 | −0.48 * | 0.30 | −0.15 | 0.11 | 0.27 | 0.46 | 1.00 |
| BLC2 | CREB1 | LEF1 | PNOC | USF1 | PIK3CB | RPS6KB1 | MITF | MAPK14 | NURR1 | ATRN | KIT | ICAM1 | PDYN | PENK | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BLC2 | 1.00 | ||||||||||||||
| CREB1 | −0.40 | 1.00 | |||||||||||||
| LEF1 | −0.38 | 0.65 * | 1.00 | ||||||||||||
| PNOC | 0.08 | −0.38 | −0.53 | 1.00 | |||||||||||
| USF1 | −0.36 | −0.13 | −0.11 | 0.81 * | 1.00 | ||||||||||
| PIK3CB | 0.05 | 0.36 | 0.15 | −0.81 * | −0.02 | 1.00 | |||||||||
| RPS6KB1 | 0.05 | 0.34 | −0.34 | 0.11 | 0.23 | 0.18 | 1.00 | ||||||||
| MITF | 0.34 | −0.42 | −0.10 | 0.40 | −0.31 | −0.64 * | 0.04 | 1.00 | |||||||
| MAPK14 | 0.26 | −0.11 | 0.17 | 0.45 | −0.26 | −0.51 | −0.23 | 0.75 ** | 1.00 | ||||||
| NURR1 | 0.36 | −0.09 | −0.01 | 0.05 | 0.17 | −0.04 | 0.58 | 0.55 * | 0.23 | 1.00 | |||||
| ATRN | 0.11 | −0.40 | −0.28 | 0.40 | 0.09 | −0.31 | 0.16 | 0.58 * | 0.36 | 0.49 | 1.00 | ||||
| KIT | −0.23 | 0.06 | 0.05 | −0.21 | −0.17 | 0.15 | −0.71 * | −0.27 | 0.19 | −0.51 | −0.35 | 1.00 | |||
| ICAM1 | 0.10 | 0.29 | 0.27 | −0.42 | −0.23 | 0.15 | −0.24 | −0.37 | −0.22 | 0.09 | −0.33 | 0.34 | 1.00 | ||
| PDYN | −0.39 | −0.89 ** | −0.51 | 0.39 | 0.57 | −0.18 | 0.02 | 0.53 | 0.31 | 0.02 | 0.19 | −0.13 | −0.76 * | 1.00 | |
| PENK | 0.46 | −0.07 | −0.33 | 0.04 | −0.22 | −0.06 | 0.22 | 0.05 | −0.03 | 0.18 | 0.08 | −0.10 | 0.43 | −0.25 | 1.00 |
| BLC2 | CREB1 | LEF1 | PNOC | USF1 | PIK3CB | RPS6KB1 | MITF | MAPK14 | NURR1 | ATRN | KIT | ICAM1 | PDYN | PENK | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BLC2 | 1.00 | ||||||||||||||
| CREB1 | 0.16 | 1.00 | |||||||||||||
| LEF1 | −0.32 | 0.44 | 1.00 | ||||||||||||
| PNOC | −0.28 | 0.62 | 0.05 | 1.00 | |||||||||||
| USF1 | 0.42 | 0.12 | 0.22 | 0.02 | 1.00 | ||||||||||
| PIK3CB | 0.40 | 0.29 | 0.36 | 0.08 | 0.38 | 1.00 | |||||||||
| RPS6KB1 | −0.08 | 0.15 | −0.21 | −0.33 | −0.05 | −0.34 | 1.00 | ||||||||
| MITF | 0.23 | −0.18 | −0.15 | −0.43 | 0.22 | 0.53 ** | −0.12 | 1.00 | |||||||
| MAPK14 | 0.29 | 0.19 | 0.37 | 0.28 | 0.40 | 0.36 | −0.51 * | 0.36 | 1.00 | ||||||
| NURR1 | 0.07 | 0.38 | 0.13 | −0.05 | −0.14 | −0.08 | −0.23 | −0.25 | −0.14 | 1.00 | |||||
| ATRN | 0.43 | −0.48 | −0.10 | −0.09 | 0.14 | 0.50 | −0.16 | 0.01 | 0.54 | −0.46 | 1.00 | ||||
| KIT | 0.26 | −0.40 | 0.10 | 0.14 | −0.27 | 0.27 | −0.56 | −0.25 | 0.51 | −0.01 | 0.57 | 1.00 | |||
| ICAM1 | 0.13 | −0.28 | 0.02 | −0.32 | −0.04 | −0.26 | 0.10 | −0.22 | −0.30 | −0.10 | −0.36 | −0.03 | 1.00 | ||
| PDYN | −0.15 | 0.28 | −0.22 | 0.30 | 0.00 | −0.14 | −0.21 | −0.31 | −0.09 | 0.11 | 0.08 | −0.09 | 0.09 | 1.00 | |
| PENK | 0.13 | −0.35 | 0.23 | −0.28 | 0.14 | 0.33 | −0.09 | 0.34 | 0.40 | −0.41 | 0.06 | 0.37 | 0.22 | −0.73 * | 1.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loite, U.; Raam, L.; Reimann, E.; Reemann, P.; Prans, E.; Traks, T.; Vasar, E.; Silm, H.; Kingo, K.; Kõks, S. The Expression Pattern of Genes Related to Melanogenesis and Endogenous Opioids in Psoriasis. Int. J. Mol. Sci. 2021, 22, 13056. https://doi.org/10.3390/ijms222313056
Loite U, Raam L, Reimann E, Reemann P, Prans E, Traks T, Vasar E, Silm H, Kingo K, Kõks S. The Expression Pattern of Genes Related to Melanogenesis and Endogenous Opioids in Psoriasis. International Journal of Molecular Sciences. 2021; 22(23):13056. https://doi.org/10.3390/ijms222313056
Chicago/Turabian StyleLoite, Ulvi, Liisi Raam, Ene Reimann, Paula Reemann, Ele Prans, Tanel Traks, Eero Vasar, Helgi Silm, Külli Kingo, and Sulev Kõks. 2021. "The Expression Pattern of Genes Related to Melanogenesis and Endogenous Opioids in Psoriasis" International Journal of Molecular Sciences 22, no. 23: 13056. https://doi.org/10.3390/ijms222313056
APA StyleLoite, U., Raam, L., Reimann, E., Reemann, P., Prans, E., Traks, T., Vasar, E., Silm, H., Kingo, K., & Kõks, S. (2021). The Expression Pattern of Genes Related to Melanogenesis and Endogenous Opioids in Psoriasis. International Journal of Molecular Sciences, 22(23), 13056. https://doi.org/10.3390/ijms222313056







