DNA Repair Genes as Drug Candidates for Early Breast Cancer Onset in Latin America: A Systematic Review
Abstract
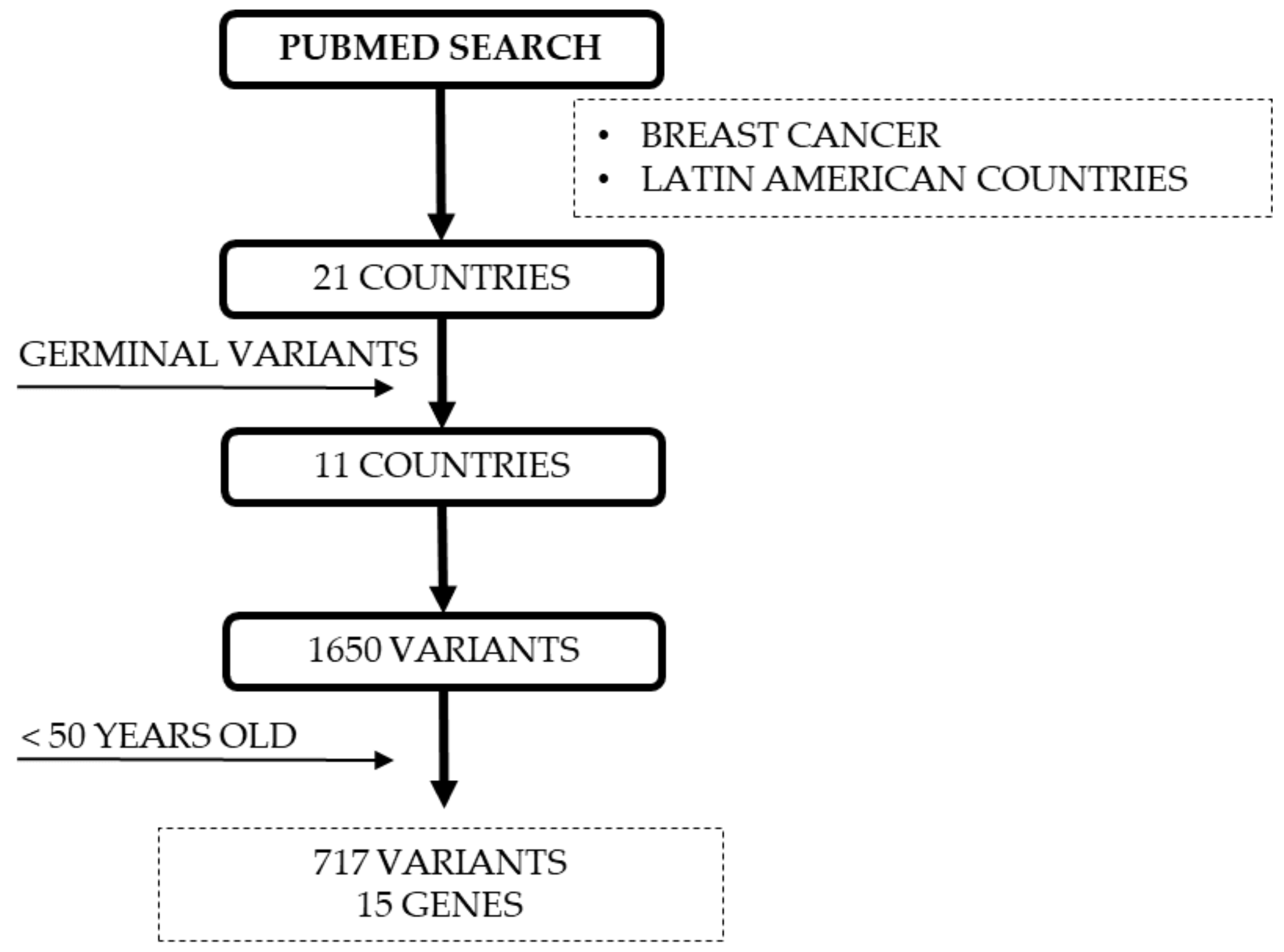
1. Introduction
2. Results
2.1. Tumor Samples Data for YWBC
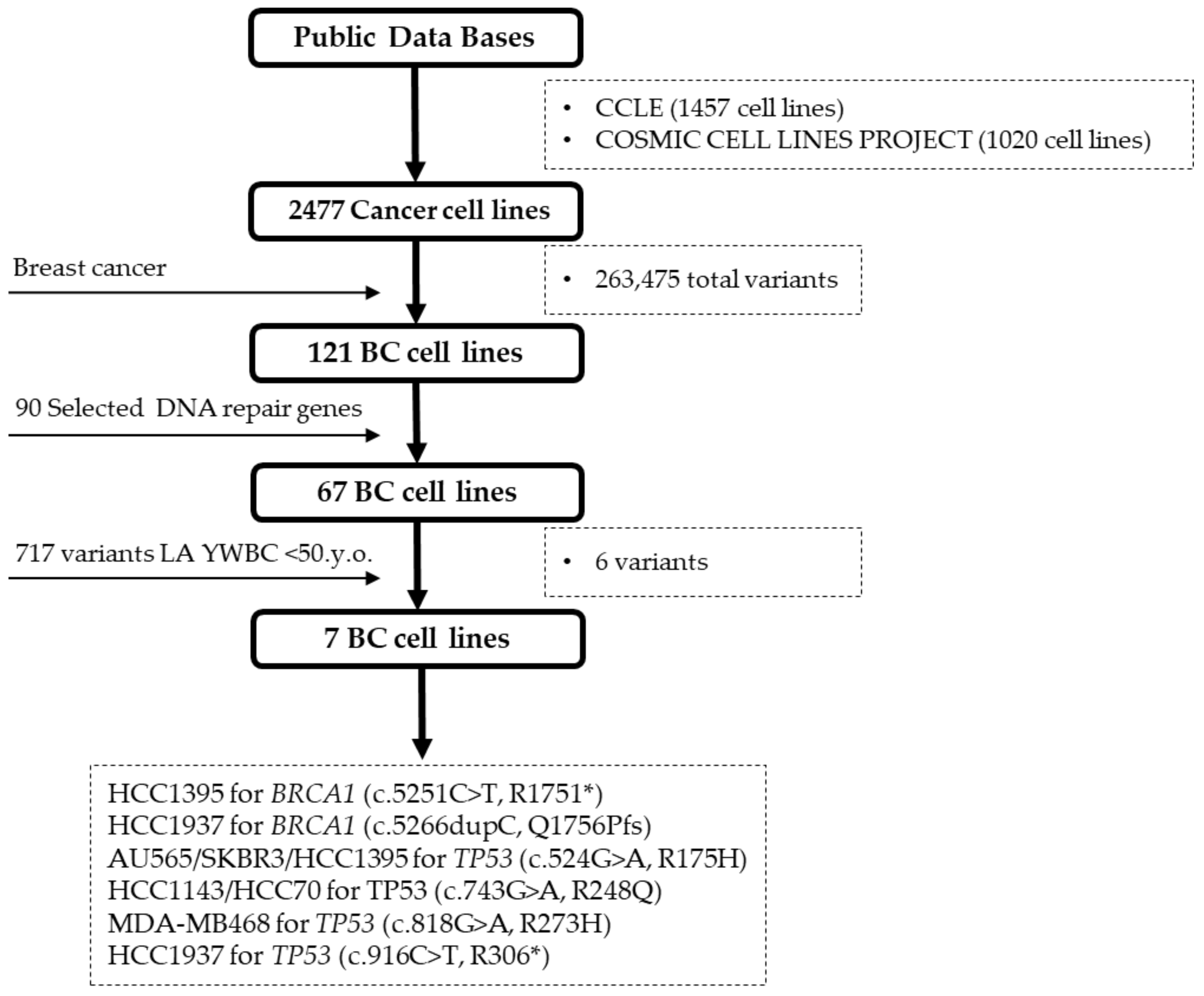
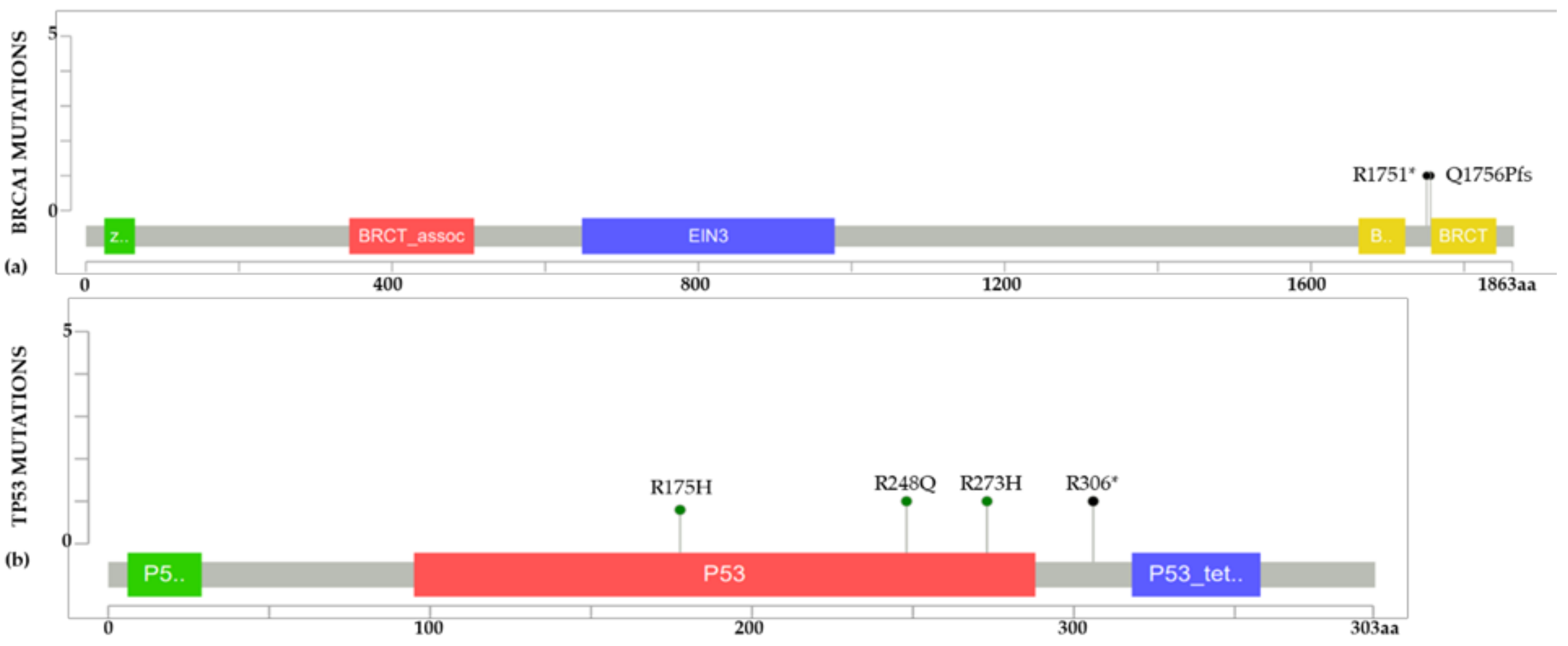
2.2. BC Cell Line Variants and DNA Repair Genes
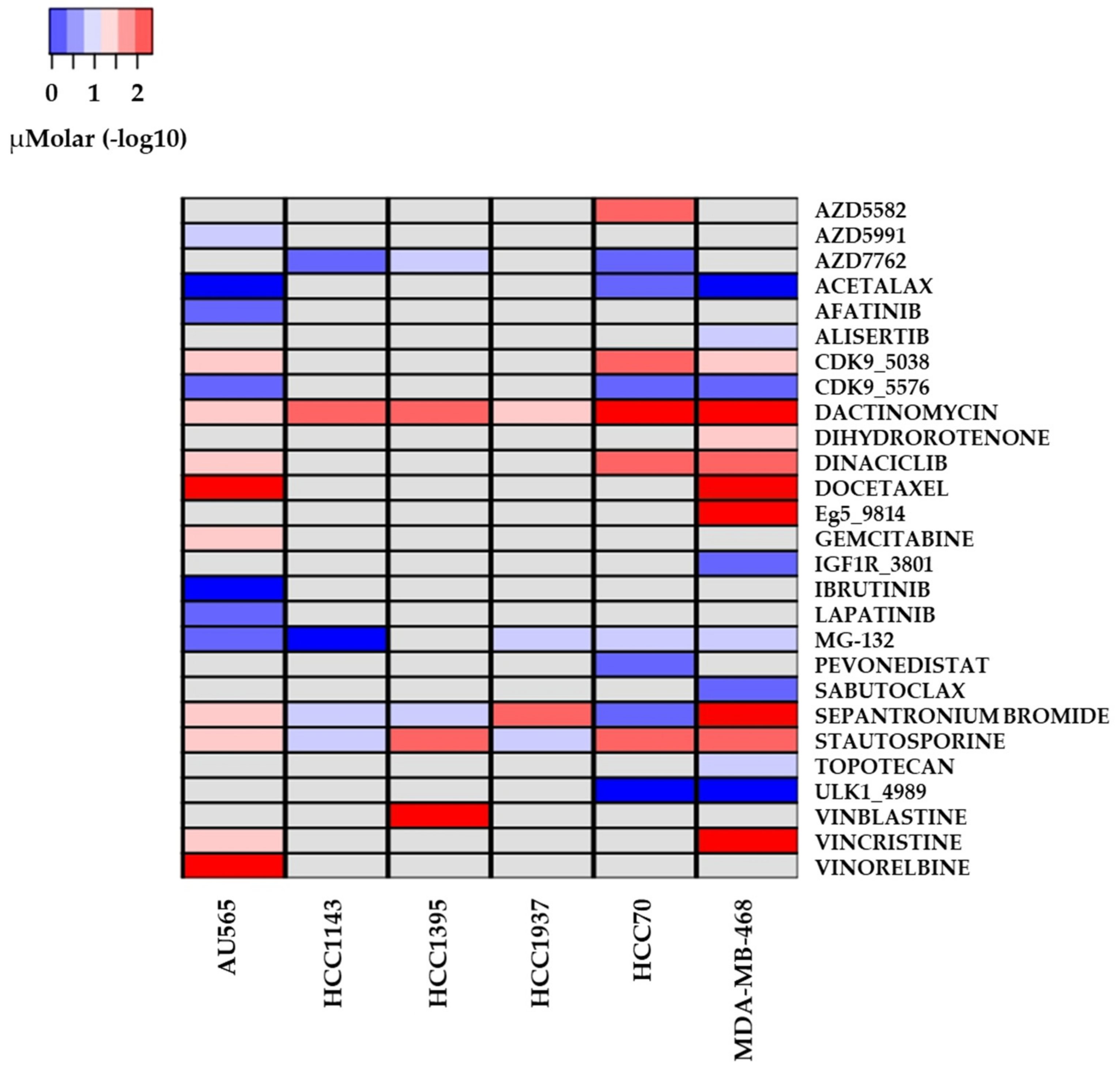
2.3. Drug Sensitivity and Cell Lines with Altered Repair Mechanisms
3. Discussion
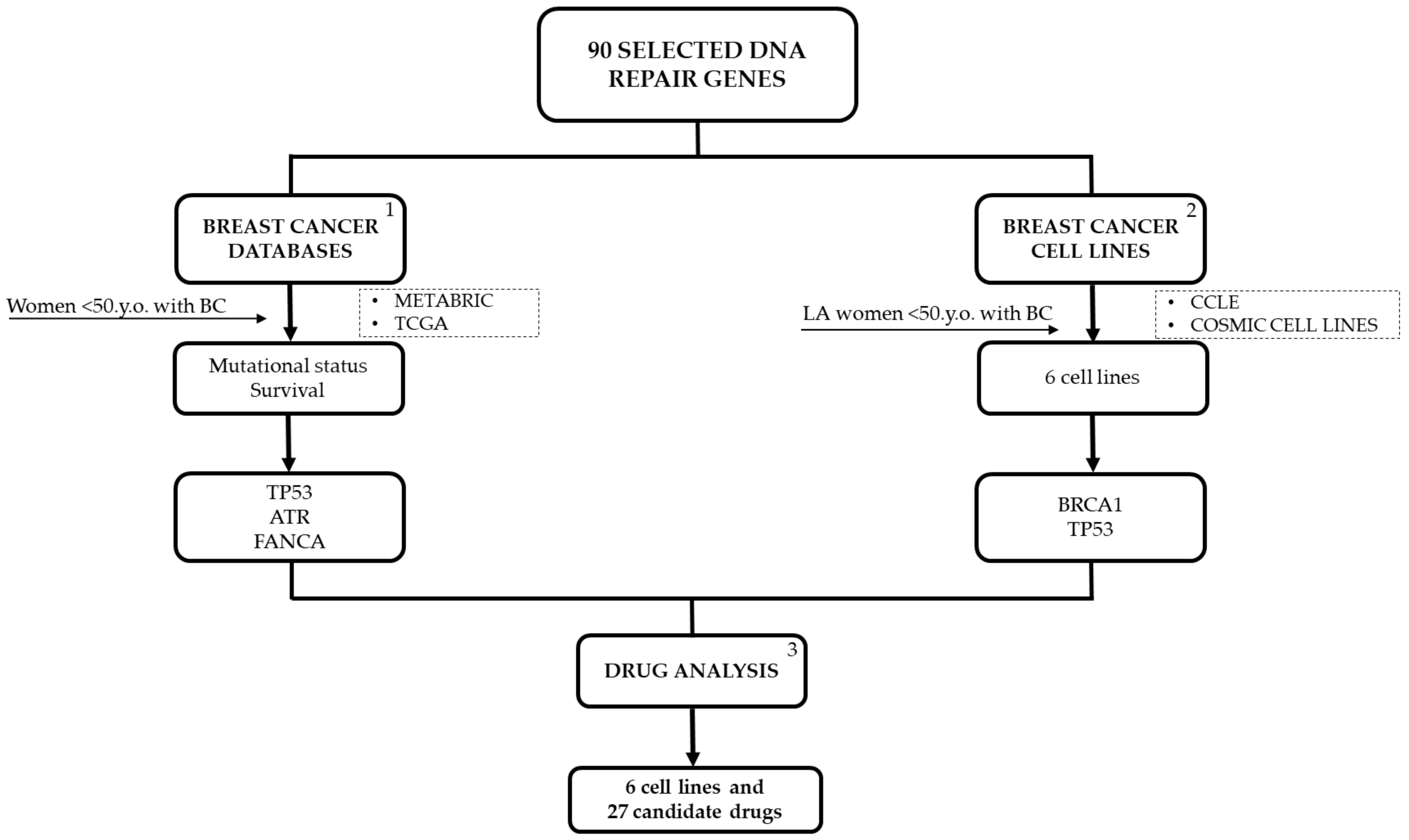
3.1. Importance of Combining Database Information and Cell Lines
3.2. Analysis of Pathogenic Variants of YWBC in LA and Public Databases
3.3. Drugs with Reported Sensitivity in BC Cell Lines
4. Materials and Methods
4.1. DNA Repair Genes of Interest
4.2. Young Women with BC Tumor Sample Data
4.3. BC Cell Lines Selection and Mutation Analysis for DNA Repair Genes
4.4. Drug Variant Sensitivity
4.5. Reported DNA Repair Variants for YWBC from LA
5. Conclusions
6. Take Home Messages
- Of our selection of 90 genes participating in DNA repair mechanisms, no representativeness of variants reported in LA was observed for YWBC <50 y.o., compared to variants from other populations.
- Cell line database analyses resulted in few variants representation of YWBC from LA countries in BC cell lines.
- More studies for LA population are needed to approach DNA repair genes and mechanisms for YWBC besides BRCA1, BRCA2, and TP53 to generate cell lines that represent variants for this population.
- Our analyses resulted in different candidate drugs, nevertheless, not all of these drugs are validated in clinical practice.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries: Global Cancer Statistics 2018. Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Merino Bonilla, J.A.; Torres Tabanera, M.; Ros Mendoza, L.H. Breast Cancer in the 21st Century: From Early Detection to New Therapies. Radiologia 2017, 59, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Ogiya, R.; Sagara, Y.; Niikura, N.; Freedman, R.A. Impact of Subtype on Survival of Young Patients with Stage IV Breast Cancer. Clin. Breast Cancer 2019, 19, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, S.M.; Greaney, M.L.; Patenaude, A.F.; Partridge, A.H. Factors Affecting Surgical Decisions in Newly Diagnosed Young Women with Early-Stage Breast Cancer. J. Adolesc. Young Adult Oncol. 2019, 8, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.H.; Anders, C.K.; Litton, J.K.; Ruddy, K.J.; Bleyer, A. Breast cancer in adolescents and young adults. Pediatr. Blood Cancer 2018, 65, e27397. [Google Scholar] [CrossRef]
- Mealey, N.E.; O’Sullivan, D.E.; Pader, J.; Ruan, Y.; Wang, E.; Quan, M.L.; Brenner, D.R. Mutational landscape differences between young-onset and older-onset breast cancer patients. BMC Cancer 2020, 20, 212. [Google Scholar] [CrossRef]
- Poggio, F.; Lambertini, M.; Bighin, C.; Conte, B.; Blondeaux, E.; D’Alonzo, A.; Dellepiane, C.; Boccardo, F.M.; Del Mastro, L. Management of young women with early breast cancer. ESMO Open 2018, 3, e000458. [Google Scholar] [CrossRef]
- Peña-Chilet, M.; Martínez, M.T.; Pérez-Fidalgo, J.A.; Peiró-Chova, L.; Oltra, S.S.; Tormo, E.; Alonso-Yuste, E.; Martinez-Delgado, B.; Eroles, P.; Climent, J.; et al. MicroRNA profile in very young women with breast cancer. BMC Cancer 2014, 14, 529. [Google Scholar] [CrossRef]
- Anastasiadi, Z.; Lianos, G.D.; Ignatiadou, E.; Harissis, H.V.; Mitsis, M. Breast cancer in young women: An overview. Updates Surg. 2017, 69, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Hironaka-Mitsuhashi, A.; Tsuda, H.; Yoshida, M.; Shimizu, C.; Asaga, S.; Hojo, T.; Tamura, K.; Kinoshita, T.; Ushijima, T.; Hiraoka, N.; et al. Invasive breast cancers in adolescent and young adult women show more aggressive immunohistochemical and clinical features than those in women aged 40–44 years. Breast Cancer 2018, 26, 386–396. [Google Scholar] [CrossRef]
- Lee, K.J.; Piett, C.G.; Andrews, J.F.; Mann, E.; Nagel, Z.D.; Gassman, N.R. Defective base excision repair in the response to DNA damaging agents in triple negative breast cancer. PLoS ONE 2019, 14, e0223725. [Google Scholar] [CrossRef] [PubMed]
- Trenner, A.; Sartori, A.A. Harnessing DNA Double-Strand Break Repair for Cancer Treatment. Front. Oncol. 2019, 9, 1388. [Google Scholar] [CrossRef]
- Gilmore, E.; McCabe, N.; Kennedy, R.D.; Parkes, E.E. DNA Repair Deficiency in Breast Cancer: Opportunities for Immunotherapy. Available online: https://www.hindawi.com/journals/jo/2019/4325105/ (accessed on 4 July 2019).
- Kleiblova, P.; Stolarova, L.; Krizova, K.; Lhota, F.; Hojny, J.; Zemankova, P.; Havranek, O.; Vocka, M.; Cerna, M.; Lhotova, K.; et al. Identification of deleterious germline CHEK2 mutations and their association with breast and ovarian cancer. Int. J. Cancer 2019, 145, 1782–1797. [Google Scholar] [CrossRef]
- Gómez-Flores-Ramos, L.; Castro-Sanchez, A.; Peña-Curiel, O.; Mohar-Betancourt, A. Molecular Biology in Young Women With Breast Cancer: From Tumor Gene Expression To Dna Mutations. Rev. Investig. Clin. 2017, 69, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Grešner, P.; Jablonska, E.; Gromadzińska, J. Rad51 paralogs and the risk of unselected breast cancer: A case-control study. PLoS ONE 2020, 15, e0226976. [Google Scholar] [CrossRef]
- Gentles, L.; Goranov, B.; Matheson, E.; Herriott, A.; Kaufmann, A.; Hall, S.; Mukhopadhyay, A.; Drew, Y.; Curtin, N.J.; O’Donnell, R. Exploring the Frequency of Homologous Recombination DNA Repair Dysfunction in Multiple Cancer Types. Cancers 2019, 11, 354. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, S. Targeting DNA Double-Strand Break (DSB) Repair to Counteract Tumor Radio-resistance. Curr. Drug Targets 2019, 20, 891–902. [Google Scholar] [CrossRef] [PubMed]
- Riaz, N.; Blecua, P.; Lim, R.S.; Shen, R.; Higginson, D.; Weinhold, N.; Norton, L.; Weigelt, B.; Powell, S.N.; Reis-Filho, J.S. Pan-cancer analysis of bi-allelic alterations in homologous recombination DNA repair genes. Nat. Commun. 2017, 8, 857. [Google Scholar] [CrossRef] [PubMed]
- Sonnenblick, A.; Zick, A.; Maoz, M.; Cohen, S.; Kadouri, L.; Peretz, T.; Hubert, A. Defects in homologous recombination repair genes are associated with good prognosis and clinical sensitivity to DNA-damaging agents in pancreatic cancer: A case report. Mol. Clin. Oncol. 2018, 8, 683–685. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ung, M.H.; Cantor, S.; Cheng, C. Computational Investigation of Homologous Recombination DNA Repair Deficiency in Sporadic Breast Cancer. Sci. Rep. 2017, 7, 15742. [Google Scholar] [CrossRef]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Iorio, F.; Knijnenburg, T.A.; Vis, D.J.; Bignell, G.R.; Menden, M.P.; Schubert, M.; Aben, N.; Gonçalves, E.; Barthorpe, S.; Lightfoot, H.; et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016, 166, 740–754. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, M.; Xi, J.; Yang, J.; Li, A. A novel heterogeneous network-based method for drug response prediction in cancer cell lines. Sci. Rep. 2018, 8, 3355. [Google Scholar] [CrossRef]
- Jastrzebski, K.; Thijssen, B.; Kluin, R.J.; De Lint, K.; Majewski, I.J.; Beijersbergen, R.L.; Wessels, L.F. Integrative Modeling Identifies Key Determinants of Inhibitor Sensitivity in Breast Cancer Cell Lines. Cancer Res. 2018, 78, 4396–4410. [Google Scholar] [CrossRef] [PubMed]
- Yao, F.; Tonekaboni, S.A.M.; Safikhani, Z.; Smirnov, P.; El-Hachem, N.; Freeman, M.; Manem, V.S.K.; Haibe-Kains, B. Tissue specificity of in vitro drug sensitivity. J. Am. Med. Inform. Assoc. 2017, 25, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Sundaresan, V.; Zhou, L.; Kahveci, T. Integrating Domain Specific Knowledge and Network Analysis to Predict Drug Sensitivity of Cancer Cell Lines. PLoS ONE 2016, 11, e0162173. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Liu, C.; Zheng, X.; Li, Y. Comprehensive anticancer drug response prediction based on a simple cell line-drug complex network model. BMC Bioinform. 2019, 20, 44. [Google Scholar] [CrossRef] [PubMed]
- Olsson, E.; Winter, C.; George, A.; Chen, Y.; Törngren, T.; Bendahl, P.-O.; Borg, A.; Gruvberger-Saal, S.K.; Saal, L.H. Mutation Screening of 1,237 Cancer Genes across Six Model Cell Lines of Basal-Like Breast Cancer. PLoS ONE 2015, 10, e0144528. [Google Scholar] [CrossRef]
- Lodovichi, S.; Mercatanti, A.; Cervelli, T.; Galli, A. Computational analysis of data from a genome-wide screening identifies new PARP1 functional interactors as potential therapeutic targets. Oncotarget 2019, 10, 2722–2737. [Google Scholar] [CrossRef]
- Liu, Q.; Yao, S.; Zhao, H.; Hu, Q.; Kwan, M.L.; Roh, J.M.; Ambrosone, C.B.; Kushi, L.H.; Liu, S.; Zhu, Q. Early-onset triple-negative breast cancer in multiracial/ethnic populations: Distinct trends of prevalence of truncation mutations. Cancer Med. 2019, 8, 1845–1853. [Google Scholar] [CrossRef]
- Urbina-Jara, L.K.; Martinez, R.; Martinez-Ledesma, E.; Aguilar, D.; Villarreal-Garza, C.; Ortiz-Lopez, R.; Jara, U.; Ledesma, M.; Garza, V.; Lopez, O.; et al. Landscape of Germline Mutations in DNA Repair Genes for Breast Cancer in Latin America: Opportunities for PARP-Like Inhibitors and Immunotherapy. Genes 2019, 10, 786. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The CBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the CBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Spratt, D.E.; Chan, T.; Waldron, L.; Speers, C.; Feng, F.Y.; Ogunwobi, O.; Osborne, J.R. Racial/Ethnic Disparities in Genomic Sequencing. JAMA Oncol. 2016, 2, 1070–1074. [Google Scholar] [CrossRef] [PubMed]
- Wojtyla, C.; Bertuccio, P.; Ciebiera, M.; La Vecchia, C. Breast Cancer Mortality in the Americas and Australasia over the Period 1980–2017 with Predictions for 2025. Biology 2021, 10, 814. [Google Scholar] [CrossRef]
- Dutil, J.; Golubeva, V.; Pacheco-Torres, A.L.; Diaz-Zabala, H.J.; Matta, J.L.; Monteiro, A. The spectrum of BRCA1 and BRCA2 alleles in Latin America and the Caribbean: A clinical perspective. Breast Cancer Res. Treat. 2015, 154, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.; Orozco, A.; Shao, K.; Albanez, A.; Ortiz, J.; Cao, B.; Wang, L.; Barreda, L.; Alvarez, C.S.; Garland, L.; et al. Germline Variants in Hereditary Breast Cancer Genes Are Associated with Early Age at Diagnosis and Family History in Guatemalan Breast Cancer. Breast Cancer Res. Treat. 2021, 189, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Moyer, T.C.; Holland, A.J. Generation of a conditional analog-sensitive kinase in human cells using CRISPR/Cas9-mediated genome engineering. Methods Cell Biol. 2015, 129, 19–36. [Google Scholar] [CrossRef]
- Gonzalez-Salinas, F.; Rojo, R.; Martinez-Amador, C.; Herrera-Gamboa, J.; Trevino, V. Transcriptomic and cellular analyses of CRISPR/Cas9-mediated edition of FASN show inhibition of aggressive characteristics in breast cancer cells. Biochem. Biophys. Res. Commun. 2020, 529, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Brachova, P.; Thiel, K.W.; Leslie, K.K. The Consequence of Oncomorphic TP53 Mutations in Ovarian Cancer. Int. J. Mol. Sci. 2013, 14, 19257–19275. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Langerød, A.; Carrieri, P.; Bergh, J.; Klaar, S.; Eyfjord, J.; Theillet, C.; Rodriguez, C.; Lidereau, R.; Bièche, I.; et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin. Cancer Res. 2006, 12, 1157–1167. [Google Scholar] [CrossRef]
- Wani, T.; Surendran, S.; Mishra, V.S.; Chaturvedi, J.; Chowdhury, G.; Chakrabarty, A. Adaptation to chronic exposure to sepantronium bromide (YM155), a prototypical survivin suppressant is due to persistent DNA damage-response in breast cancer cells. Oncotarget 2018, 9, 33589–33600. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mazzio, E.A.; Lewis, C.A.; Elhag, R.; Soliman, K.F. Effects of Sepantronium Bromide (YM-155) on the Whole Transcriptome of MDA-MB-231 Cells: Highlight on Impaired ATR/ATM Fanconi Anemia DNA Damage Response. Cancer Genom. Proteom. 2018, 15, 249–264. [Google Scholar] [CrossRef]
- Rajapakse, V.N.; Luna, A.; Yamade, M.; Loman, L.; Varma, S.; Sunshine, M.; Iorio, F.; Sousa, F.G.; Elloumi, F.; Aladjem, M.I.; et al. CellMinerCDB for Integrative Cross-Database Genomics and Pharmacogenomics Analyses of Cancer Cell Lines. iScience 2018, 10, 247–264. [Google Scholar] [CrossRef]
- Morrison, B.L.; Mullendore, M.E.; Stockwin, L.H.; Borgel, S.; Hollingshead, M.G.; Newton, D.L. Oxyphenisatin acetate (NSC 59687) triggers a cell starvation response leading to autophagy, mitochondrial dysfunction, and autocrine TNF α-mediated apoptosis. Cancer Med. 2013, 2, 687–700. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, E.J.; Adam, A.; Aquila, B.M.; Castriotta, L.M.; Cook, D.; Hattersley, M.; Hird, A.W.; Huntington, C.; Kamhi, V.M.; Laing, N.M.; et al. Discovery of a Novel Class of Dimeric Smac Mimetics as Potent IAP Antagonists Resulting in a Clinical Candidate for the Treatment of Cancer (AZD5582). J. Med. Chem. 2013, 56, 9897–9919. [Google Scholar] [CrossRef] [PubMed]
- Polanski, R.; Vincent, J.; Polanska, U.M.; Petreus, T.; Tang, E.K.Y. Caspase-8 activation by TRAIL monotherapy predicts responses to IAPi and TRAIL combination treatment in breast cancer cell lines. Cell Death Dis. 2015, 6, e1893. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Park, J.-S.; Lee, C.; Kim, H.-K.; Kim, D.; Son, J.B.; Ko, E.; Cho, J.-H.; Kim, N.-D.; Nan, H.-Y.; Kim, C.-Y.; et al. Suppression of the metastatic spread of breast cancer by DN10764 (AZD7762)-mediated inhibition of AXL signaling. Oncotarget 2016, 7, 83308–83318. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Min, D.-J.; He, S.; Green, J.E. Birinapant (TL32711) Improves Responses to GEM/AZD7762 Combination Therapy in Triple-negative Breast Cancer Cell Lines. Anticancer. Res. 2016, 36, 2649–2657. [Google Scholar] [PubMed]
- Alsamman, K.; El-Masry, O.S. Staurosporine Alleviates Cisplatin Chemoresistance in Human Cancer Cell Models by Suppressing the Induction of SQSTM1/P62. Oncol. Rep. 2018, 40, 2157–2162. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Lalani, R.; Maiti, K.; Banerjee, S.; Patel, V.; Bhowmick, S.; Misra, A. Optimization and efficacy study of synergistic vincristine coloaded liposomal doxorubicin against breast and lung cancer. Nanomedicine 2020, 15, 2585–2607. [Google Scholar] [CrossRef]
- Ghosh, S.; Lalani, R.; Maiti, K.; Banerjee, S.; Bhatt, H.; Bobde, Y.S.; Patel, V.; Biswas, S.; Bhowmick, S.; Misra, A. Synergistic co-loading of vincristine improved chemotherapeutic potential of pegylated liposomal doxorubicin against triple negative breast cancer and non-small cell lung cancer. Nanomed. Nanotechnol. Biol. Med. 2020, 31, 102320. [Google Scholar] [CrossRef]
- Falchook, G.; Coleman, R.L.; Roszak, A.; Behbakht, K.; Matulonis, U.; Ray-Coquard, I.; Sawrycki, P.; Duska, L.R.; Tew, W.; Ghamande, S.; et al. Alisertib in Combination With Weekly Paclitaxel in Patients With Advanced Breast Cancer or Recurrent Ovarian Cancer. JAMA Oncol. 2019, 5, e183773. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Mohammad, A.S.; Saralkar, P.; Sprowls, S.A.; Vickers, S.D.; John, D.; Tallman, R.M.; Lucke-Wold, B.P.; Jarrell, K.E.; Pinti, M.; et al. Investigational chemotherapy and novel pharmacokinetic mechanisms for the treatment of breast cancer brain metastases. Pharmacol. Res. 2018, 132, 47–68. [Google Scholar] [CrossRef]
- Zhang, W.; Xia, D.; Li, Z.; Zhou, T.; Chen, T.; Wu, Z.; Zhou, W.; Li, Z.; Li, L.; Xu, J. Aurora-A/ERK1/2/MTOR Axis Promotes Tumor Progression in Triple-Negative Breast Cancer and Dual-Targeting Aurora-A/MTOR Shows Synthetic Lethality. Cell Death Dis. 2019, 10, 606. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, B.; He, M.; Chang, J.; Li, J.; Shan, L.; Wang, H.; Hong, W.; Luo, D.; Song, Y.; et al. Pilot study of docetaxel combined with lobaplatin or gemcitabine for recurrent and metastatic breast cancer. Medicine 2019, 98, e18513. [Google Scholar] [CrossRef] [PubMed]
- Caparica, R.; Bruzzone, M.; Poggio, F.; Ceppi, M.; de Azambuja, E.; Lambertini, M. Anthracycline and taxane-based chemotherapy versus docetaxel and cyclophosphamide in the adjuvant treatment of HER2-negative breast cancer patients: A systematic review and meta-analysis of randomized controlled trials. Breast Cancer Res. Treat. 2018, 174, 27–37. [Google Scholar] [CrossRef]
- Lin, S.-Q.; Jia, F.-J.; Zhang, C.-Y.; Liu, F.-Y.; Ma, J.-H.; Han, Z.; Xie, W.-D.; Li, X. Actinomycin V Suppresses Human Non-Small-Cell Lung Carcinoma A549 Cells by Inducing G2/M Phase Arrest and Apoptosis via the p53-Dependent Pathway. Mar. Drugs 2019, 17, 572. [Google Scholar] [CrossRef] [PubMed]
- Das, T.; Nair, R.R.; Green, R.; Padhee, S.; Howell, M.; Banerjee, J.; Mohapatra, S.S. Actinomycin D Down-regulates SOX2 Expression and Induces Death in Breast Cancer Stem Cells. Anticancer. Res. 2017, 37, 1655–1663. [Google Scholar] [CrossRef]
- Sinha, B.K.; Tokar, E.J.; Bushel, P.R. Elucidation of Mechanisms of Topotecan-Induced Cell Death in Human Breast MCF-7 Cancer Cells by Gene Expression Analysis. Front. Genet. 2020, 11, 775. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wang, Q.; Zhang, Y.; Sun, W.; Zhang, S.; Li, Y.; Wang, J.; Bao, Y. Functional daidzein enhances the anticancer effect of topotecan and reverses BCRP-mediated drug resistance in breast cancer. Pharmacol. Res. 2019, 147, 104387. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Yagüe, E.; Zhao, J.; Wang, L.; Bai, J.; Yang, Q.; Pan, T.; Zhao, H.; Liu, J.; Zhang, J. Sabutoclax, pan-active BCL-2 protein family antagonist, overcomes drug resistance and eliminates cancer stem cells in breast cancer. Cancer Lett. 2018, 423, 47–59. [Google Scholar] [CrossRef]
- Criscitiello, C.; Viale, G.; Esposito, A.; Curigliano, G. Dinaciclib for the treatment of breast cancer. Expert Opin. Investig. Drugs 2014, 23, 1305–1312. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.F.; Cruz, C.; Greifenberg, A.K.; Dust, S.; Stover, D.; Chi, D.; Primack, B.; Cao, S.; Bernhardy, A.J.; Coulson, R.; et al. CDK12 Inhibition Reverses De Novo and Acquired PARP Inhibitor Resistance in BRCA Wild-Type and Mutated Models of Triple-Negative Breast Cancer. Cell Rep. 2016, 17, 2367–2381. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, Y.; Zhang, C.; Chu, J.; Wu, Y.; Li, Y.; Liu, J.; Li, Q.; Li, S.; Shi, Q.; et al. Tamoxifen-resistant breast cancer cells are resistant to DNA-damaging chemotherapy because of upregulated BARD1 and BRCA1. Nat. Commun. 2018, 9, 1595. [Google Scholar] [CrossRef] [PubMed]
- Nie, L.; Wei, Y.; Zhang, F.; Hsu, Y.-H.; Chan, L.-C.; Xia, W.; Ke, B.; Zhu, C.; Deng, R.; Tang, J.; et al. CDK2-mediated site-specific phosphorylation of EZH2 drives and maintains triple-negative breast cancer. Nat. Commun. 2019, 10, 5114. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, B.; Musolino, A.; Llop-Guevara, A.; Serra, V.; De Silva, P.; Hlavata, Z.; Sangiolo, D.; Willard-Gallo, K.; Solinas, C. Homologous Recombination Repair Deficiency and the Immune Response in Breast Cancer: A Literature Review. Transl. Oncol. 2020, 13, 410–422. [Google Scholar] [CrossRef]
- Chen, C.-C.; Feng, W.; Lim, P.X.; Kass, E.M.; Jasin, M. Homology-Directed Repair and the Role of BRCA1, BRCA2, and Related Proteins in Genome Integrity and Cancer. Annu. Rev. Cancer Biol. 2018, 2, 313–336. [Google Scholar] [CrossRef] [PubMed]
- Dietlein, F.; Thelen, L.; Reinhardt, H.C. Cancer-specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends Genet. 2014, 30, 326–339. [Google Scholar] [CrossRef] [PubMed]
- Christou, C.M.; Kyriacou, K. BRCA1 and Its Network of Interacting Partners. Biology 2013, 2, 40–63. [Google Scholar] [CrossRef] [PubMed]
- Tanner, M.; Kapanen, A.I.; Junttila, T.; Raheem, O.; Grenman, S.; Elo, J.; Elenius, K.; Isola, J. Characterization of a novel cell line established from a patient with Herceptin-resistant breast cancer. Mol. Cancer Ther. 2004, 3, 1585–1592. [Google Scholar] [PubMed]
- Friedland, J.C.; Smith, N.L.; Sang, J.; Acquaviva, J.; He, S.; Zhang, C.; Proia, D.A. Targeted inhibition of Hsp90 by ganetespib is effective across a broad spectrum of breast cancer subtypes. Investig. New Drugs 2013, 32, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Tanida, T.; Matsuda, K.I.; Yamada, S.; Hashimoto, T.; Kawata, M. Estrogen-related Receptor β Reduces the Subnuclear Mobility of Estrogen Receptor α and Suppresses Estrogen-dependent Cellular Function. J. Biol. Chem. 2015, 290, 12332–12345. [Google Scholar] [CrossRef] [PubMed]
- Yamane, M.; Nishiki, M.; Kataoka, T.; Kishi, N.; Amano, K.; Nakagawa, K.; Okumichi, T.; Naito, M.; Ito, A.; Ezaki, H. Establishment and characterization of new cell line (YMB-1) derived from human breast carcinoma. In Hiroshima J. Med. Sci.; 1984; 33, pp. 715–720. Available online: https://pubmed.ncbi.nlm.nih.gov/6534930/ (accessed on 13 April 2020). [PubMed]
- O’Brien, N.; Conklin, D.; Beckmann, R.; Luo, T.; Chau, K.; Thomas, J.; Mc Nulty, A.; Marchal, C.; Kalous, O.; von Euw, E.; et al. Preclinical Activity of Abemaciclib Alone or in Combination with Antimitotic and Targeted Therapies in Breast Cancer. Mol. Cancer Ther. 2018, 17, 897–907. [Google Scholar] [CrossRef]
- Jiang, G.; Zhang, S.; Yazdanparast, A.; Li, M.; Pawar, A.V.; Liu, Y.; Inavolu, S.M.; Cheng, L. Comprehensive Comparison of Molecular Portraits between Cell Lines and Tumors in Breast Cancer. BMC Genom. 2016, 17, 525. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Flores-Ramos, L.; Dean, M.; Jones, K.; Wang, M.; Villarreal-Garza, C.; Álvarez-Gómez, R.M.; Wegman-Ostrosky, T.; Reynoso-Noverón, N.; Fragoso-Ontiveros, V.; Sánchez-Zamorano, L.; et al. Clinically Relevant Germline Mutations in a Cohort of Young Women with Breast Cancer: A Comprehensive Analysis of Hereditary-Cancer Genes from Whole-Exome Sequencing. 2020. Available online: https://www.preprints.org/manuscript/202008.0718/v1 (accessed on 21 November 2021).
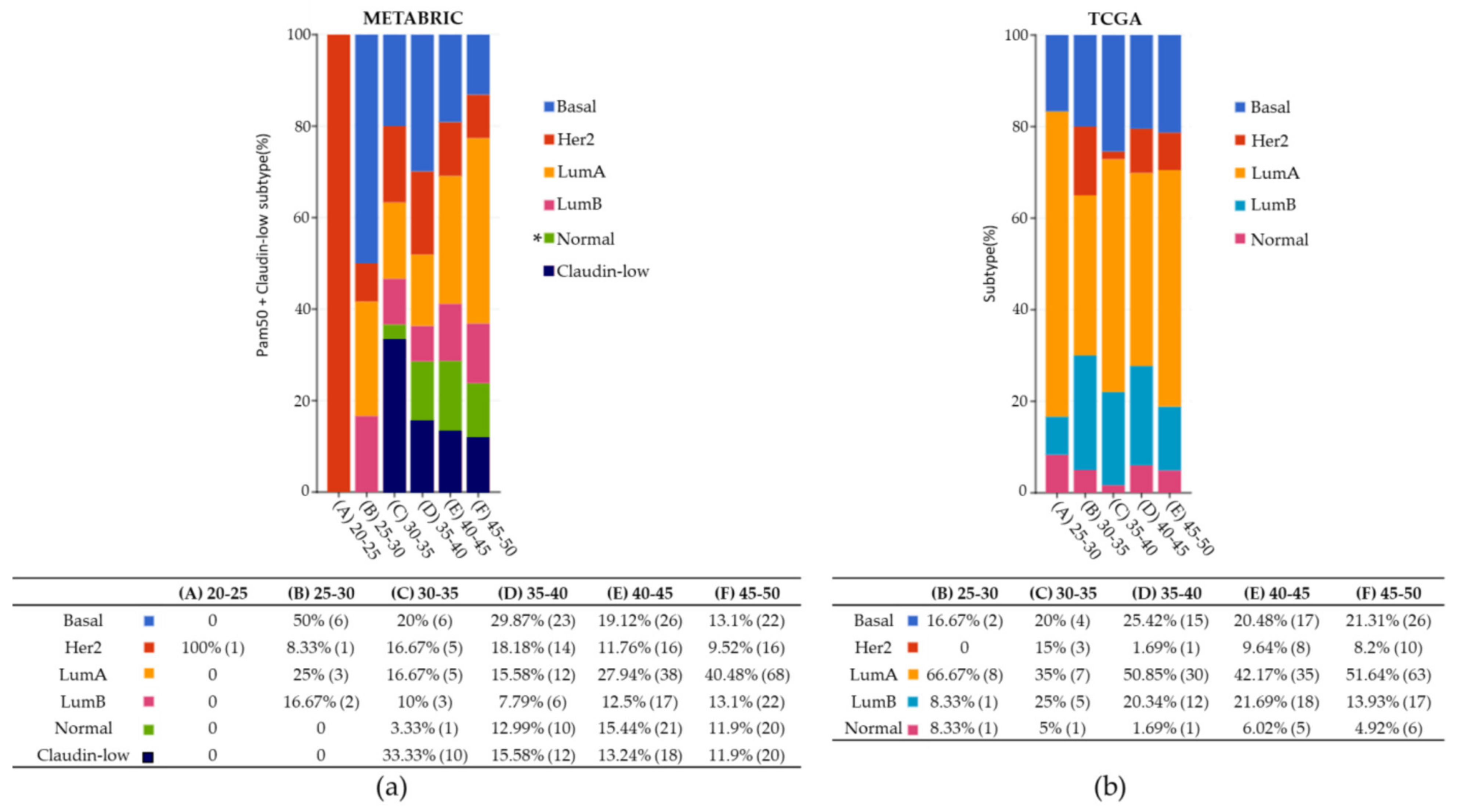
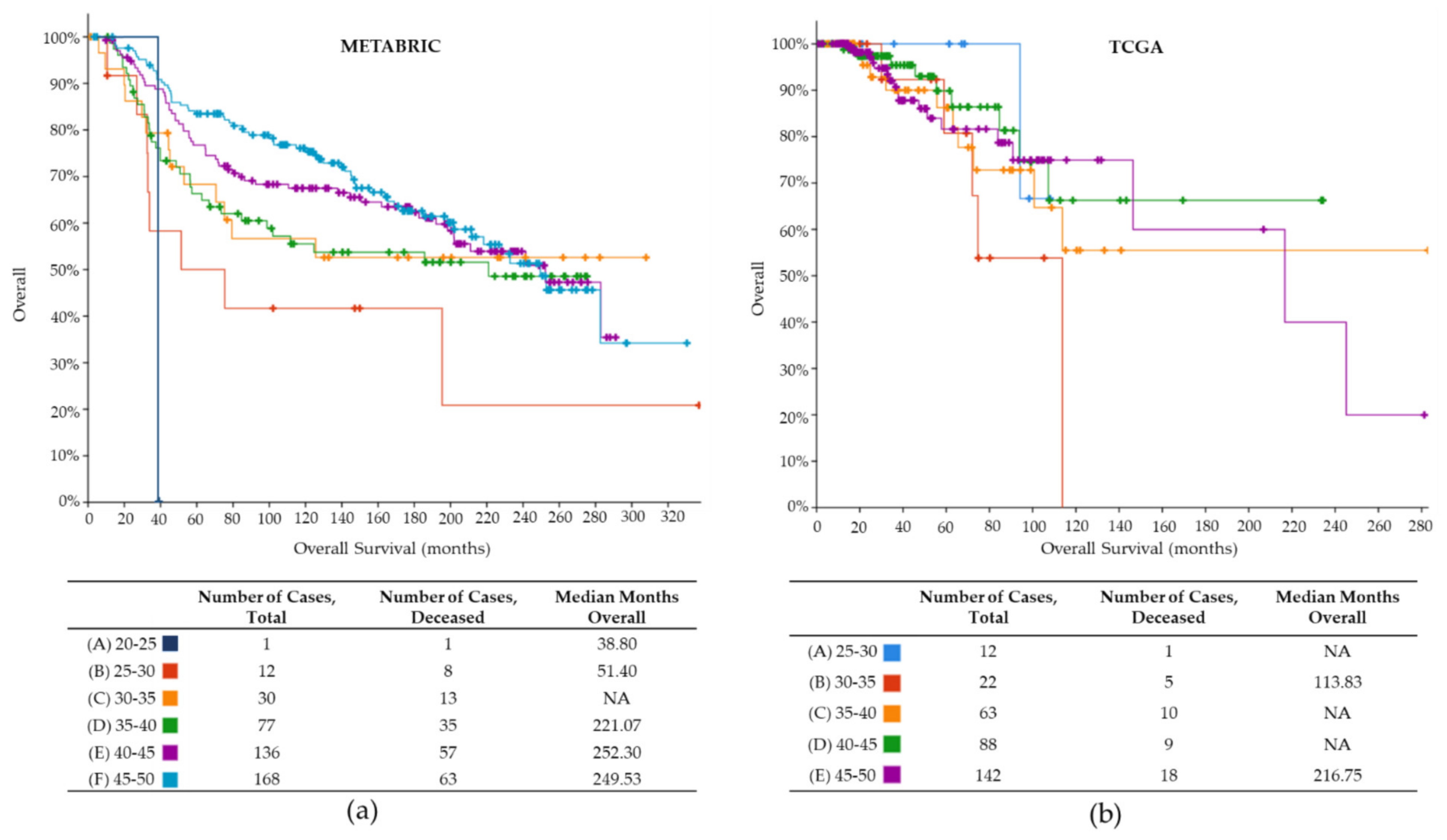
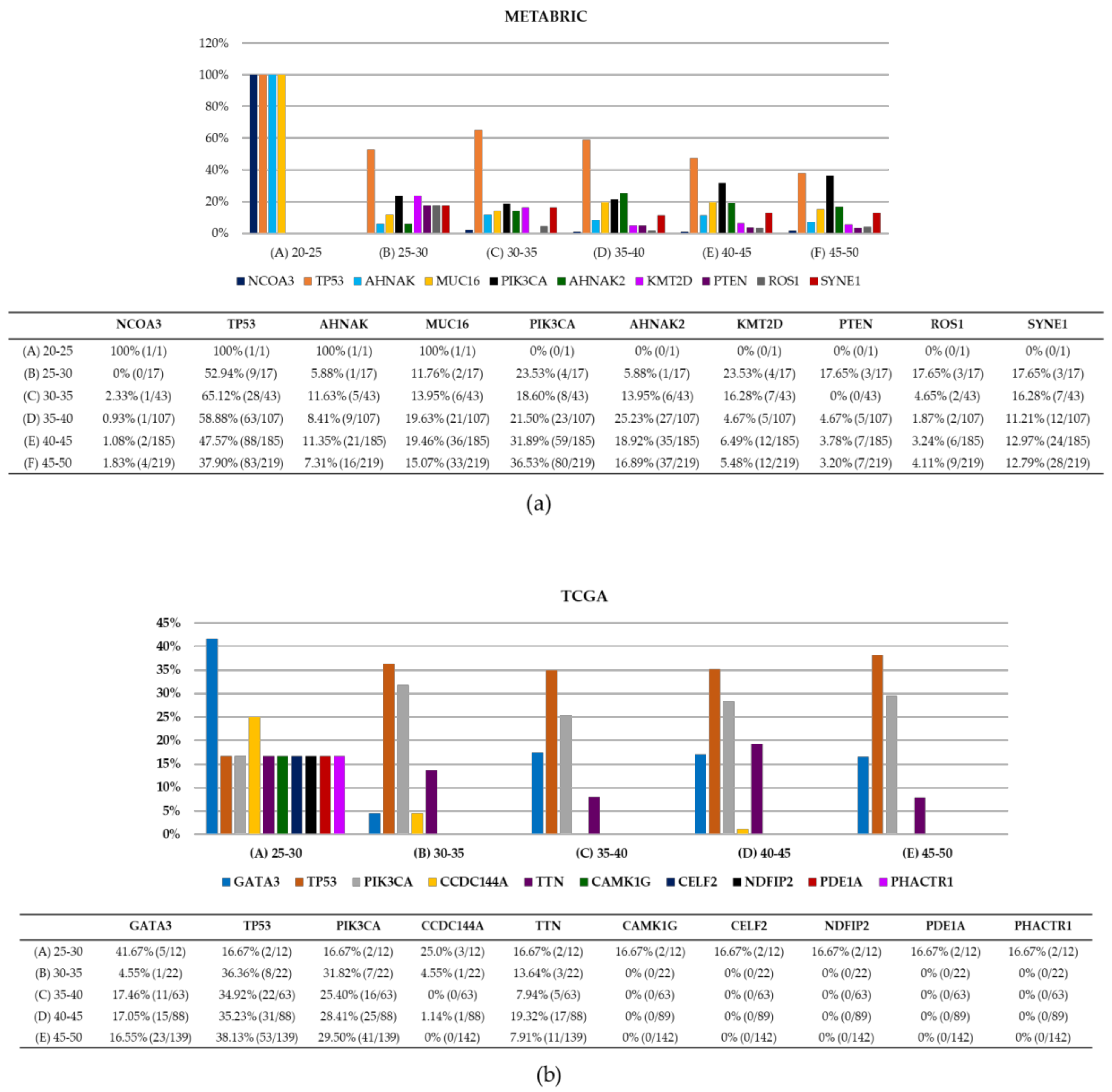
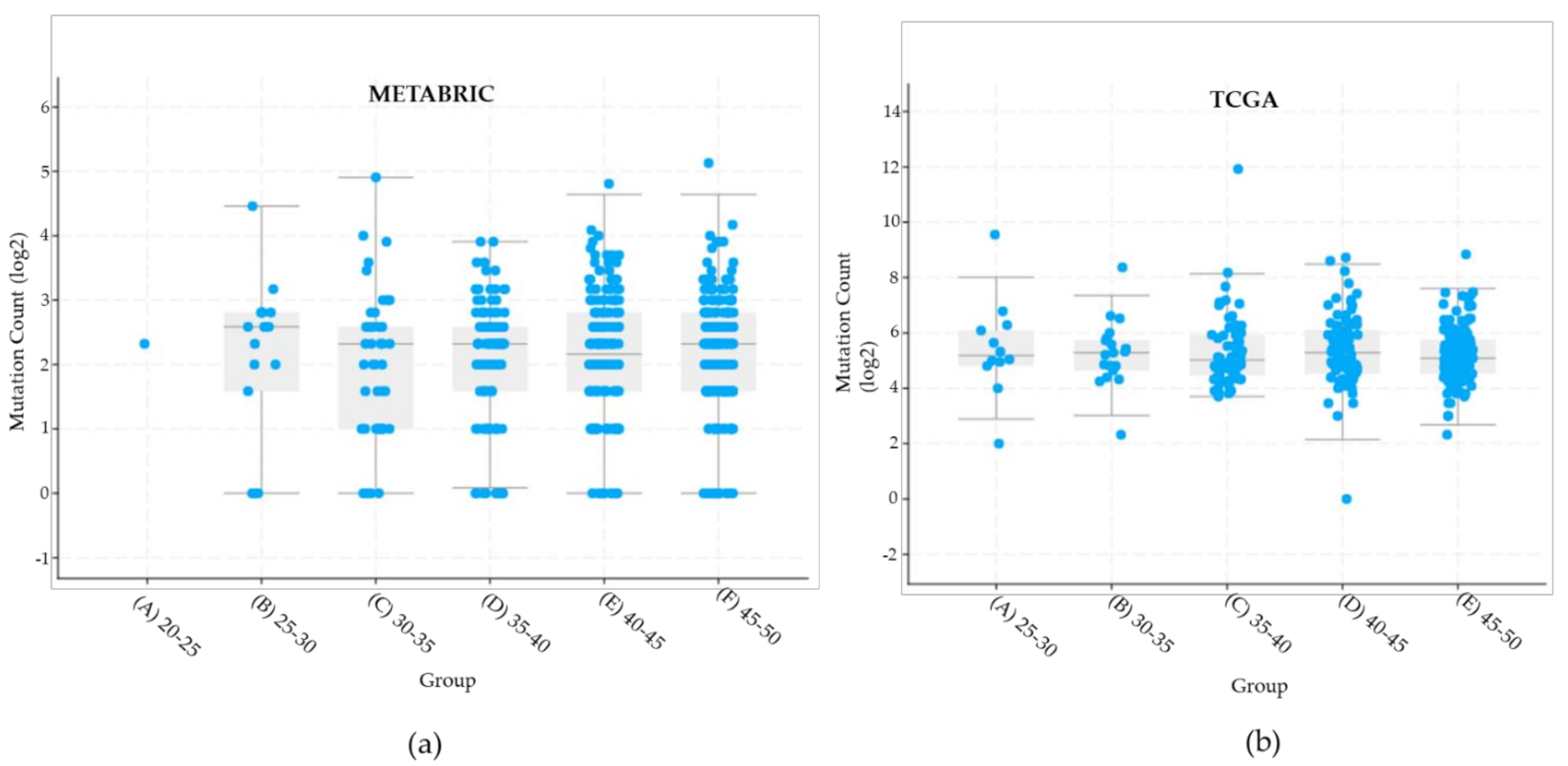

| Clinical Characteristics | METABRIC | TCGA |
|---|---|---|
| Total samples clinical data | 567 | 292 |
| Mean age at diagnosis (years) | 42.4 | 42.4 |
| Youngest age at diagnosis (years) | 21.9 | 26.0 |
| Mean OS (months) | 134.1 | 40.4 |
| Lowest OS (months) | 1.4 | 0.0 |
| Highest OS (months) | 337 | 283 |
| BC Subtype | ||
| Lum A | 22.2% | 44.9% |
| Basal | 14.6% | 20.2% |
| Claudine-low | 10.6% | 0 |
| HER2 | 9.3% | 5.5% |
| Lum B | 8.8% | 16.8% |
| Normal | 9.2% | 4.1% |
| NA | 25.3% | 8.5% |
| Total samples | 567 | 292 |
| METABRIC | TCGA | |
|---|---|---|
| Total samples (all ages) | 2509 samples | 1084 samples |
| Total variants all samples, all genes | 17,272 variants | 130,495 variants |
| Patients <50 years old | ||
| Total samples | 567 samples | 292 samples |
| Total variants all genes | 3839 variants | 1693 variants |
| Top 5 mutated genes | TP53, PIK3CA, MUC16, SYNEI1, and AHNAK2 | TP53, PRKDC, ATM, BRCA2 and BRCA1 |
| Reported DNA repair variants | 420 variants | 269 variants |
| Reported DNA repair genes | 11 genes | 90 genes |
| Samples with DNA repair variants | 314 samples | 122 samples |
| Top reported DNA repair genes | TP53, ATR, and FANCA | TP53, ATM, and POLQ |
| Gene with most variants | TP53 (275 variants) | TP53 (80 variants) |
| Most frequent variant | TP53 p.R175H (12 samples) | TP53 p.R175H (6 samples) |
| Sample with most reported DNA repair variants | MTS-T1284, 4 variants in APC, ATR, BRCA1, and FANCA | TCGA-EW-A2FV-01, 23 variants in 23 genes |
| Drug Name | Cell Lines | Putative Target | Pathway Name |
|---|---|---|---|
| Acetalax | A565, HCC70, MDA-MB-468 | - | Unclassified |
| Afatinib | A565 | ERBB2, EGFR | EGFR signaling |
| Alisertib | MDA-MB-468 | AURKA | Mitosis |
| AZD5582 | HCC70 | XIAP, cIAP | Apoptosis regulation |
| AZD5991 | A565 | MCL1 | Apoptosis regulation |
| AZD7762 | HCC1143, HCC1395, HCC70, | CHEK1, CHEK2 | Cell cycle |
| CDK9_5038 | A565, HCC70, MDA-MB-468 | CDK9 | Cell cycle |
| CDK9_5576 | A565, HCC70, MDA-MB-468 | CDK9 | Cell cycle |
| Dactinomycin | A565, HCC1143, HCC1395, HCC1937, HCC70, MDA-MB-468 | RNA polymerase | Other |
| Dihydrorotenone | MDA-MB-468 | - | Unclassified |
| Dinaciclib | A565, HCC70, MDA-MB-468 | CDK1, CDK2, CDK5, CDK9 | Cell cycle |
| Docetaxel | A565, MDA-MB-468 | Microtubule stabilizer | Mitosis |
| Eg5_9814 | MDA-MB-468 | KSP11 | Other |
| Gemcitabine | A565 | Pyrimidine antimetabolite | DNA replication |
| Ibrutinib | A565 | BTK | Other, kinases |
| IGF1R_3801 | MDA-MB-468 | IGFR1 | IGF1R signaling |
| Lapatinib | A565 | EGFR, ERBB2 | EGFR signaling |
| MG-132 | A565, HCC1143, HCC1937, HCC70, MDA-MB-468 | Proteasome, CAPN1 | Protein stability and degradation |
| Pevonedistat | HCC70 | NAE | Other |
| Sabutoclax | MDA-MB-468 | BCL2, BCL-XL, BFL1, MCL1 | Apoptosis regulation |
| Sepantronium bromide | HCC1143, HCC1395, HCC1937, HCC70, MDA-MB-468 | BIRC5 | Apoptosis regulation |
| Staurosporine | A565, HCC1143, HCC1937, HCC70, MDA-MB-468 | Broad-spectrum kinase inhibitor | RTK signaling |
| Topotecan | MDA-MB-468 | - | DNA replication |
| ULK1_4989 | HCC70, MDA-MB-468 | ULK1 | Other, kinases |
| Vinblastine | HCC1395 | Microtubule destabilizer | Mitosis |
| Vincristine | A565, MDA-MB-468 | - | Mitosis |
| Vinorelbine | A565 | Microtubule destabilizer | Mitosis |
| APEX1 | CHEK2 | LIG4 | POLD2 | SSBP1 |
| ATM | CTiP | MDC1 | POLE | STK11 |
| ATR | DNTT | MLH1 | POLH | STRA13 |
| BARD1 | ERCC3 | MLH3 | POLQ | TIMELESS |
| BLM | ERCC6 | MRE11A | PP4C | TOP2A |
| BRCA1 | EXO1 | MSH2 | PRKDC | TOP3A |
| BRCA2 | FAM175A | MSH3 | RAD50 | TOPBP1 |
| BRIP1 | FANCA | MSH6 | RAD51 | TP53 |
| CCNA2 | FANCB | MUTYH | RAD51C | TP53BP1 |
| CCNB1 | FANCC | NBN | RAD51D | TRIP13 |
| CCNB2 | FANCD2 | NEIL2 | RAD52 | UBE2T |
| CDC25A | FANCI | NHEJ1 | RECQL4 | UIMC1 |
| CDK1 | FANCL | PALB2 | REV3L | WRN |
| CDK12 | FANCM | PARP1 | RIF1 | XPA |
| CDK4 | GADD45B | PARP2 | RPA1 | XPC |
| CDKN2A | GEN1 | PARPBP | RPA2 | XRCC1 |
| CENPS | H2AFX | PCNA | RRM2 | XRCC5 |
| CHEK1 | HDAC2 | PMS2 | SMC1A | XRCC6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urbina-Jara, L.K.; Martinez-Ledesma, E.; Rojas-Martinez, A.; Rodriguez-Recio, F.R.; Ortiz-Lopez, R. DNA Repair Genes as Drug Candidates for Early Breast Cancer Onset in Latin America: A Systematic Review. Int. J. Mol. Sci. 2021, 22, 13030. https://doi.org/10.3390/ijms222313030
Urbina-Jara LK, Martinez-Ledesma E, Rojas-Martinez A, Rodriguez-Recio FR, Ortiz-Lopez R. DNA Repair Genes as Drug Candidates for Early Breast Cancer Onset in Latin America: A Systematic Review. International Journal of Molecular Sciences. 2021; 22(23):13030. https://doi.org/10.3390/ijms222313030
Chicago/Turabian StyleUrbina-Jara, Laura Keren, Emmanuel Martinez-Ledesma, Augusto Rojas-Martinez, Francisco Ricardo Rodriguez-Recio, and Rocio Ortiz-Lopez. 2021. "DNA Repair Genes as Drug Candidates for Early Breast Cancer Onset in Latin America: A Systematic Review" International Journal of Molecular Sciences 22, no. 23: 13030. https://doi.org/10.3390/ijms222313030
APA StyleUrbina-Jara, L. K., Martinez-Ledesma, E., Rojas-Martinez, A., Rodriguez-Recio, F. R., & Ortiz-Lopez, R. (2021). DNA Repair Genes as Drug Candidates for Early Breast Cancer Onset in Latin America: A Systematic Review. International Journal of Molecular Sciences, 22(23), 13030. https://doi.org/10.3390/ijms222313030







