RGS4 RNA Secondary Structure Mediates Staufen2 RNP Assembly in Neurons
Abstract
:1. Introduction
2. Results
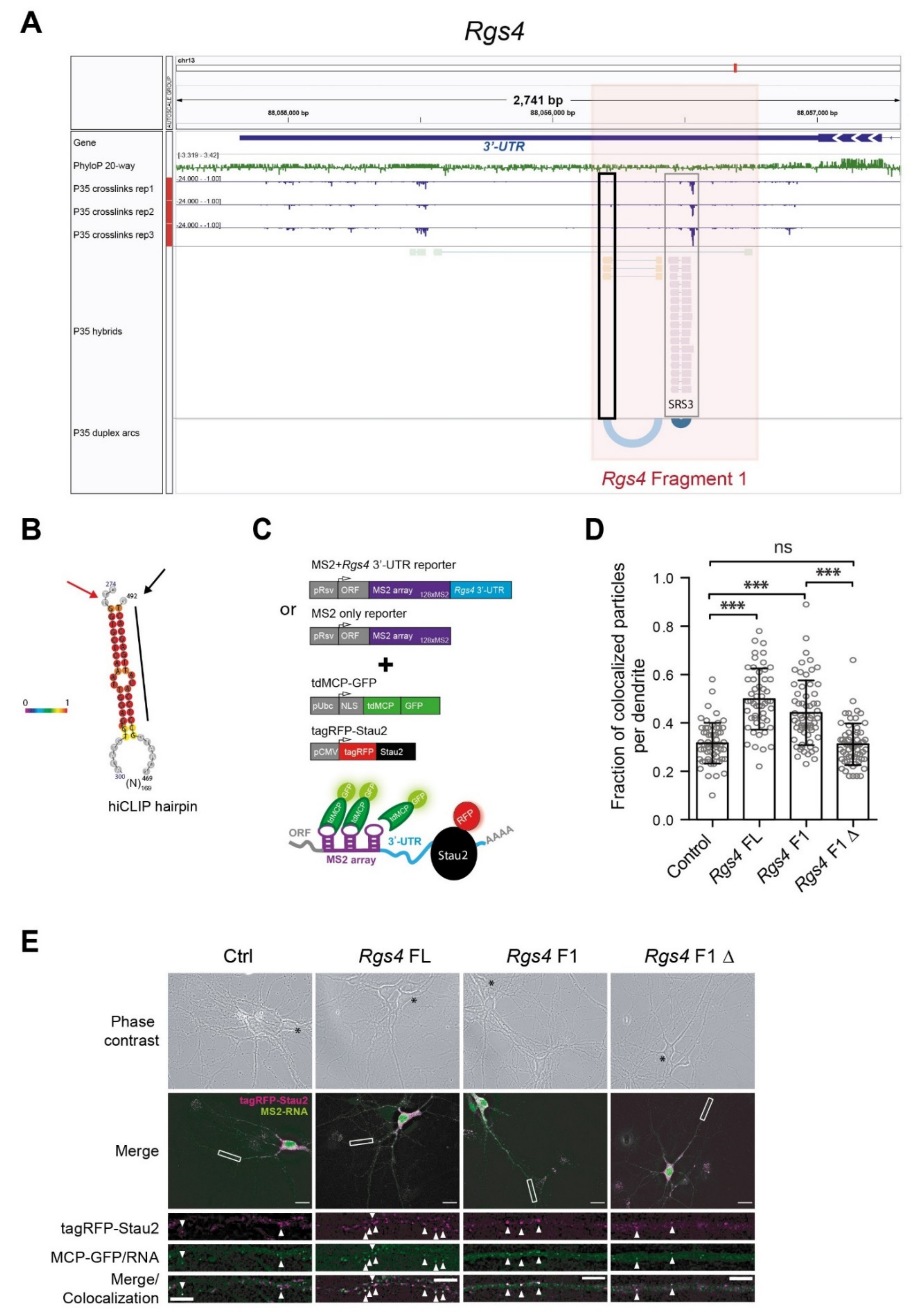
2.1. Stau2 Binds RNA Structures in the 3′-UTR In Vivo
2.2. A Defined Long-Range RNA Structure Recruits Stau2
2.3. RNA Structure Conveys Stau2-Dependent Dendritic mRNA Localization
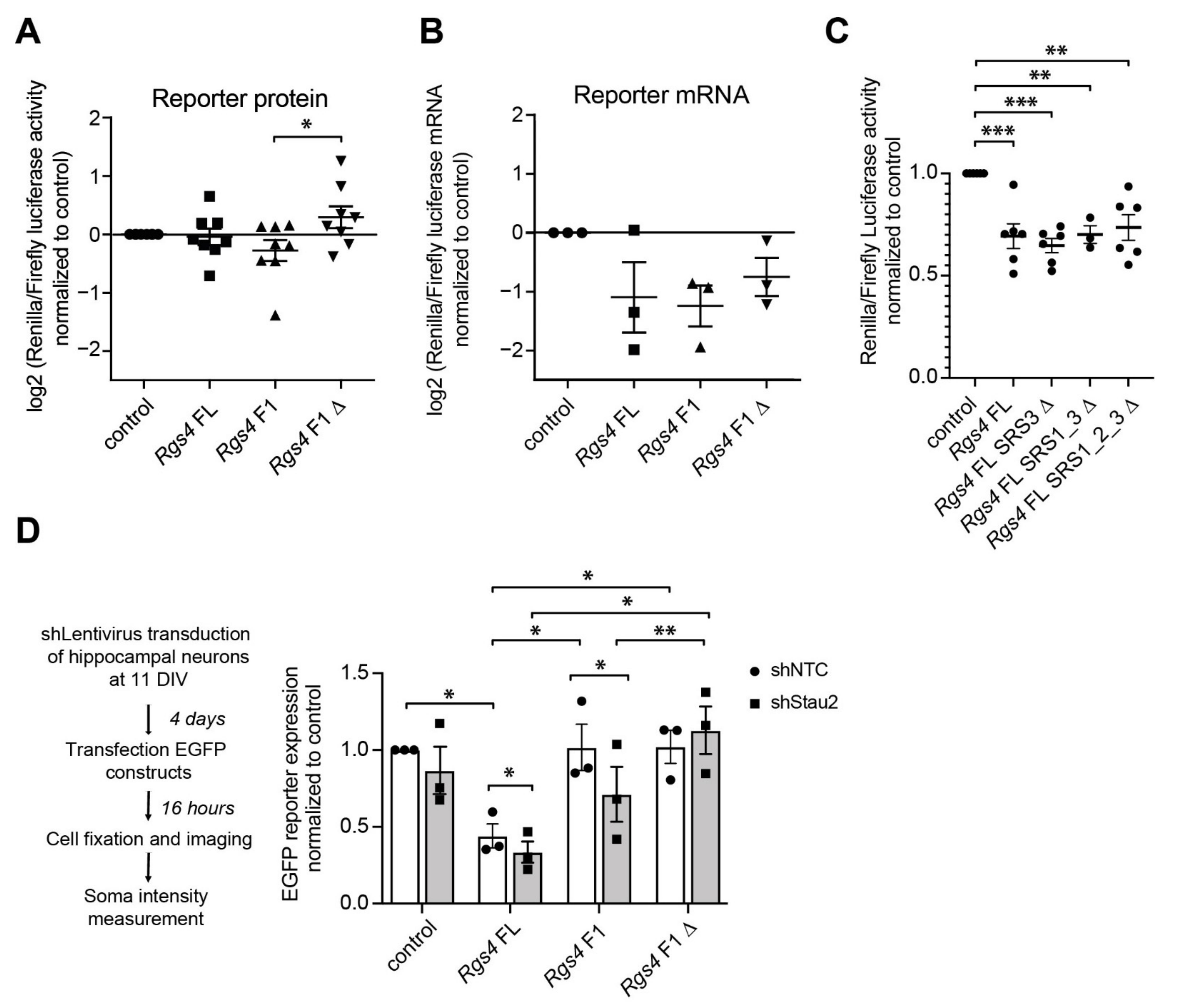
2.4. Rgs4 RNA Structure Contributes to Protein Expression
3. Discussion
4. Materials and Methods
4.1. Plasmids
4.2. Neuronal Cell Culture, Treatment, Transduction and Transfection
4.3. Lentivirus Production
4.4. RNA Extraction, cDNA Synthesis and qPCR
4.5. Bioinformatics
4.6. Rgs4 RNA Secondary Structure Prediction
4.7. Luciferase Assay
4.8. Imaging-Based GFP Expression Assay in Primary Neurons
4.9. Imaging and Data Analysis
4.10. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kiebler, M.A.; Bassell, G.J. Neuronal RNA Granules: Movers and Makers. Neuron 2006, 51, 685–690. [Google Scholar] [CrossRef] [Green Version]
- Köhrmann, M.; Luo, M.; Kaether, C.; DesGroseillers, L.; Dotti, G.C.; Kiebler, M.A. Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol. Biol. Cell 1999, 10, 2945–2953. [Google Scholar] [CrossRef]
- Kiebler, M.A.; Scheiffele, P.; Ule, J. What, where, and when: The importance of post-transcriptional regulation in the brain. Front. Neurosci. 2013, 7, 192. [Google Scholar] [CrossRef] [Green Version]
- Holt, C.E.; Bullock, S.L. Subcellular mRNA Localization in Animal Cells and Why It Matters. Science 2009, 326, 1212–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schieweck, R.; Ninkovic, J.; Kiebler, M.A. RNA-binding proteins balance brain function in health and disease. Physiol. Rev. 2021, 101, 1309–1370. [Google Scholar] [CrossRef] [PubMed]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, M.B.; Strein, C.; Davey, E.N.; Humphreys, T.D.; Preiss, T.; Steinmetz, M.L.; et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 149, 1393–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langdon, E.M.; Qiu, Y.; Niaki, A.G.; McLaughlin, G.A.; Weidmann, C.A.; Gerbich, T.M.; Smith, J.A.; Crutchley, J.M.; Termini, C.M.; Weeks, K.M.; et al. mRNA structure determines specificity of a polyQ-driven phase separation. Science 2018, 360, 922–927. [Google Scholar] [CrossRef] [Green Version]
- Sanchez de Groot, N.; Armaos, A.; Graña-Montes, R.; Alriquet, M.; Calloni, G.; Vabulas, M.R.; Tartaglia, G.G. RNA structure drives interaction with proteins. Nat. Commun. 2019, 10, 3246. [Google Scholar] [CrossRef]
- Tauber, D.; Tauber, G.; Parker, R. Mechanisms and Regulation of RNA Condensation in RNP Granule Formation. Trends Biochem. Sci. 2020, 45, 764–778. [Google Scholar] [CrossRef]
- Bullock, S.L.; Ringel, I.; Ish-Horowicz, D.; Lukavsky, P.J. A′-form RNA helices are required for cytoplasmic mRNA transport in Drosophila. Nat. Struct. Mol. Biol. 2010, 17, 703–709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, V.L.; Mitra, S.; Harris, R.; Buxbaum, A.R.; Lionnet, T.; Brenowitz, M.; Girvin, M.; Levy, M.; Almo, S.C.; Singer, R.H.; et al. Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev. 2012, 26, 43–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, K.C.; Ephrussi, A. mRNA Localization: Gene Expression in the Spatial Dimension. Cell 2009, 136, 719–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heraud-Farlow, J.E.; Kiebler, M.A. The multifunctional Staufen proteins: Conserved roles from neurogenesis to synaptic plasticity. Trends Neurosci. 2014, 37, 470–479. [Google Scholar] [CrossRef] [Green Version]
- Park, E.; Gleghorn, M.L.; Maquat, L.E. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc. Natl. Acad. Sci. USA 2012, 110, 405–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebeau, G.; Miller, L.C.; Tartas, M.; McAdam, R.; Laplante, I.; Badeaux, F.; DesGroseillers, L.; Sossin, W.S.; Lacaille, J.-C. Staufen 2 regulates mGluR long-term depression and Map1b mRNA distribution in hippocampal neurons. Learn. Mem. 2011, 18, 314–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, S.J.; Meulemans, D.; Vazquez, L.; Colaco, N.; Schuman, E. A Role for a Rat Homolog of Staufen in the Transport of RNA to Neuronal Dendrites. Neuron 2001, 32, 463–475. [Google Scholar] [CrossRef] [Green Version]
- Zimyanin, V.L.; Belaya, K.; Pecreaux, J.; Gilchrist, M.J.; Clark, A.; Davis, I.; Johnston, D.S. In Vivo Imaging of oskar mRNA Transport Reveals the Mechanism of Posterior Localization. Cell 2008, 134, 843–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, K.E.; Segura, I.; Gaspar, I.; Scheuss, V.; Illig, C.; Ammer, G.; Hutten, S.; Basyuk, E.; Fernández-Moya, S.M.; Ehses, J.; et al. Live cell imaging reveals 3′-UTR dependent mRNA sorting to synapses. Nat. Commun. 2019, 10, 3178. [Google Scholar] [CrossRef] [Green Version]
- Heraud-Farlow, J.E.; Sharangdhar, T.; Li, X.; Pfeifer, P.; Tauber, S.; Orozco, D.; Hörmann, A.; Thomas, S.; Bakosova, A.; Farlow, A.R.; et al. Staufen2 regulates neuronal target RNAs. Cell Rep. 2013, 5, 1511–1518. [Google Scholar] [CrossRef] [Green Version]
- Sharangdhar, T.; Sugimoto, Y.; Heraud-Farlow, J.; Fernández-Moya, S.M.; Ehses, J.; de Los Mozos, I.R.; Ule, J.; Kiebler, M.A. A retained intron in the 3′-UTR of Calm3 mRNA mediates its Staufen2- and activity-dependent localization to neuronal dendrites. EMBO Rep. 2017, 18, 1762–1774. [Google Scholar] [CrossRef]
- Laver, J.D.; Li, X.; Ancevicius, K.; Westwood, J.T.; Smibert, C.A.; Morris, Q.D.; Lipshitz, H.D. Genome-wide analysis of Staufen-associated mRNAs identifies secondary structures that confer target specificity. Nucleic Acids Res. 2013, 41, 9438–9460. [Google Scholar] [CrossRef] [Green Version]
- Ricci, E.P.; Kucukural, A.; Cenik, C.; Mercier, B.C.; Singh, G.; Heyer, E.; Ashar-Patel, A.; Peng, L.; Moore, M.J. Staufen1 senses overall transcript secondary structure to regulate translation. Nat. Struct. Mol. Biol. 2013, 21, 26–35. [Google Scholar] [CrossRef] [Green Version]
- Lazzaretti, D.; Bandholz-Cajamarca, L.; Emmerich, C.; Schaaf, K.; Basquin, C.; Irion, U.; Bono, F. The crystal structure of Staufen1 in complex with a physiological RNA sheds light on substrate selectivity. Life Sci. Alliance 2018, 1, e201800187. [Google Scholar] [CrossRef]
- Heber, S.; Gáspár, I.; Tants, J.-N.; Günther, J.; Moya, S.M.F.; Janowski, R.; Ephrussi, A.; Sattler, M.; Niessing, D. Staufen2-mediated RNA recognition and localization requires combinatorial action of multiple domains. Nat. Commun. 2019, 10, 1659. [Google Scholar] [CrossRef] [Green Version]
- Poblete, S.; Guzman, H. Structural 3D Domain Reconstruction of the RNA Genome from Viruses with Secondary Structure Models. Viruses 2021, 13, 1555. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, Y.; Vigilante, A.; Darbo, E.; Zirra, A.; Militti, C.; D’Ambrogio, A.; Luscombe, N.M.; Ule, J. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen. Nature 2015, 519, 491–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cajigas, I.J.; Tushev, G.; Will, T.J.; Dieck, S.T.; Fuerst, N.; Schuman, E.M. The Local Transcriptome in the Synaptic Neuropil Revealed by Deep Sequencing and High-Resolution Imaging. Neuron 2012, 74, 453–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berman, D.M.; Wilkie, T.M.; Gilman, A.G. GAIP and RGS4 Are GTPase-Activating Proteins for the Gi Subfamily of G Protein α Subunits. Cell 1996, 86, 445–452. [Google Scholar] [CrossRef] [Green Version]
- Grillet, N.; Pattyn, A.; Contet, C.; Kieffer, B.L.; Goridis, C.; Brunet, J.-F. Generation and Characterization of Rgs4 Mutant Mice. Mol. Cell. Biol. 2005, 25, 4221–4228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertrand, E.; Chartrand, P.; Schaefer, M.; Shenoy, S.; Singer, R.H.; Long, R.M. Localization of ASH1 mRNA Particles in Living Yeast. Mol. Cell 1998, 2, 437–445. [Google Scholar] [CrossRef] [Green Version]
- Roden, C.; Gladfelter, A.S. RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 2020, 22, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Tübing, F.; Vendra, G.; Mikl, M.; Macchi, P.; Thomas, S.; Kiebler, M.A. Dendritically Localized Transcripts Are Sorted into Distinct Ribonucleoprotein Particles That Display Fast Directional Motility along Dendrites of Hippocampal Neurons. J. Neurosci. 2010, 30, 4160–4170. [Google Scholar] [CrossRef] [Green Version]
- Floor, S.N.; Doudna, J.A. Tunable protein synthesis by transcript isoforms in human cells. eLife 2016, 5, e10921. [Google Scholar] [CrossRef]
- Kertesz, M.; Wan, Y.; Mazor, E.; Rinn, J.; Nutter, R.C.; Chang, H.Y.; Segal, E. Genome-wide measurement of RNA secondary structure in yeast. Nature 2010, 467, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Rouskin, S.; Zubradt, M.; Washietl, S.; Kellis, M.; Weissman, J.S. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 2013, 505, 701–705. [Google Scholar] [CrossRef] [Green Version]
- Courel, M.; Clément, Y.; Bossevain, C.; Foretek, D.; Cruchez, O.V.; Yi, Z.; Bénard, M.; Benassy, M.-N.; Kress, M.; Vindry, C.; et al. GC content shapes mRNA storage and decay in human cells. eLife 2019, 8, e49708. [Google Scholar] [CrossRef] [PubMed]
- Masliah, G.; Barraud, P.; Allain, F.H.-T. RNA recognition by double-stranded RNA binding domains: A matter of shape and sequence. Experientia 2012, 70, 1875–1895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schieweck, R.; Riedemann, T.; Forné, I.; Harner, M.; Bauer, K.E.; Rieger, D.; Ang, F.Y.; Hutten, S.; Demleitner, A.F.; Popper, B.; et al. Pumilio2 and Staufen2 selectively balance the synaptic proteome. Cell Rep. 2021, 35, 109279. [Google Scholar] [CrossRef]
- Berkovits, B.D.; Mayr, C. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature 2015, 522, 363–367. [Google Scholar] [CrossRef] [Green Version]
- Ehses, J.; Fernández-Moya, M.; Schröger, L.; Kiebler, M.A. Synergistic regulation of Rgs4 mRNA by HuR and miR-26/RISC in neurons. RNA Biol. 2021, 18, 988–998. [Google Scholar] [CrossRef]
- Van Treeck, B.; Protter, D.S.W.; Matheny, T.; Khong, A.; Link, C.D.; Parker, R. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl. Acad. Sci. USA 2018, 115, 2734–2739. [Google Scholar] [CrossRef] [Green Version]
- Youn, J.Y.; Dunham, W.H.; Hong, S.J.; Knight, J.D.R.; Bashkurov, M.; Chen, G.I.; Bagci, H.; Rathod, B.; MacLeod, G.; Eng, S.W.M.; et al. High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Mol. Cell 2018, 69, 517–532.e11. [Google Scholar] [CrossRef] [PubMed]
- Berger, S.M.; Fernández-Lamo, I.; Schönig, K.; Moya, S.M.F.; Ehses, J.; Schieweck, R.; Clementi, S.; Enkel, T.; Grothe, S.; Halbach, O.V.B.U.; et al. Forebrain-specific, conditional silencing of Staufen2 alters synaptic plasticity, learning, and memory in rats. Genome Biol. 2017, 18, 222. [Google Scholar] [CrossRef] [PubMed]
- Popper, B.; Demleitner, A.; Bolivar, V.; Kusek, G.; Snyder-Keller, A.; Schieweck, R.; Temple, S.; Kiebler, M.A. Staufen2 deficiency leads to impaired response to novelty in mice. Neurobiol. Learn. Mem. 2018, 150, 107–115. [Google Scholar] [CrossRef]
- Schilling, M.; Prusty, A.B.; Boysen, B.; Oppermann, F.S.; Riedel, Y.L.; Husedzinovic, A.; Rasouli, H.; König, A.; Ramanathan, P.; Reymann, J.; et al. TOR signaling regulates liquid phase separation of the SMN complex governing snRNP biogenesis. Cell Rep. 2021, 35, 109277. [Google Scholar] [CrossRef]
- Weng, Y.-L.; Wang, X.; An, R.; Cassin, J.; Vissers, C.; Liu, Y.; Liu, Y.; Xu, T.; Wang, X.; Wong, S.Z.H.; et al. Epitranscriptomic m6A Regulation of Axon Regeneration in the Adult Mammalian Nervous System. Neuron 2018, 97, 313–325.e6. [Google Scholar] [CrossRef] [Green Version]
- Ries, R.J.; Zaccara, S.; Klein, P.; Olarerin-George, A.; Namkoong, S.; Pickering, B.F.; Patil, D.P.; Kwak, H.; Lee, J.H.; Jaffrey, S.R. m6A enhances the phase separation potential of mRNA. Nature 2019, 571, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Goetze, B.; Tuebing, F.; Xie, Y.; Dorostkar, M.; Thomas, S.; Pehl, U.; Boehm, S.; Macchi, P.; Kiebler, M. The brain-specific double-stranded RNA-binding protein Staufen2 is required for dendritic spine morphogenesis. J. Cell Biol. 2006, 172, 221–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venny, 2.1, An Interactive Tool for Comparing Lists with Venn’s Diagrams. BioinfoGP, CNB-CSIC: Madrid, Spanish, 2007–2015. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 12 October 2020).
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruber, A.; Lorenz, R.; Bernhart, S.H.F.; Neuböck, R.; Hofacker, I.L. The Vienna RNA Websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef] [Green Version]
- Goetze, B.; Grunewald, B.; Baldassa, S.; Kiebler, M. Chemically controlled formation of a DNA/calcium phosphate coprecipitate: Application for transfection of mature hippocampal neurons. J. Neurobiol. 2004, 60, 517–525. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org/ (accessed on 12 October 2020).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer: New York, NY, USA, 2009. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Moya, S.M.; Ehses, J.; Bauer, K.E.; Schieweck, R.; Chakrabarti, A.M.; Lee, F.C.Y.; Illig, C.; Luscombe, N.M.; Harner, M.; Ule, J.; et al. RGS4 RNA Secondary Structure Mediates Staufen2 RNP Assembly in Neurons. Int. J. Mol. Sci. 2021, 22, 13021. https://doi.org/10.3390/ijms222313021
Fernández-Moya SM, Ehses J, Bauer KE, Schieweck R, Chakrabarti AM, Lee FCY, Illig C, Luscombe NM, Harner M, Ule J, et al. RGS4 RNA Secondary Structure Mediates Staufen2 RNP Assembly in Neurons. International Journal of Molecular Sciences. 2021; 22(23):13021. https://doi.org/10.3390/ijms222313021
Chicago/Turabian StyleFernández-Moya, Sandra M., Janina Ehses, Karl E. Bauer, Rico Schieweck, Anob M. Chakrabarti, Flora C. Y. Lee, Christin Illig, Nicholas M. Luscombe, Max Harner, Jernej Ule, and et al. 2021. "RGS4 RNA Secondary Structure Mediates Staufen2 RNP Assembly in Neurons" International Journal of Molecular Sciences 22, no. 23: 13021. https://doi.org/10.3390/ijms222313021
APA StyleFernández-Moya, S. M., Ehses, J., Bauer, K. E., Schieweck, R., Chakrabarti, A. M., Lee, F. C. Y., Illig, C., Luscombe, N. M., Harner, M., Ule, J., & Kiebler, M. A. (2021). RGS4 RNA Secondary Structure Mediates Staufen2 RNP Assembly in Neurons. International Journal of Molecular Sciences, 22(23), 13021. https://doi.org/10.3390/ijms222313021





