Unraveling the Spatiotemporal Distribution of VPS13A in the Mouse Brain
Abstract
:1. Introduction
2. Results
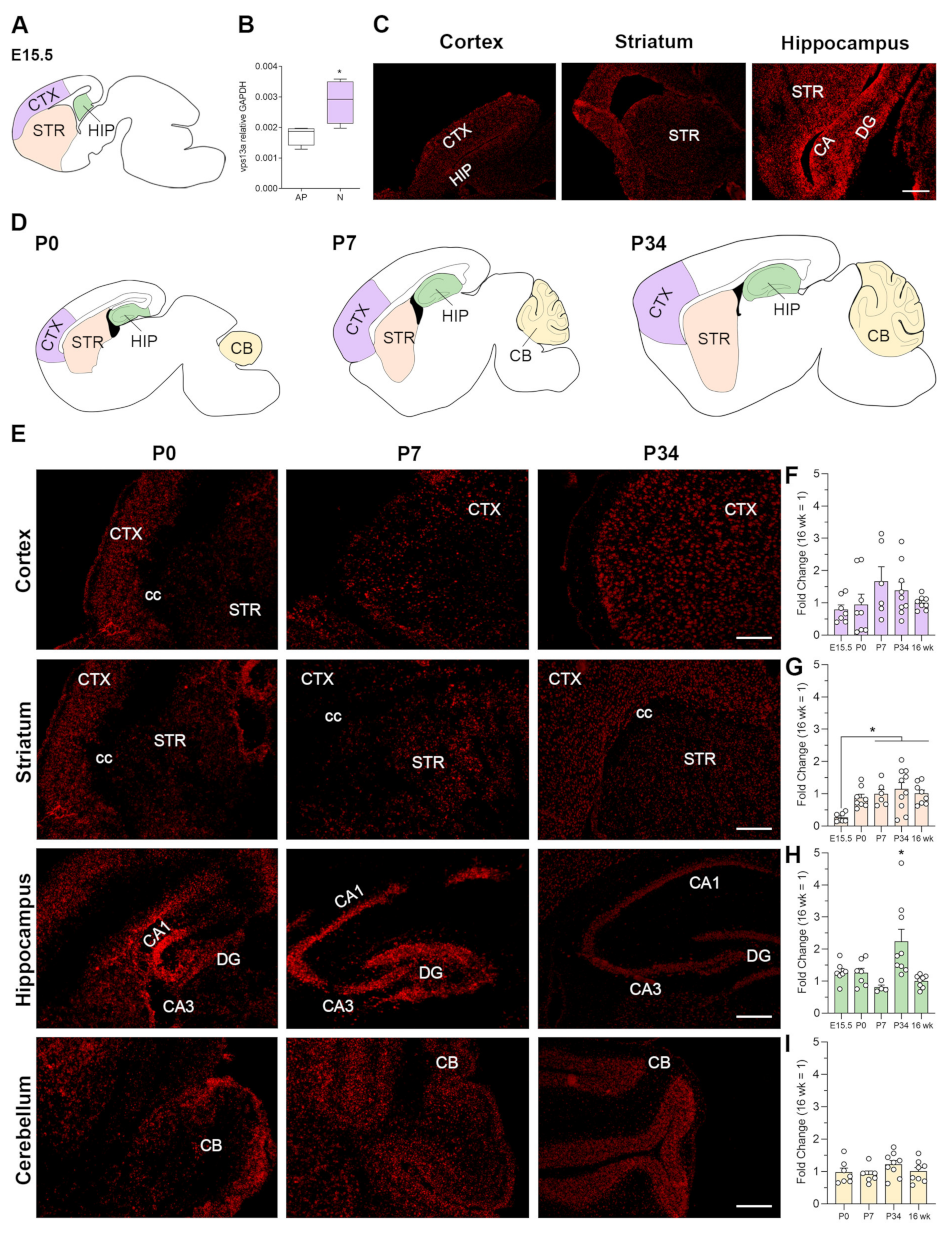
2.1. VPS13A Expression Starts from Embryonic Stage and Is Stable over Time
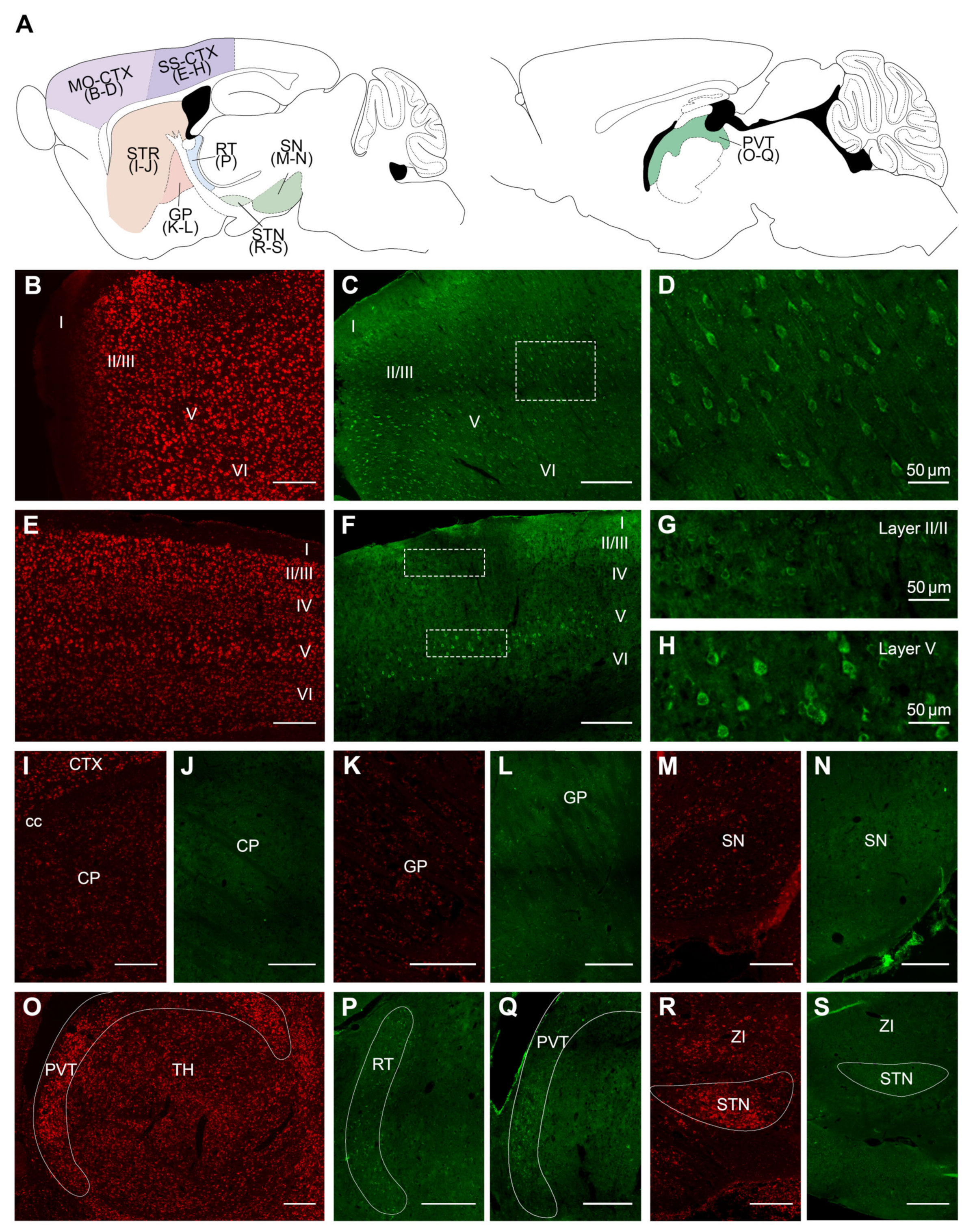
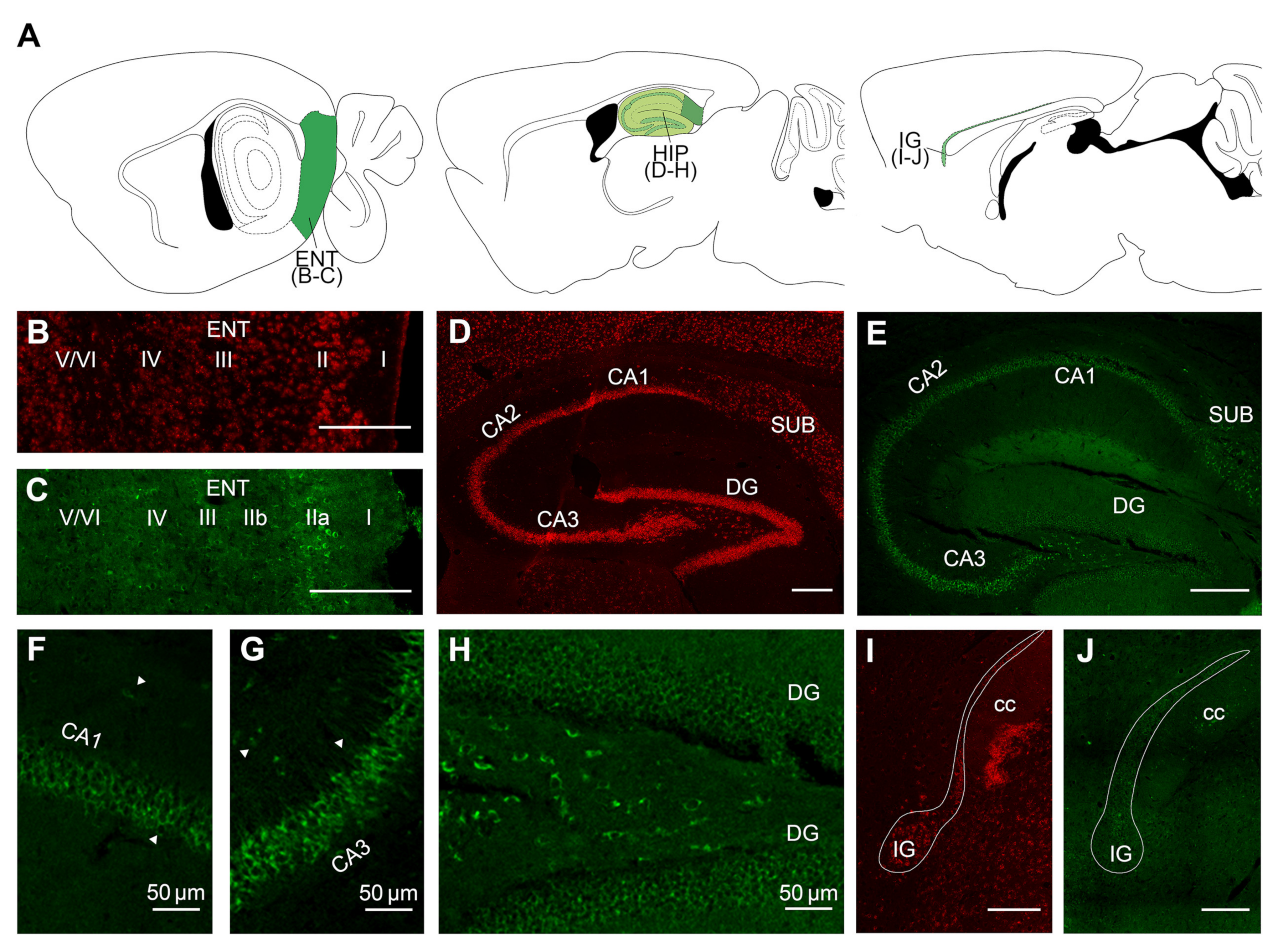
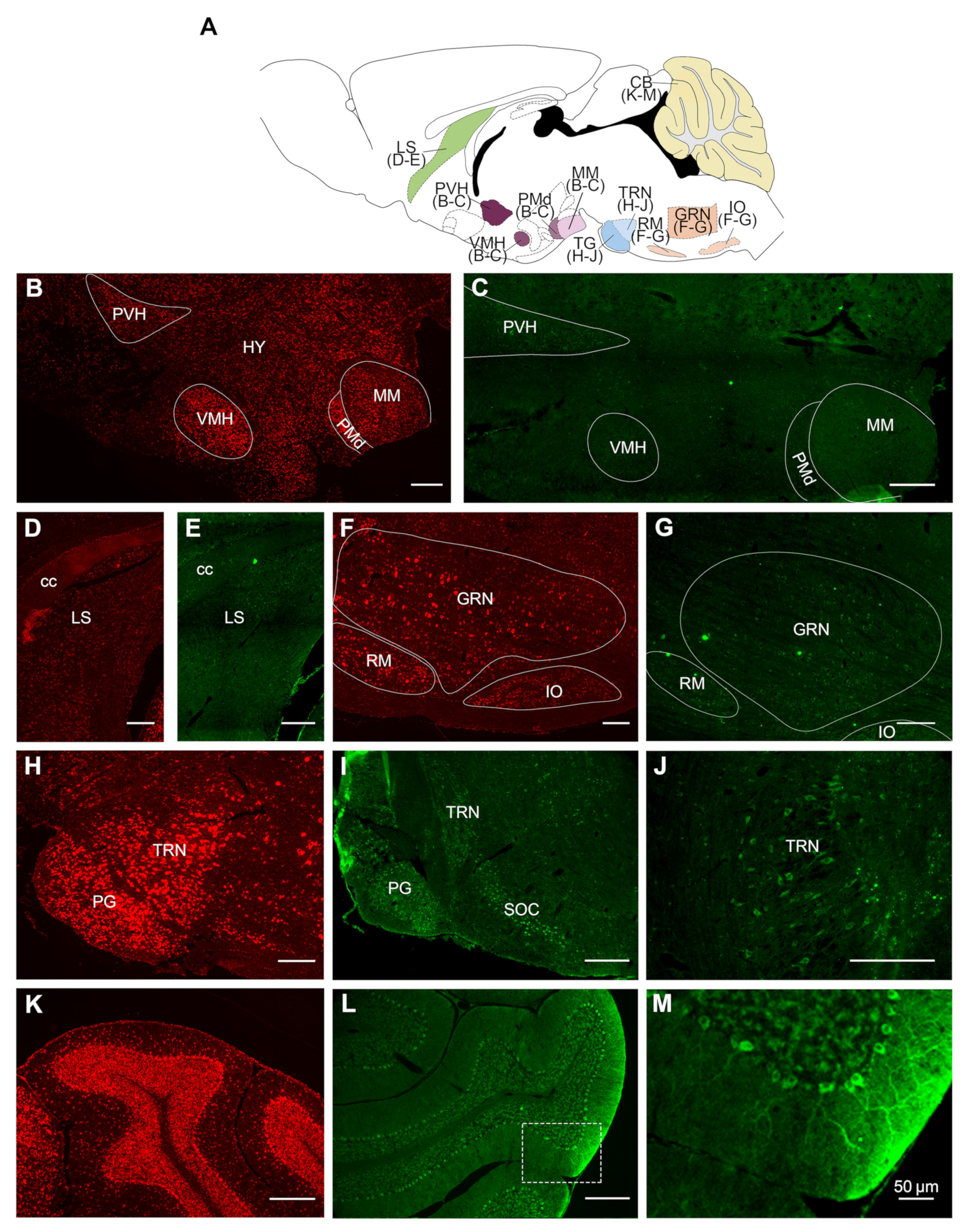
2.2. Overall Distribution of VPS13A mRNA and Protein in the Adult Mouse Brain
2.3. VPS13A Is Enriched in Glutamatergic, GABAergic, and Cholinergic Neurons
2.4. VPS13A Is Present Mainly in the Soma of Neurons and Co-Localizes with Both ER and Mitochondria
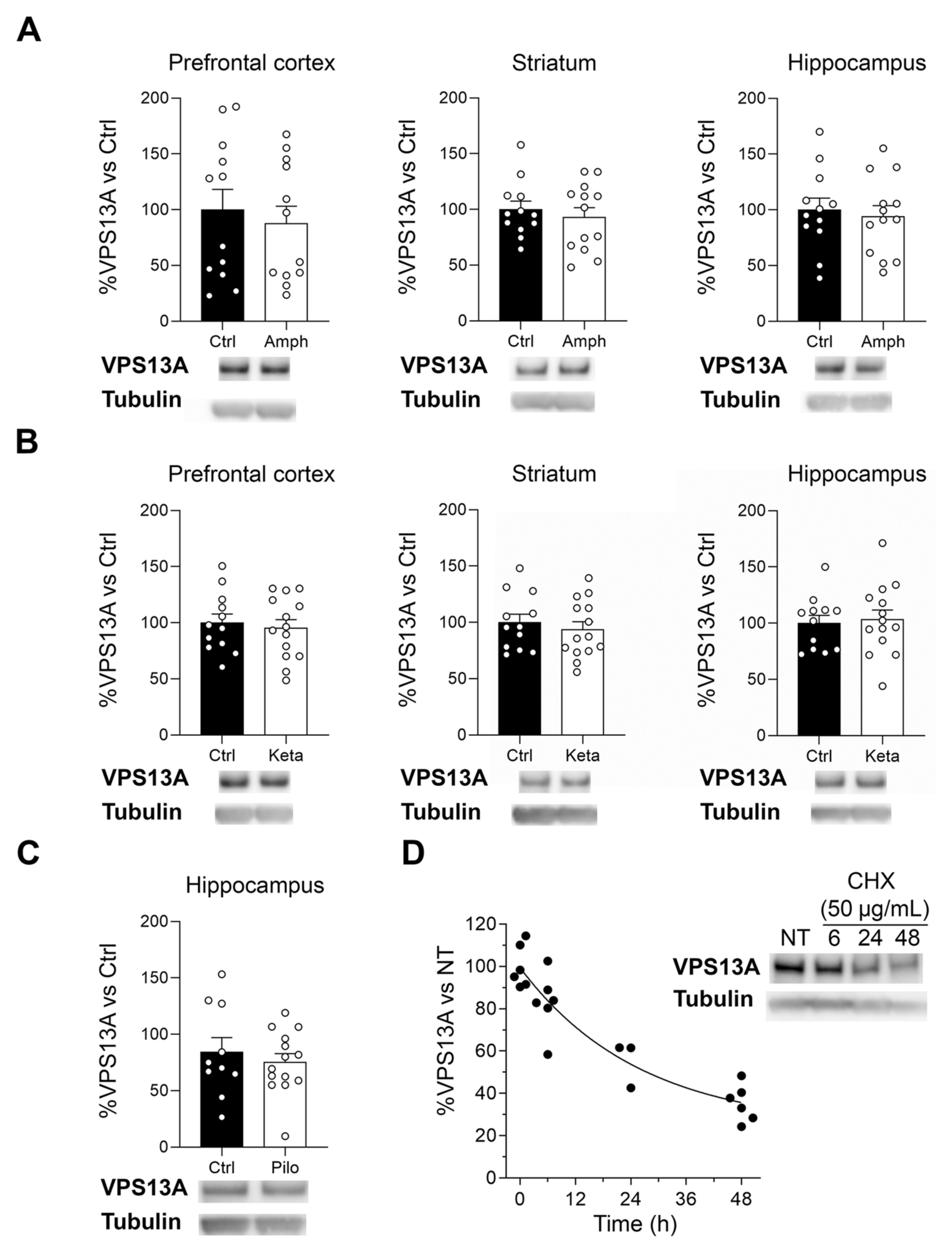
2.5. Manipulation of the Dopaminergic, Glutamatergic, and Cholinergic Circuits Does Not Modify VPS13A Levels
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Tissue Sampling
4.3. Fluorescence In Situ Hybridization
4.4. Immunohistochemistry
4.5. Quantitative Real-Time PCR
4.6. Synaptosomal Fractionation and Western Blotting
4.7. Primary Cell Cultures
4.8. Immunocytochemistry and Sub-Cellular Localization Analysis
4.9. Amphetamine, Ketamine and Pilocarpine Treatment
4.10. Analysis of VPS13A Protein Stability
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Danek, A.; Walker, R. Neuroacanthocytosis. Curr. Opin. Neurol. 2005, 18, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Velayos-Baeza, A.; Dobson-Stone, C.; Rampoldi, L.; Bader, B.; Walker, R.H.; Danek, A.; Monaco, A.P. Chorea-Acanthocytosis. Curated Ref. Collect. Neurosci. Biobehav. Psychol. 2019; pp. 217–219. Available online: https://pubmed.ncbi.nlm.nih.gov/20301561/\ (accessed on 29 November 2021).
- Walker, R.H.; Danek, A. “Neuroacanthocytosis”—Overdue for a Taxonomic Update. Tremor Other Hyperkinetic Mov. 2021, 11, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, D.J.; Walker, R.H. Functional neuroimaging and chorea: A systematic review. J. Clin. Mov. Disord. 2017, 4, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ichiba, M.; Nakamura, M.; Kusumoto, A.; Mizuno, E.; Kurano, Y.; Matsuda, M.; Kato, M.; Agemura, A.; Tomemori, Y.; Muroya, S.; et al. Clinical and molecular genetic assessment of a chorea-acanthocytosis pedigree. J. Neurol. Sci. 2007, 263, 124–132. [Google Scholar] [CrossRef]
- Walterfang, M.; Looi, J.C.L.; Styner, M.; Walker, R.H.; Danek, A.; Niethammer, M.; Evans, A.; Kotschet, K.; Rodrigues, G.R.; Hughes, A.; et al. Shape alterations in the striatum in chorea-acanthocytosis. Psychiatry Res. Neuroimaging 2011, 192, 29–36. [Google Scholar] [CrossRef] [Green Version]
- Connolly, B.; Hazrati, L.; Lang, A. Neuropathological findings in chorea-acanthocytosis: New insights into mechanisms underlying parkinsonism and seizures. Acta Neuropathol. 2014, 127, 613–615. [Google Scholar] [CrossRef]
- Rinne, J.O.; Daniel, S.E.; Scaravilli, F.; Pires, M.; Harding, A.E.; Marsden, C.D. The neuropathological features of neuroacanthocytosis. Mov. Disord. 1994, 9, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Glaß, H.; Neumann, P.; Pal, A.; Reinhardt, P.; Storch, A.; Sterneckert, J.; Hermann, A. Combined Dendritic and Axonal Deterioration Are Responsible for Motoneuronopathy in Patient-Derived Neuronal Cell Models of Chorea-Acanthocytosis. Int. J. Mol. Sci. 2020, 21, 1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankaitis, V.A.; Johnson, L.M.; Emr, S.D. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc. Natl. Acad. Sci. USA 1986, 83, 9075–9079. [Google Scholar] [CrossRef] [Green Version]
- John Peter, A.; Herrmann, B.; Antunes, D.; Rapaport, D.; Dimmer, K.; Kornmann, B. Vps13-Mcp1 interact at vacuole-mitochondria interfaces and bypass ER-mitochondria contact sites. J. Cell Biol. 2017, 216, 3219–3229. [Google Scholar] [CrossRef] [Green Version]
- Lang, A.; John Peter, A.; Walter, P.; Kornmann, B. ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J. Cell Biol. 2015, 210, 883–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leonzino, M.; Reinisch, K.M.; De Camilli, P. Insights into VPS13 properties and function reveal a new mechanism of eukaryotic lipid transport. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 159003. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Thorsness, M.K.; Policastro, R.; McGoldrick, L.L.; Hollingsworth, N.M.; Thorsness, P.E.; Neiman, A.M. Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Mol. Biol. Cell 2016, 27, 2435–2449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeshaw, W.; van der Zwaag, M.; Pinto, F.; Lahaye, L.; Faber, A.; Gómez-Sánchez, R.; Dolga, A.; Poland, C.; Monaco, A.; van IJzendoorn, S.; et al. Human VPS13A is associated with multiple organelles and influences mitochondrial morphology and lipid droplet motility. Elife 2019, 8, 43561. [Google Scholar] [CrossRef] [PubMed]
- Lupo, F.; Tibaldi, E.; Matte, A.; Sharma, A.K.; Brunati, A.M.; Alper, S.L.; Zancanaro, C.; Benati, D.; Siciliano, A.; Bertoldi, M.; et al. A new molecular link between defective autophagy and erythroid abnormalities in chorea-acanthocytosis. Blood 2016, 128, 2976–2987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz-Braceras, S.; Calvo, R.; Escalante, R. TipC and the chorea-acanthocytosis protein VPS13A regulate autophagy in Dictyostelium and human HeLa cells. Autophagy 2015, 11, 918–927. [Google Scholar] [CrossRef] [Green Version]
- Vonk, J.J.; Yeshaw, W.M.; Pinto, F.; Faber, A.I.E.; Lahaye, L.L.; Kanon, B.; Zwaag, M. van der Velayos-Baeza, A.; Freire, R.; IJzendoorn, S.C.; et al. Drosophila Vps13 Is Required for Protein Homeostasis in the Brain. PLoS ONE 2017, 12, e0170106. [Google Scholar] [CrossRef] [PubMed]
- Munõz-Braceras, S.; Tornero-Écija, A.R.; Vincent, O.; Escalante, R. VPS13A is closely associated with mitochondria and is required for efficient lysosomal degradation. DMM Dis. Model. Mech. 2019, 12, 918–927. [Google Scholar] [CrossRef] [Green Version]
- Pelzl, L.; Elsir, B.; Sahu, I.; Bissinger, R.; Singh, Y.; Sukkar, B.; Honisch, S.; Schoels, L.; Jemaà, M.; Lang, E.; et al. Lithium Sensitivity of Store Operated Ca2+ Entry and Survival of Fibroblasts Isolated from Chorea-Acanthocytosis Patients. Cell. Physiol. Biochem. 2017, 42, 2066–2077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelzl, L.; Hauser, S.; Elsir, B.; Sukkar, B.; Sahu, I.; Singh, Y.; Höflinger, P.; Bissinger, R.; Jemaà, M.; Stournaras, C.; et al. Lithium Sensitive ORAI1 Expression, Store Operated Ca 2+ Entry and Suicidal Death of Neurons in Chorea-Acanthocytosis. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Yu, W.; Honisch, S.; Schmidt, S.; Yan, J.; Schmid, E.; Alkahtani, S.; AlKahtane, A.; Alarifi, S.; Stournaras, C.; Lang, F. Chorein Sensitive Orai1 Expression and Store Operated Ca2+ Entry in Rhabdomyosarcoma Cells. Cell. Physiol. Biochem. 2016, 40, 1141–1152. [Google Scholar] [CrossRef]
- Kurano, Y.; Nakamura, M.; Ichiba, M.; Matsuda, M.; Mizuno, E.; Kato, M.; Agemura, A.; Izumo, S.; Sano, A. In vivo distribution and localization of chorein. Biochem. Biophys. Res. Commun. 2007, 353, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Florio, M.; Albert, M.; Taverna, E.; Namba, T.; Brandl, H.; Lewitus, E.; Haffner, C.; Sykes, A.; Wong, F.; Peters, J.; et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 2015, 347, 1465–1470. [Google Scholar] [CrossRef] [PubMed]
- Vorspan, F.; Icick, R.; Mekdad, N.; Courtin, C.; Bloch, V.; Bellivier, F.; Laplanche, J.-L.; Prince, N.; Pishalin, D.; Firmo, C.; et al. Translational study of the whole transcriptome in rats and genetic polymorphisms in humans identifies LRP1B and VPS13A as key genes involved in tolerance to cocaine-induced motor disturbances. Transl. Psychiatry 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Daroff, R.; Aminoff, M. Encyclopedia of the Neurological Sciences, 2nd ed.; Elsevier: Oxford, UK, 2014. [Google Scholar]
- Koziol, L.F.; Budding, D.; Andreasen, N.; D’Arrigo, S.; Bulgheroni, S.; Imamizu, H.; Ito, M.; Manto, M.; Marvel, C.; Parker, K.; et al. Consensus paper: The cerebellum’s role in movement and cognition. Cerebellum 2014, 13, 151–177. [Google Scholar] [CrossRef]
- Knierim, J.J. The hippocampus. Curr. Biol. 2015, 25, R1116–R1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mente, K.; Kim, S.A.; Grunseich, C.; Hefti, M.M.; Crary, J.F.; Danek, A.; Karp, B.I.; Walker, R.H. Hippocampal sclerosis and mesial temporal lobe epilepsy in chorea-acanthocytosis: A case with clinical, pathologic and genetic evaluation. Neuropathol. Appl. Neurobiol. 2017, 43, 542–546. [Google Scholar] [CrossRef] [PubMed]
- Neutel, D.; Miltenberger-Miltenyi, G.; Silva, I.; de Carvalho, M. Chorea-acanthocytosis presenting as motor neuron disease. Muscle Nerve 2012, 45, 293–295. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.; Nath, K.; Acharya, M.; Kumawat, B.; Khandelwal, D.; Jain, D. Cerebellar atrophy in neuroacanthocytosis. BMJ Case Rep. 2014, 2014, bcr2014205232. [Google Scholar] [CrossRef] [Green Version]
- Katsube, T.; Shimono, T.; Ashikaga, R.; Hosono, M.; Kitagaki, H.; Murakami, T. Demonstration of cerebellar atrophy in neuroacanthocytosis of 2 siblings. AJNR. Am. J. Neuroradiol. 2009, 30, 386–388. [Google Scholar] [CrossRef] [Green Version]
- Kontoleon, P.; Ilias, I.; Matsouka, A.; Gatzonis, S.; Spiropoulou, C.; Papapetrou, P. Impaired hypothalamic endocrine function in neuroacanthocytosis. J. Clin. Neurosci. 2003, 10, 701–703. [Google Scholar] [CrossRef]
- Gradstein, L.; Danek, A.; Grafman, J.; Fitzgibbon, E. Eye movements in chorea-acanthocytosis. Invest. Ophthalmol. Vis. Sci. 2005, 46, 1979–1987. [Google Scholar] [CrossRef]
- Henkel, K.; Walterfang, M.; Velakoulis, D.; Danek, A.; Kassubek, J. Volumetric Neuroimaging in Neuroacanthocytosis. Neuroacanthocytosis Syndr. II 2008, 2008, 175–185. [Google Scholar] [CrossRef]
- Wu, Y.; Li, H.; Zhang, C.; Sun, B.; Li, D.; Wu, Y. Subthalamic nucleus deep brain stimulation in two siblings with chorea-acanthocytosis. Neurol. Sci. 2020, 41, 1623–1625. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chen, K.; Sloan, S.; Bennett, M.; Scholze, A.; O’Keeffe, S.; Phatnani, H.; Guarnieri, P.; Caneda, C.; Ruderisch, N.; et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014, 34, 11929–11947. [Google Scholar] [CrossRef]
- Lundgaard, I.; Osório, M.J.; Kress, B.; Sanggaard, S.; Nedergaard, M. White matter astrocytes in health and disease. Neuroscience 2014, 276, 161. [Google Scholar] [CrossRef] [Green Version]
- Vaidyanathan, T.; Collard, M.; Yokoyama, S.; Reitman, M.; Poskanzer, K. Cortical astrocytes independently regulate sleep depth and duration via separate GPCR pathways. Elife 2021, 10, 63329. [Google Scholar] [CrossRef]
- Rodríguez, M.J.; Martínez-Sánchez, M.; Bernal, F.; Mahy, N. Heterogeneity between hippocampal and septal astroglia as a contributing factor to differential in vivo AMPA excitotoxicity. J. Neurosci. Res. 2004, 77, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Kishida, M.; Nishizawa, Y.; Iijima, M.; Koriyama, C.; Nakamura, M.; Sano, A.; Kishida, S. Subcellular localization and putative role of VPS13A/chorein in dopaminergic neuronal cells. Biochem. Biophys. Res. Commun. 2012, 419, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Leonzino, M.; Hancock-Cerutti, W.; Horenkamp, F.A.; Li, P.Q.; Lees, J.A.; Wheeler, H.; Reinisch, K.M.; De Camilli, P. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 2018, 217, 3625–3639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gauthier, J.; Meijer, I.; Lessel, D.; Mencacci, N.; Krainc, D.; Hempel, M.; Tsiakas, K.; Prokisch, H.; Rossignol, E.; Helm, M.; et al. Recessive mutations in VPS13D cause childhood onset movement disorders. Ann. Neurol. 2018, 83, 1089–1095. [Google Scholar] [CrossRef] [PubMed]
- Lesage, S.; Drouet, V.; Majounie, E.; Deramecourt, V.; Jacoupy, M.; Nicolas, A.; Cormier-Dequaire, F.; Hassoun, S.; Pujol, C.; Ciura, S.; et al. Loss of VPS13C Function in Autosomal-Recessive Parkinsonism Causes Mitochondrial Dysfunction and Increases PINK1/Parkin-Dependent Mitophagy. Am. J. Hum. Genet. 2016, 98, 500–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, J.; Liu, S.; Zhao, Y.; Tan, X.; Aljohi, H.A.; Liu, W.; Hu, S. Identification and analysis of house-keeping and tissue-specific genes based on RNA-seq data sets across 15 mouse tissues. Gene 2016, 576, 560–570. [Google Scholar] [CrossRef]
- Kim, A.; García-García, E.; Straccia, M.; Comella-Bolla, A.; Miguez, A.; Masana, M.; Alberch, J.; Canals, J.M.; Rodríguez, M.J. Reduced Fractalkine Levels Lead to Striatal Synaptic Plasticity Deficits in Huntington’s Disease. Front. Cell. Neurosci. 2020, 14, 14. [Google Scholar] [CrossRef]
- Fernández-García, S.; Orlandi, J.G.; García-Díaz Barriga, G.A.; Rodríguez, M.J.; Masana, M.; Soriano, J.; Alberch, J. Deficits in coordinated neuronal activity and network topology are striatal hallmarks in Huntington’s disease. BMC Biol. 2020, 18, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Fernández-García, S.; Sancho-Balsells, A.; Longueville, S.; Hervé, D.; Gruart, A.; Delgado-García, J.M.; Alberch, J.; Giralt, A. Astrocytic BDNF and TrkB regulate severity and neuronal activity in mouse models of temporal lobe epilepsy. Cell Death Dis. 2020, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Trettel, F.; Rigamonti, D.; Hilditch-Maguire, P.; Wheeler, V.; Sharp, A.; Persichetti, F.; Cattaneo, E.; MacDonald, M. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum. Mol. Genet. 2000, 9, 2799–2809. [Google Scholar] [CrossRef] [PubMed]
- Xifró, X.; García-Martínez, J.M.; del Toro, D.; Alberch, J.; Pérez-Navarro, E. Calcineurin is involved in the early activation of NMDA-mediated cell death in mutant huntingtin knock-in striatal cells. J. Neurochem. 2008, 105, 1596–1612. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Song, P.; Zou, M. Inhibition of AMP-activated protein kinase α (AMPKα) by doxorubicin accentuates genotoxic stress and cell death in mouse embryonic fibroblasts and cardiomyocytes: Role of p53 and SIRT1. J. Biol. Chem. 2012, 287, 8001–8012. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Brain Structure | mRNA | Protein | Brain Structure | mRNA | Protein |
|---|---|---|---|---|---|
| Motor Cortex | Basal ganglia | ||||
| Layer I | − | − | Caudate putamen | + | + |
| Layer II/III | ++ | ++ | Fundus of striatum | + | + |
| Layer V | ++ | ++ | Globus pallidus | + | + |
| Layer VI | ++ | ++ | Bed nucleus of stria terminalis | ++ | + |
| Somatosensory Cortex | Nucleus accumbens | + | + | ||
| Layer I | − | − | Substantia nigra | + | + |
| Layer II/III | ++ | + | Subthalamic nucleus | +++ | ++ |
| Layer IV | ++ | + | Amygdaloid complex | ||
| Layer V | +++ | ++ | Basolateral amygdalar nucleus | ++ | ++ |
| Layer VI | ++ | + | Basomedial amygdalar nucleus | ++ | ++ |
| Visual Cortex | Central amygdalar nucleus | ++ | ++ | ||
| Layer I | − | − | Thalamus | ||
| Layer II/III | ++ | + | Reticular nucleus | ++ | ++ |
| Layer IV | ++ | + | Lateral dorsal nucleus | + | + |
| Layer V | ++ | ++ | Posterior complex | + | + |
| Layer VI | ++ | + | Ventral medial nucleus | + | + |
| Entorhinal Area | Ventral/Dorsal geniculate n. | + | + | ||
| Layer I | − | − | Paraventricular nucleus | ++ | ++ |
| Layer II | +++ | +++ | Medial habenula | ++ | ++ |
| Layer III | ++ | + | Nucleus of reuniens | + | + |
| Layer IV | ++ | + | Hypothalamus | ||
| Layer V/VI | ++ | + | Paraventricular hypothalamic n. | ++ | ++ |
| Hippocampal Region | Ventromedial hypothalamic n. | ++ | ++ | ||
| CA3 | +++ | ++ | Ventral premammillary nucleus | ++ | ++ |
| CA2 | +++ | ++ | Lateral mammillary nucleus | ++ | n/a |
| CA1 | ++ | + | Medial mammillary nucleus | ++ | n/a |
| Granular layer of the DG | +++ | + | Medial preoptic area | + | + |
| Polymorphic layer of the DG | + | + | Arcuate hypothalamic nucleus | + | + |
| Molecular layer of the DG | − | − | Suprachiasmatic nucleus | + | + |
| Hilus | ++ | ++ | Zona incerta | + | + |
| Postsubiculum | ++ | ++ | Septal region | ||
| Presubiculum | ++ | ++ | Lateral septal nucleus | + | + |
| Subiculum | ++ | +++ | Medial septal nucleus | + | + |
| Induseum griseum | ++ | ++ | Septohippocampal nucleus | + | + |
| Mid-Brain | Cerebellum | ||||
| Superior colliculus | + | + | Purkinje cell layer | +++ | +++ |
| Inferior colliculus | + | + | Molecular layer | + | + |
| Edinger–Westphal nucleus | ++ | ++ | Granular layer | ++ | ++ |
| Trochelar nucleus | ++ | n/a | White matter structures | ||
| Oculomotor nucleus | ++ | n/a | Corpus callosum | + | − |
| Pons | Anterior commissure | − | − | ||
| Pontine gray | +++ | +++ | Fornix system | + | + |
| Tegmental reticular nucleus | +++ | +++ | Optic tract | + | − |
| Pontine reticular nucleus | ++ | n/a | Ventral hippocampal commissure | + | − |
| Motor nucleus trigeminal | +++ | n/a | Non-neuronal tissue | ||
| Medulla | Choroid plexus | +++ | +++ | ||
| Gigantocellular reticular n. | ++ | ++ | |||
| Nucleus raphe magnus | ++ | ++ | |||
| Facial motor nucleus | +++ | n/a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-García, E.; Chaparro-Cabanillas, N.; Coll-Manzano, A.; Carreras-Caballé, M.; Giralt, A.; Del Toro, D.; Alberch, J.; Masana, M.; Rodríguez, M.J. Unraveling the Spatiotemporal Distribution of VPS13A in the Mouse Brain. Int. J. Mol. Sci. 2021, 22, 13018. https://doi.org/10.3390/ijms222313018
García-García E, Chaparro-Cabanillas N, Coll-Manzano A, Carreras-Caballé M, Giralt A, Del Toro D, Alberch J, Masana M, Rodríguez MJ. Unraveling the Spatiotemporal Distribution of VPS13A in the Mouse Brain. International Journal of Molecular Sciences. 2021; 22(23):13018. https://doi.org/10.3390/ijms222313018
Chicago/Turabian StyleGarcía-García, Esther, Nerea Chaparro-Cabanillas, Albert Coll-Manzano, Maria Carreras-Caballé, Albert Giralt, Daniel Del Toro, Jordi Alberch, Mercè Masana, and Manuel J. Rodríguez. 2021. "Unraveling the Spatiotemporal Distribution of VPS13A in the Mouse Brain" International Journal of Molecular Sciences 22, no. 23: 13018. https://doi.org/10.3390/ijms222313018
APA StyleGarcía-García, E., Chaparro-Cabanillas, N., Coll-Manzano, A., Carreras-Caballé, M., Giralt, A., Del Toro, D., Alberch, J., Masana, M., & Rodríguez, M. J. (2021). Unraveling the Spatiotemporal Distribution of VPS13A in the Mouse Brain. International Journal of Molecular Sciences, 22(23), 13018. https://doi.org/10.3390/ijms222313018






