Identification of SCN5a p.C335R Variant in a Large Family with Dilated Cardiomyopathy and Conduction Disease
Abstract
1. Introduction
2. Material and Methods
2.1. Clinical Evaluation
2.2. DNA Sequencing
2.3. Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes (iPSC-CMs)
2.4. Site-Directed Mutagenesis and Heterologous Expression in Xenopus Oocytes
2.5. Electrophysiology Techniques
2.6. Transient Transfection and Western Blotting
2.7. Statistics
3. Results
3.1. Clinical Investigation of a DCM Pedigree
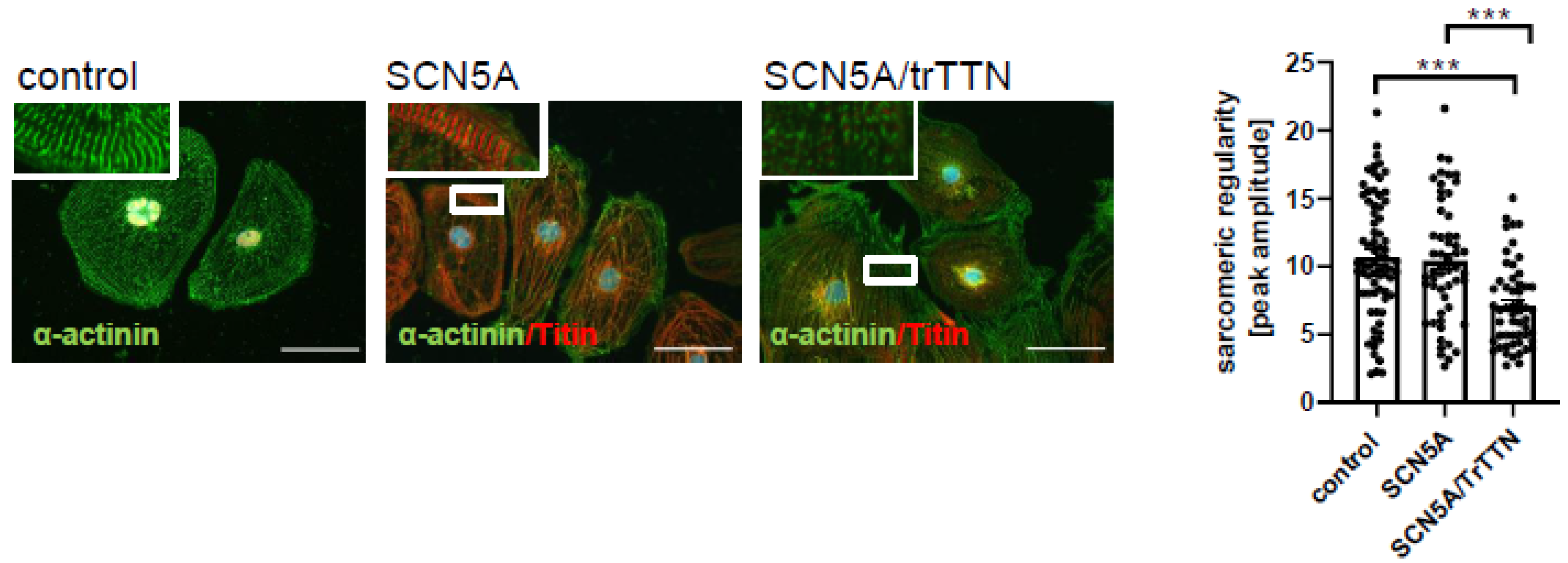
3.2. The SCN5a p.C335R Variant Is Causing a Loss-of-Function of Sodium Channel Current
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Kayvanpour, E.; Sedaghat-Hamedani, F.; Amr, A.; Lai, A.; Haas, J.; Holzer, D.B.; Frese, K.S.; Keller, A.; Jensen, K.; Katus, H.A.; et al. Genotype-phenotype associations in dilated cardiomyopathy: Metaanalysis on more than 8000 individuals. Clin. Res. Cardiol. 2017, 106, 127–139. [Google Scholar] [CrossRef]
- Kayvanpour, E.; Katus, H.A.; Meder, B. Determined to Fail—The Role of Genetic Mechanisms in Heart Failure. Curr. Heart Fail Rep. 2015, 12, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Haas, J.; Frese, K.S.; Peil, B.; Kloos, W.; Keller, A.; Nietsch, R.; Feng, Z.; Müller, S.; Kayvanpour, E.; Vogel, B.; et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur. Heart J. 2015, 36, 1123–1135. [Google Scholar] [CrossRef]
- Hershberger, R.E.; Hedges, D.J.; Morales, A. Dilated cardiomyopathy: The complexity of a diverse genetic architecture. Nat. Rev. Cardiol. 2013, 10, 531–547. [Google Scholar] [CrossRef] [PubMed]
- Golbus, J.; Puckelwartz, M.J.; Dellefave-Castillo, L.; Fahrenbach, J.; Nelakuditi, V.; Pesce, L.L.; Pytel, P.; McNally, E.M. Targeted analysis of whole genome sequence data to diagnose genetic cardiomyopathy. Circ. Cardiovasc. Genet. 2014, 7, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Gosselin-Badaroudine, P.; Keller, D.I.; Huang, H.; Pouliot, V.; Chatelier, A.; Osswald, S.; Brink, M.; Chahine, M. A proton leak current through the cardiac sodium channel is linked to mixed arrhythmia and the dilated cardiomyopathy phenotype. PLoS ONE 2012, 7, e38331. [Google Scholar]
- Liu, M.; Yang, K.C.; Dudley, S.C., Jr. Cardiac sodium channel mutations: Why so many phenotypes? Nat. Rev. Cardiol. 2014, 11, 607–615. [Google Scholar] [CrossRef]
- Moreau, A.; Gosselin-Badaroudine, P.; Mercier, A.; Burger, B.; Keller, D.I.; Chahine, M. A leaky voltage sensor domain of cardiac sodium channels causes arrhythmias associated with dilated cardiomyopathy. Sci. Rep. 2018, 8, 13804. [Google Scholar] [CrossRef]
- Borchert, T.; Hübscher, D.; Guessoum, C.I.; Lam, T.D.; Ghadri, J.R.; Schellinger, I.N.; Tiburcy, M.; Liaw, N.Y.; Li, Y.; Haas, J.; et al. Catecholamine-Dependent beta-Adrenergic Signaling in a Pluripotent Stem Cell Model of Takotsubo Cardiomyopathy. J. Am. Coll. Cardiol. 2017, 70, 975–991. [Google Scholar] [CrossRef]
- Huang, H.; Ding, D.-B.; Fan, L.-L.; Jin, J.-Y.; Li, J.-J.; Guo, S.; Chen, Y.-Q.; Xiang, R. Whole-exome sequencing identifies a Novel SCN5A mutation (C335R) in a Chinese family with arrhythmia. Cardiol. Young 2018, 28, 688–691. [Google Scholar] [CrossRef]
- Van Malderen, S.C.; Daneels, D.; Kerkhove, D.; Peeters, U.; Theuns, D.A.; Droogmans, S.; Van Camp, G.; Weytjens, C.; Biervliet, M.; Bonduelle, M.-L.; et al. Prolonged Right Ventricular Ejection Delay in Brugada Syndrome Depends on the Type of SCN5A Variant- Electromechanical Coupling Through Tissue Velocity Imaging as a Bridge Between Genotyping and Phenotyping. Circ. J. 2017, 82, 53–61. [Google Scholar] [CrossRef]
- Moretti, M.; Merlo, M.; Barbati, G.; Di Lenarda, A.; Brun, F.; Pinamonti, B.; Gregori, D.; Mestroni, L.; Sinagra, G. Prognostic impact of familial screening in dilated cardiomyopathy. Eur. J. Heart Fail. 2010, 12, 922–927. [Google Scholar] [CrossRef]
- Leung, G.K.C.; Luk, H.M.; Tang, V.H.M.; Gao, W.W.; Mak, C.C.Y.; Yu, M.H.C.; Wong, H.S.W.; Chu, Y.W.Y.; Yang, W.L.; Ma, A.C.H.; et al. Integrating Functional Analysis in the Next-Generation Sequencing Diagnostic Pipeline of RASopathies. Sci. Rep. 2018, 8, 2421. [Google Scholar] [CrossRef]
- Sedaghat-Hamedani, F.; Katus, H.A.; Meder, B. Precision medicine for cardiovascular disease: Learning lessons from cardiomyopathies. Herz 2018, 43, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Jordan, E.; Peterson, L.; Ai, T.; Asatryan, B.; Bronicki, L.; Brown, E.; Celeghin, R.; Edwards, M.; Fan, J.; Ingles, J.; et al. Evidence-Based Assessment of Genes in Dilated Cardiomyopathy. Circultion 2021, 144, 7–19. [Google Scholar] [CrossRef]
- Detta, N.; Frisso, G.; Salvatore, F. The multi-faceted aspects of the complex cardiac Nav1.5 protein in membrane function and pathophysiology. Biochim. Biophys. Acta 2015, 1854, 1502–1509. [Google Scholar] [CrossRef]
- Swan, H.; Amarouch, M.Y.; Leinonen, J.; Marjamaa, A.; Kucera, J.P.; Laitinen-Forsblom, P.J.; Lahtinen, A.M.; Palotie, A.; Kontula, K.; Toivonen, L.; et al. Gain-of-function mutation of the SCN5A gene causes exercise-induced polymorphic ventricular arrhythmias. Circ. Cardiovasc. Genet. 2014, 7, 771–781. [Google Scholar] [CrossRef]
- Cheng, J.; Morales, A.; Siegfried, J.D.; Li, D.; Norton, N.; Song, J.; Gonzalez-Quintana, J.; Makielski, J.C.; Hershberger, R.E. SCN5A rare variants in familial dilated cardiomyopathy decrease peak sodium current depending on the common polymorphism H558R and common splice variant Q1077del. Clin. Transl. Sci. 2010, 3, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Olson, T.M.; Michels, V.V.; Ballew, J.D.; Reyna, S.P.; Karst, M.L.; Herron, K.J.; Horton, S.C.; Rodeheffer, R.J.; Anderson, J.L. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA 2005, 293, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Glazer, A.M.; Wada, Y.; Li, B.; Muhammad, A.; Kalash, O.R.; O’Neill, M.J.; Shields, T.; Hall, L.; Short, L.; Blair, M.A.; et al. High-Throughput Reclassification of SCN5A Variants. Am. J. Hum. Genet. 2020, 107, 111–123. [Google Scholar] [CrossRef]
- Bezzina, C.R.; Remme, C.A. Dilated cardiomyopathy due to sodium channel dysfunction: What is the connection? Circ. Arrhythm. Electrophysiol. 2008, 1, 80–82. [Google Scholar] [CrossRef][Green Version]
- Brignole, M.; Auricchio, A.; Baron-Esquivias, G.; Bordachar, P.; Boriani, G.; Breithardt, O.; Cleland, J.; Deharo, J.; Delgado, V.; Elliott, P.M.; et al. 2013 ESC Guidelines on cardiac pacing and cardiac resynchronization therapy: The Task Force on cardiac pacing and resynchronization therapy of the European Society of Cardiology (ESC). Developed in collaboration with the European Heart Rhythm Association (EHRA). Eur. Heart J. 2013, 34, 2281–2329. [Google Scholar] [PubMed]
- Shy, D.; Gillet, L.; Abriel, H. Cardiac sodium channel NaV1.5 distribution in myocytes via interacting proteins: The multiple pool model. Biochim. Biophys. Acta 2013, 1833, 886–894. [Google Scholar] [CrossRef] [PubMed]
- Jansweijer, J.A.; Nieuwhof, K.; Russo, F.; Hoorntje, E.T.; Jongbloed, J.D.H.; Deprez, R.H.L.; Postma, A.; Bronk, M.; Van Rijsingen, I.A.W.; De Haij, S.; et al. Truncating titin mutations are associated with a mild and treatable form of dilated cardiomyopathy. Eur. J. Heart Fail. 2017, 19, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Herman, D.S.; Lam, L.; Taylor, M.R.; Wang, L.; Teekakirikul, P.; Christodoulou, D.; Conner, L.; DePalma, S.R.; McDonough, B.; Sparks, E.; et al. Truncations of titin causing dilated cardiomyopathy. N. Engl. J. Med. 2012, 366, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Jansen, M.; Baas, A.F.; van Spaendonck-Zwarts, K.Y.; Ummels, A.S.; Wijngaard, A.V.D.; Jongbloed, J.D.; van Slegtenhorst, M.A.; Deprez, R.H.L.; Wessels, M.W.; Michels, M.; et al. Mortality Risk Associated With Truncating Founder Mutations in Titin. Circ. Genom. Precis. Med. 2019, 12, e002436. [Google Scholar] [CrossRef]
- Corden, B.; Jarman, J.; Whiffin, N.; Tayal, U.; Buchan, R.; Sehmi, J.; Harper, A.; Midwinter, W.; Lascelles, K.; Markides, V.; et al. Association of Titin-Truncating Genetic Variants With Life-threatening Cardiac Arrhythmias in Patients With Dilated Cardiomyopathy and Implanted Defibrillators. JAMA Netw. Open 2019, 2, e196520. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.M.; Lorenzini, M.; Cicerchia, M.; Ochoa, J.P.; Hey, T.M.; Molina, M.S.; Restrepo-Cordoba, M.A.; Ferro, M.D.; Stolfo, D.; Johnson, R.; et al. Clinical Phenotypes and Prognosis of Dilated Cardiomyopathy Caused by Truncating Variants in the TTN Gene. Circ. Heart Fail. 2020, 13, e006832. [Google Scholar] [CrossRef]
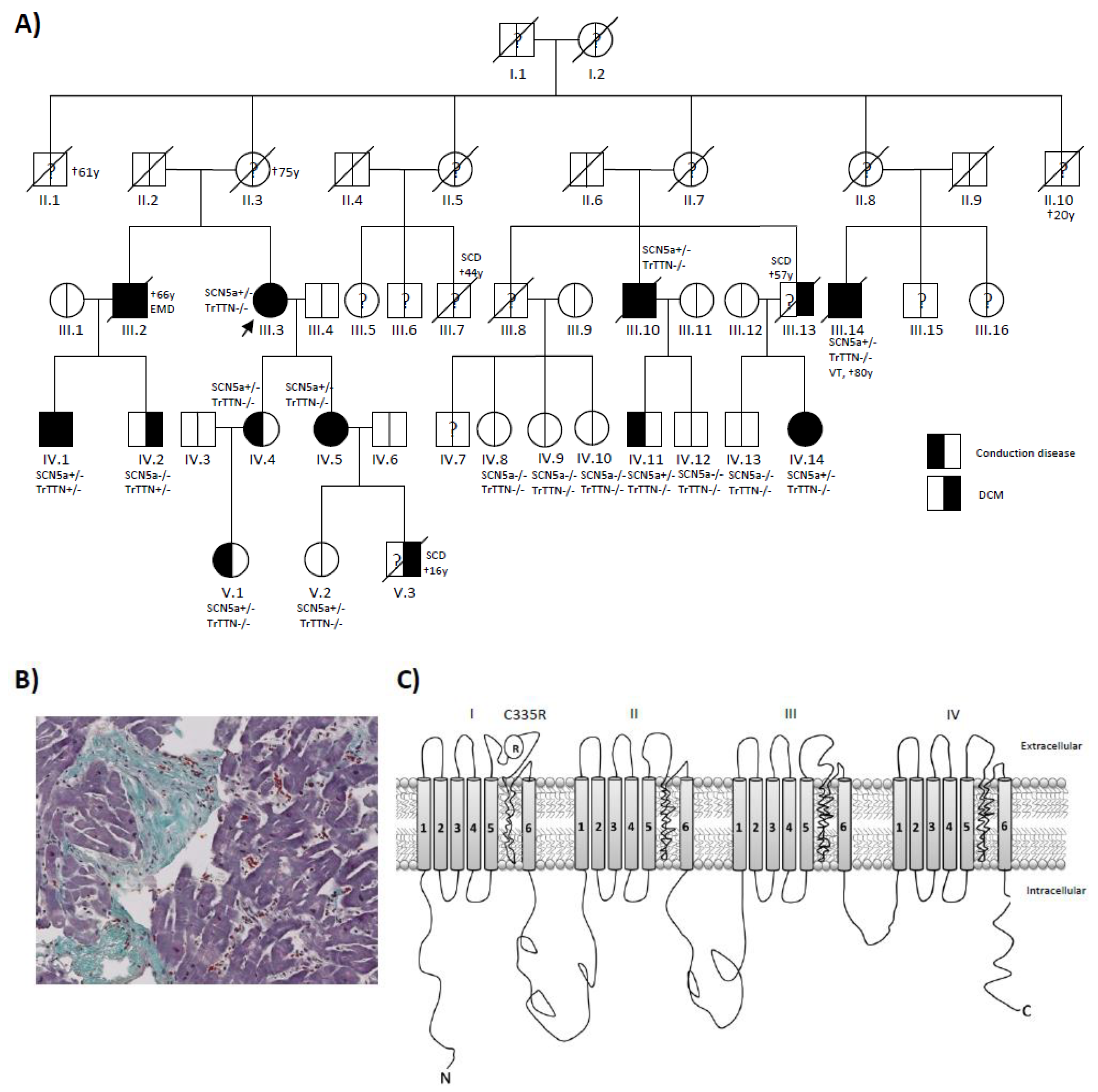


| ID | Age | Gender | DCM | Conduction Disease | SCN5a: C335R | TTNtv | Age at Onset | NYHA | EF | Rhythm | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|
| III.2 | 66 | M | Yes | RBBB | NA | NA | 60 | II | 15 | AF, PM | CRT-D, †66y:electromechanical dissociation |
| III. 3 | 78 | F | Yes | LBBB, SSS | +/− | −/− | 59 | III | 35 | AF, PM | CRT-D |
| III.10 | 79 | M | Yes | LBBB, SSS | +/− | −/− | 63 | III | 30 | AF, PM | CRT-D, †79y |
| III.14 | 80 | M | Yes | LBBB, SSS, AVB-III° | +/− | −/− | 64 | III | 20 | AF, PM | VT 70y, CRT-D, ICD therapy 72 and 78y, †80y |
| IV.1 | 35 | M | Yes | LBBB, AVB-III° | +/− | +/− | 24 | I | 48 | SR | CPR 34y AVB-III°, PM |
| IV.2 | 33 | M | Yes | No | −/− | +/− | 33 | I | 45 | SR | |
| IV.4 | 55 | F | No | AV-I° | +/− | −/− | 49 | I | 55 | SR | |
| IV.5 | 44 | F | Yes | LAH | +/− | −/− | 44 | I | 40 | SR | |
| IV.8 | 36 | F | No | No | −/− | −/− | - | I | 60 | SR | |
| IV.9 | 45 | F | No | No | −/− | −/− | - | I | 59 | SR | |
| IV.10 | 44 | F | No | No | −/− | −/− | - | I | 60 | SR | |
| IV.11 | 45 | M | No | RBBB | +/− | −/− | 45 | I | 56 | SR | |
| IV.12 | 49 | M | No | No | −/− | −/− | - | I | 60 | SR | |
| IV.13 | 64 | M | No | No | −/− | −/− | - | I | 62 | SR | |
| IV.14 | 61 | F | Yes | LBBB | +/− | −/− | 57 | I | 50 | SR | |
| V.1 | 19 | F | No | AV-I° | +/− | −/− | 19 | I | 59 | SR | x2 Syncope 22y |
| V.2 | 18 | F | No | No | −/− | −/− | - | Î | 60 | SR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sedaghat-Hamedani, F.; Rebs, S.; El-Battrawy, I.; Chasan, S.; Krause, T.; Haas, J.; Zhong, R.; Liao, Z.; Xu, Q.; Zhou, X.; et al. Identification of SCN5a p.C335R Variant in a Large Family with Dilated Cardiomyopathy and Conduction Disease. Int. J. Mol. Sci. 2021, 22, 12990. https://doi.org/10.3390/ijms222312990
Sedaghat-Hamedani F, Rebs S, El-Battrawy I, Chasan S, Krause T, Haas J, Zhong R, Liao Z, Xu Q, Zhou X, et al. Identification of SCN5a p.C335R Variant in a Large Family with Dilated Cardiomyopathy and Conduction Disease. International Journal of Molecular Sciences. 2021; 22(23):12990. https://doi.org/10.3390/ijms222312990
Chicago/Turabian StyleSedaghat-Hamedani, Farbod, Sabine Rebs, Ibrahim El-Battrawy, Safak Chasan, Tobias Krause, Jan Haas, Rujia Zhong, Zhenxing Liao, Qiang Xu, Xiaobo Zhou, and et al. 2021. "Identification of SCN5a p.C335R Variant in a Large Family with Dilated Cardiomyopathy and Conduction Disease" International Journal of Molecular Sciences 22, no. 23: 12990. https://doi.org/10.3390/ijms222312990
APA StyleSedaghat-Hamedani, F., Rebs, S., El-Battrawy, I., Chasan, S., Krause, T., Haas, J., Zhong, R., Liao, Z., Xu, Q., Zhou, X., Akin, I., Zitron, E., Frey, N., Streckfuss-Bömeke, K., & Kayvanpour, E. (2021). Identification of SCN5a p.C335R Variant in a Large Family with Dilated Cardiomyopathy and Conduction Disease. International Journal of Molecular Sciences, 22(23), 12990. https://doi.org/10.3390/ijms222312990









