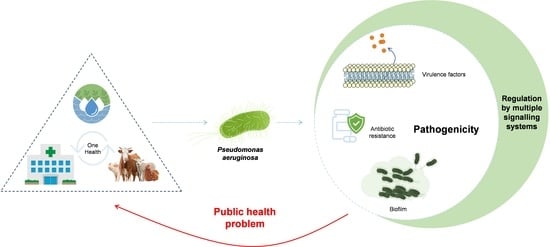
Genomic and Metabolic Characteristics of the Pathogenicity in Pseudomonas aeruginosa
Abstract
:1. Introduction
2. Pseudomonas aeruginosa
2.1. Bacterial Pathogenesis
2.2. Clinical Impact of Pseudomonas aeruginosa
3. Genome Structure in Pseudomonas aeruginosa
4. Metabolic Pathways Associated with Pathogenicity
4.1. Virulence Factors
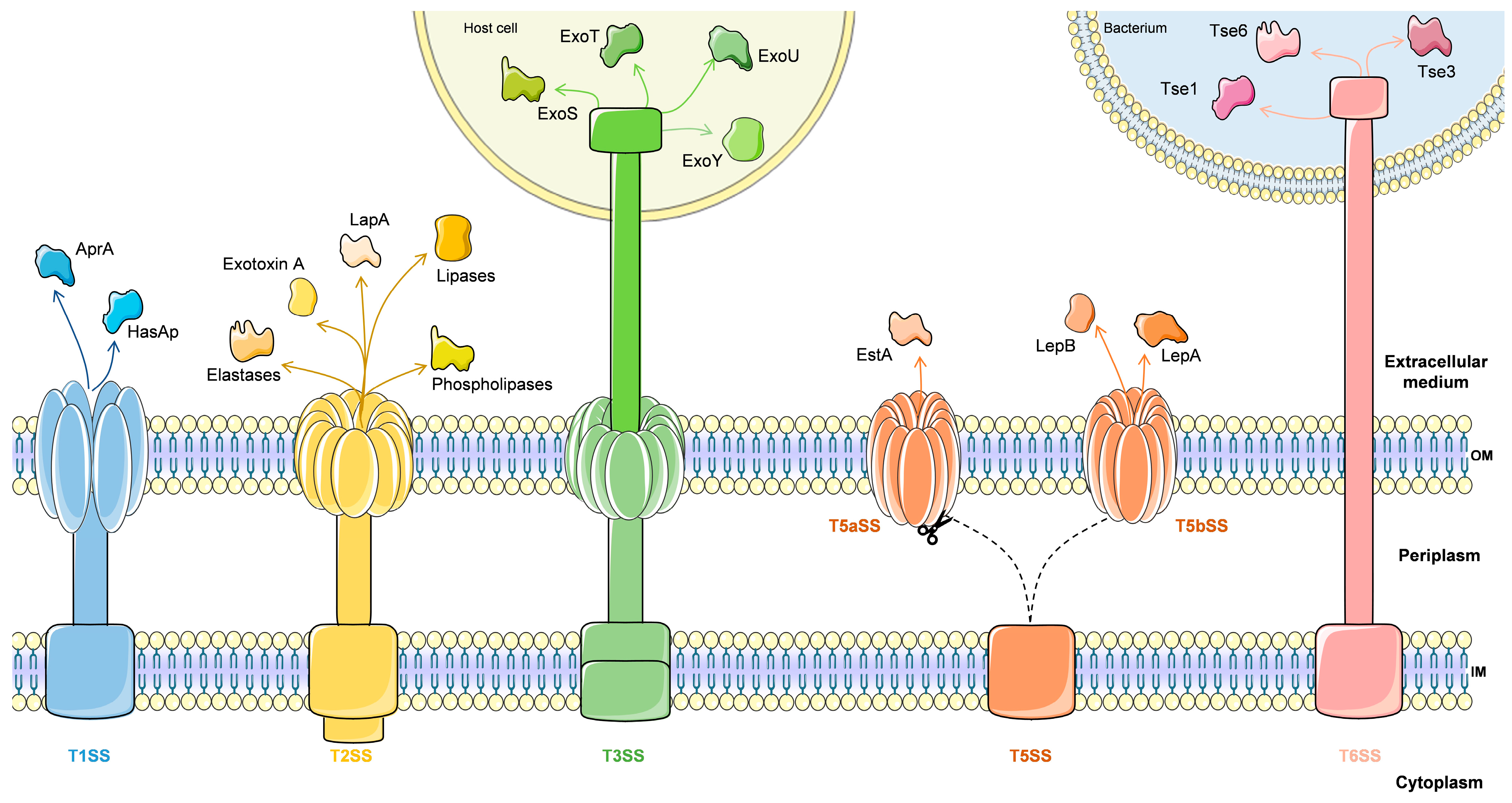
4.1.1. Secretion Systems
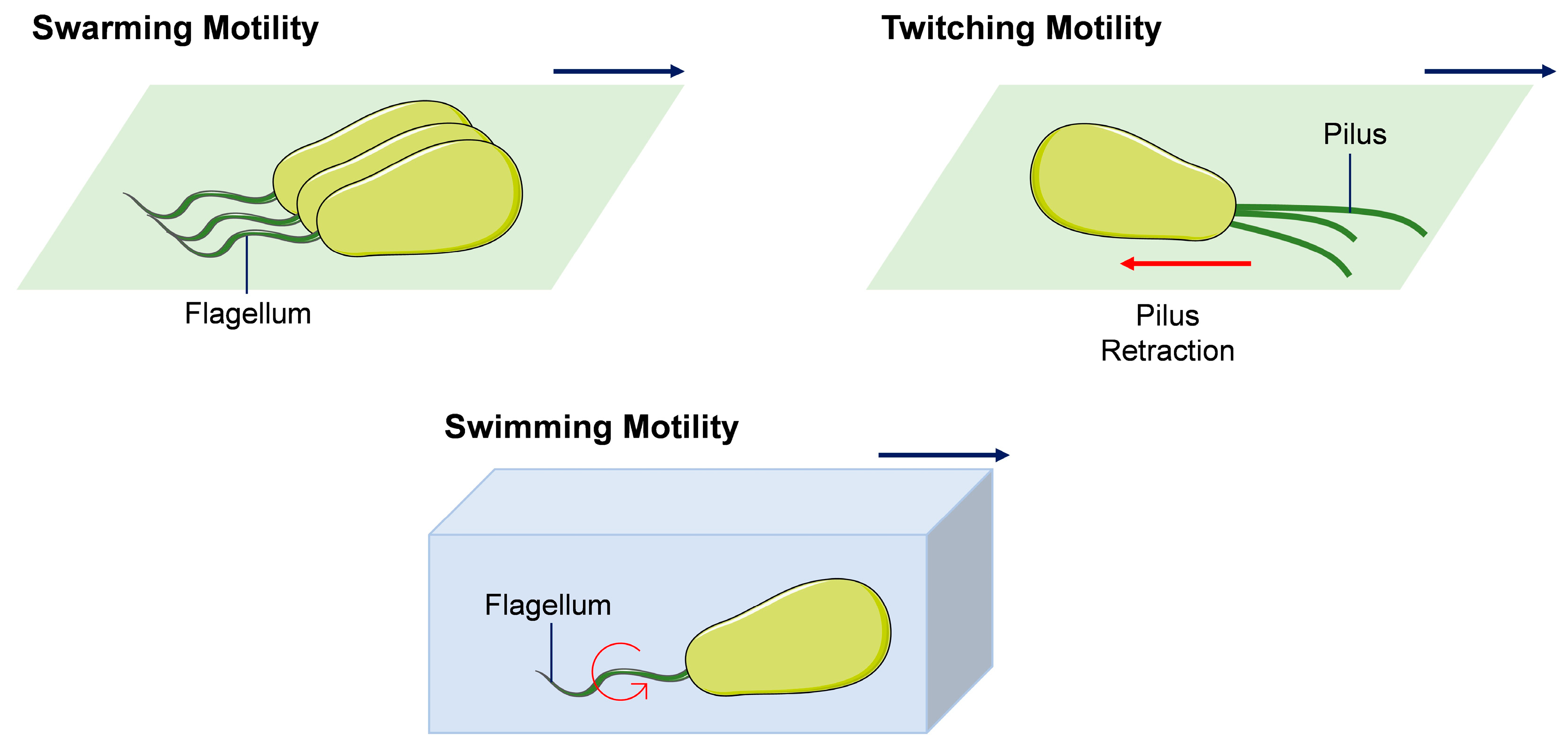
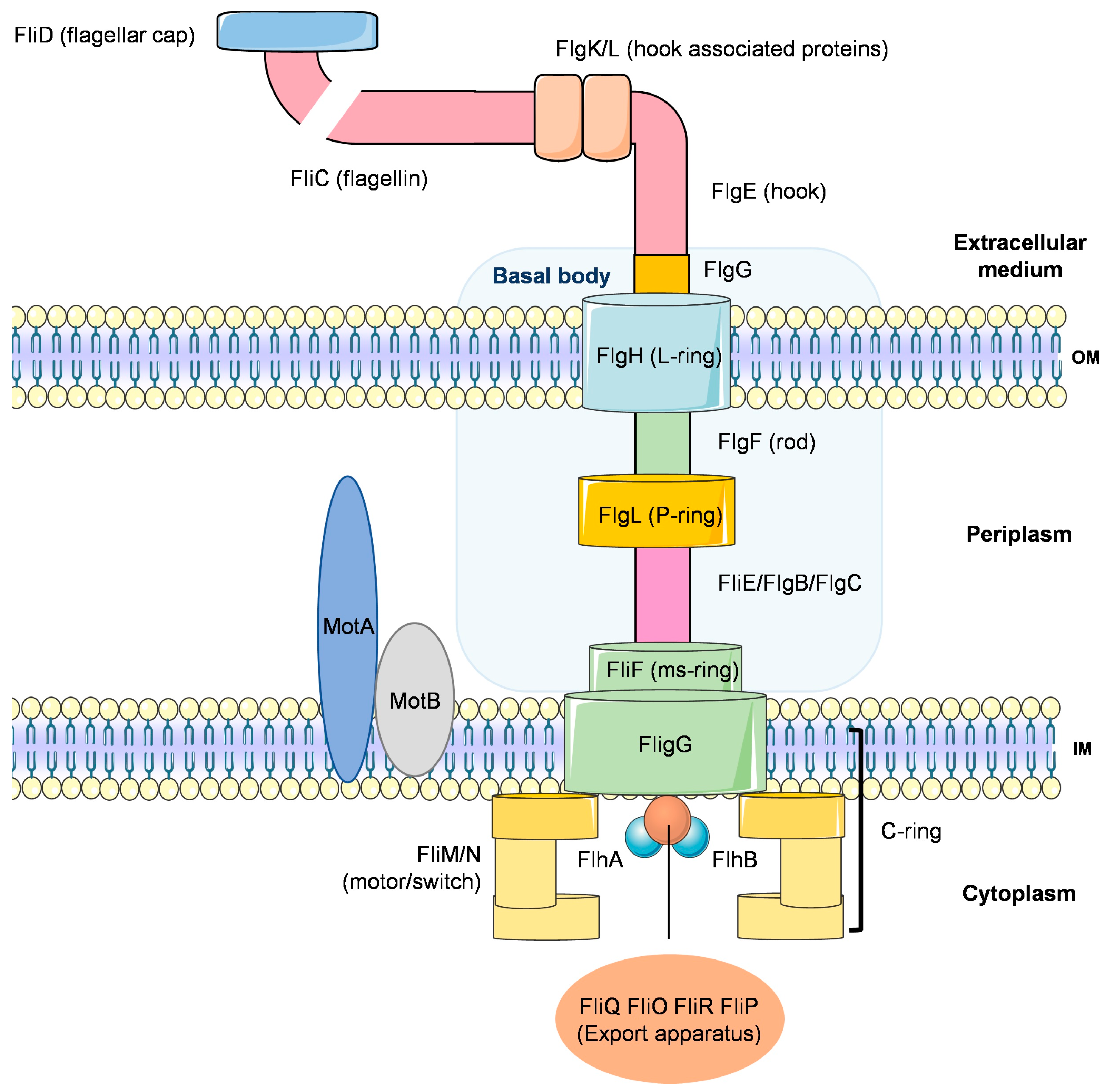
4.1.2. Motility
4.1.3. Secondary Metabolites
4.1.4. Multiple Signalling Systems in the Regulation of Virulence Genes
Quorum Sensing System
GacS/GacA, Two-Component Systems
Nucleotide Signals
4.2. Biofilm
4.3. Antibiotic Resistance
4.3.1. Intrinsic Antibiotic Resistance
4.3.2. Acquired Resistance
4.3.3. Adaptive Resistance
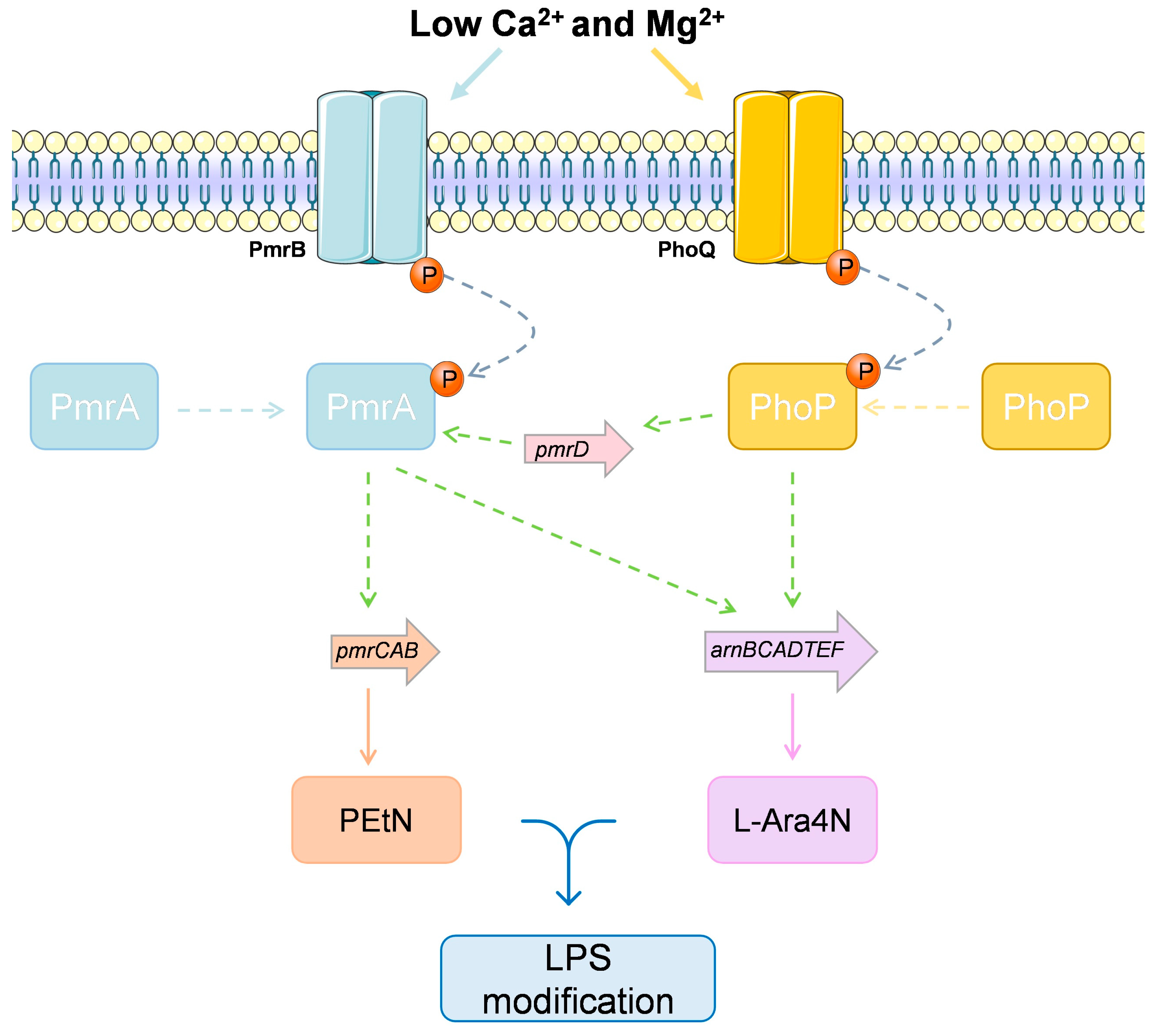
4.3.4. The Case of Multiple Colistin Response Mechanisms
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization (WHO). Available online: http://www.who.int/ (accessed on 8 May 2021).
- De M Campos, J.C.; Antunes, L.C.; Ferreira, R.B. Global priority pathogens: Virulence, antimicrobial resistance and prospective treatment options. Future Microbiol. 2020, 15, 649–677. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Jin, Y.; Bai, F.; Jin, S. Pseudomonas aeruginosa. In Molecular Medical Microbiology; Elsevier: Amsterdam, The Netherlands, 2015; pp. 753–767. [Google Scholar]
- Hauser, A.R. The type III secretion system of Pseudomonas aeruginosa: Infection by injection. Nat. Rev. Microbiol. 2009, 7, 654–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frieri, M.; Kumar, K.; Boutin, A. Antibiotic resistance. J. Infect. Public Health 2017, 10, 369–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aloush, V.; Navon-Venezia, S.; Seigman-Igra, Y.; Cabili, S.; Carmeli, Y. Multidrug-resistant Pseudomonas aeruginosa: Risk factors and clinical impact. Antimicrob Agents Chemother. 2006, 50, 43–48. [Google Scholar] [CrossRef] [Green Version]
- Spiers, A.J.; Buckling, A.; Rainey, P.B. The causes of Pseudomonas diversity. Microbiology 2000, 146 Pt 10, 2345–2350. [Google Scholar] [CrossRef] [Green Version]
- Migula, W. System der Bakterien: Handbuch der Morphologie, Entwicklungsgeschichte und Systematik der Bakterien; Fischer: Jena, Germany, 1900; Volume 2. [Google Scholar]
- Palleroni, N.J. The Pseudomonas story. Environ. Microbiol. 2010, 12, 1377–1383. [Google Scholar] [CrossRef] [PubMed]
- Ramos, J.-L. Pseudomonas: Volume 1 Genomics, Life Style and Molecular Architecture; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Pirnay, J.P.; Matthijs, S.; Colak, H.; Chablain, P.; Bilocq, F.; Van Eldere, J.; De Vos, D.; Zizi, M.; Triest, L.; Cornelis, P. Global Pseudomonas aeruginosa biodiversity as reflected in a Belgian river. Environ. Microbiol. 2005, 7, 969–980. [Google Scholar] [CrossRef]
- Mena, K.D.; Gerba, C.P. Risk assessment of Pseudomonas aeruginosa in water. Rev. Environ. Contam. Toxicol. 2009, 201, 71–115. [Google Scholar]
- Remold, S.K.; Brown, C.K.; Farris, J.E.; Hundley, T.C.; Perpich, J.A.; Purdy, M.E. Differential habitat use and niche partitioning by Pseudomonas species in human homes. Microb. Ecol. 2011, 62, 505–517. [Google Scholar] [CrossRef]
- Crone, S.; Vives-Florez, M.; Kvich, L.; Saunders, A.M.; Malone, M.; Nicolaisen, M.H.; Martinez-Garcia, E.; Rojas-Acosta, C.; Catalina Gomez-Puerto, M.; Calum, H.; et al. The environmental occurrence of Pseudomonas aeruginosa. APMIS 2020, 128, 220–231. [Google Scholar] [CrossRef]
- Khan, H.A.; Baig, F.K.; Mehboob, R. Nosocomial infections: Epidemiology, prevention, control and surveillance. Asian Pac. J. Trop. Biomed. 2017, 7, 478–482. [Google Scholar] [CrossRef]
- Abd, H.; Wretlind, B.; Saeed, A.; Idsund, E.; Hultenby, K.; Sandstrom, G. Pseudomonas aeruginosa utilises its type III secretion system to kill the free-living amoeba Acanthamoeba castellanii. J. Eukaryot. Microbiol. 2008, 55, 235–243. [Google Scholar] [CrossRef]
- Kung, V.L.; Ozer, E.A.; Hauser, A.R. The accessory genome of Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2010, 74, 621–641. [Google Scholar] [CrossRef] [Green Version]
- Silveira, M.C.; Albano, R.M.; Asensi, M.D.; Carvalho-Assef, A.P.D.A. Genetics; Evolution. Description of genomic islands associated to the multidrug-resistant Pseudomonas aeruginosa clone ST277. Infect. Genet. Evol. 2016, 42, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Battle, S.E.; Rello, J.; Hauser, A.R. Genomic islands of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2009, 290, 70–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrison, E.M.; Carter, M.E.; Luck, S.; Ou, H.Y.; He, X.; Deng, Z.; O’Callaghan, C.; Kadioglu, A.; Rajakumar, K. Pathogenicity islands PAPI-1 and PAPI-2 contribute individually and synergistically to the virulence of Pseudomonas aeruginosa strain PA14. Infect. Immun. 2010, 78, 1437–1446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granato, E.T.; Ziegenhain, C.; Marvig, R.L.; Kümmerli, R. Low spatial structure and selection against secreted virulence factors attenuates pathogenicity in Pseudomonas aeruginosa. ISME J. 2018, 12, 2907–2918. [Google Scholar] [CrossRef] [Green Version]
- Leone, I.; Chirillo, M.G.; Raso, T.; Zucca, M.; Savoia, D. Phenotypic and genotypic characterization of Pseudomonas aeruginosa from cystic fibrosis patients. Eur. J. Clin. Microbiol. Infect. Dis. 2008, 27, 1093–1099. [Google Scholar] [CrossRef]
- Palmer, G.C.; Whiteley, M. Metabolism and Pathogenicity of Pseudomonas aeruginosa Infections in the Lungs of Individuals with Cystic Fibrosis. Microbiol. Spectr. 2015, 3, 4. [Google Scholar] [CrossRef]
- Parkins, M.D.; Somayaji, R.; Waters, V.J. Epidemiology, Biology, and Impact of Clonal Pseudomonas aeruginosa Infections in Cystic Fibrosis. Clin. Microbiol. Rev. 2018, 31, e00019-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mesaros, N.; Nordmann, P.; Plesiat, P.; Roussel-Delvallez, M.; Van Eldere, J.; Glupczynski, Y.; Van Laethem, Y.; Jacobs, F.; Lebecque, P.; Malfroot, A.; et al. Pseudomonas aeruginosa: Resistance and therapeutic options at the turn of the new millennium. Clin. Microbiol. Infect. 2007, 13, 560–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrara, A.M. Potentially multidrug-resistant non-fermentative Gram-negative pathogens causing nosocomial pneumonia. Int. J. Antimicrob. Agents 2006, 27, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Haenni, M.; Hocquet, D.; Ponsin, C.; Cholley, P.; Guyeux, C.; Madec, J.Y.; Bertrand, X. Population structure and antimicrobial susceptibility of Pseudomonas aeruginosa from animal infections in France. BMC Vet. Res. 2015, 11, 9. [Google Scholar] [CrossRef] [Green Version]
- Cabassi, C.S.; Sala, A.; Santospirito, D.; Alborali, G.L.; Carretto, E.; Ghibaudo, G.; Taddei, S. Activity of AMP2041 against human and animal multidrug resistant Pseudomonas aeruginosa clinical isolates. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 17. [Google Scholar] [CrossRef] [Green Version]
- Hogardt, M.; Heesemann, J. Microevolution of Pseudomonas aeruginosa to a chronic pathogen of the cystic fibrosis lung. Curr. Top. Microbiol. Immunol. 2013, 358, 91–118. [Google Scholar] [CrossRef]
- Sommer, L.M.; Johansen, H.K.; Molin, S. Antibiotic resistance in Pseudomonas aeruginosa and adaptation to complex dynamic environments. Microb. Genom. 2020, 6, e000370. [Google Scholar] [CrossRef]
- Diggle, S.P.; Whiteley, M. Microbe Profile: Pseudomonas aeruginosa: Opportunistic pathogen and lab rat. Microbiology 2020, 166, 30–33. [Google Scholar] [CrossRef]
- Klockgether, J.; Cramer, N.; Wiehlmann, L.; Davenport, C.F.; Tummler, B. Pseudomonas aeruginosa Genomic Structure and Diversity. Front. Microbiol. 2011, 2, 150. [Google Scholar] [CrossRef] [Green Version]
- Stover, C.K.; Pham, X.Q.; Erwin, A.; Mizoguchi, S.; Warrener, P.; Hickey, M.; Brinkman, F.; Hufnagle, W.; Kowalik, D.; Lagrou, M.J.N. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef]
- Ozer, E.A.; Allen, J.P.; Hauser, A.R. Characterization of the core and accessory genomes of Pseudomonas aeruginosa using bioinformatic tools Spine and AGEnt. BMC Genom. 2014, 15, 737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chevalier, S.; Bouffartigues, E.; Bodilis, J.; Maillot, O.; Lesouhaitier, O.; Feuilloley, M.G.; Orange, N.; Dufour, A.; Cornelis, P. Structure, function and regulation of Pseudomonas aeruginosa porins. FEMS Microbiol. Rev. 2017, 41, 698–722. [Google Scholar] [CrossRef]
- Valot, B.; Guyeux, C.; Rolland, J.Y.; Mazouzi, K.; Bertrand, X.; Hocquet, D. What It Takes to Be a Pseudomonas aeruginosa ? The Core Genome of the Opportunistic Pathogen Updated. PLoS ONE 2015, 10, e0126468. [Google Scholar] [CrossRef]
- Freschi, L.; Bertelli, C.; Jeukens, J.; Moore, M.P.; Kukavica-Ibrulj, I.; Emond-Rheault, J.G.; Hamel, J.; Fothergill, J.L.; Tucker, N.P.; McClean, S.; et al. Genomic characterisation of an international Pseudomonas aeruginosa reference panel indicates that the two major groups draw upon distinct mobile gene pools. FEMS Microbiol. Lett. 2018, 365, fny120. [Google Scholar] [CrossRef] [Green Version]
- Freschi, L.; Vincent, A.T.; Jeukens, J.; Emond-Rheault, J.G.; Kukavica-Ibrulj, I.; Dupont, M.J.; Charette, S.J.; Boyle, B.; Levesque, R.C. The Pseudomonas aeruginosa Pan-Genome Provides New Insights on Its Population Structure, Horizontal Gene Transfer, and Pathogenicity. Genome Biol. Evol. 2019, 11, 109–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathee, K.; Narasimhan, G.; Valdes, C.; Qiu, X.; Matewish, J.M.; Koehrsen, M.; Rokas, A.; Yandava, C.N.; Engels, R.; Zeng, E.; et al. Dynamics of Pseudomonas aeruginosa genome evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 3100–3105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spencer, D.H.; Kas, A.; Smith, E.E.; Raymond, C.K.; Sims, E.H.; Hastings, M.; Burns, J.L.; Kaul, R.; Olson, M.V. Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J. Bacteriol. 2003, 185, 1316–1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfgang, M.C.; Kulasekara, B.R.; Liang, X.; Boyd, D.; Wu, K.; Yang, Q.; Miyada, C.G.; Lory, S. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2003, 100, 8484–8489. [Google Scholar] [CrossRef] [Green Version]
- Dobrindt, U.; Hochhut, B.; Hentschel, U.; Hacker, J. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2004, 2, 414–424. [Google Scholar] [CrossRef]
- Ho Sui, S.J.; Fedynak, A.; Hsiao, W.W.; Langille, M.G.; Brinkman, F.S. The association of virulence factors with genomic islands. PLoS ONE 2009, 4, e8094. [Google Scholar] [CrossRef]
- Sawa, T.; Momiyama, K.; Mihara, T.; Kainuma, A.; Kinoshita, M.; Moriyama, K. Molecular epidemiology of clinically high-risk Pseudomonas aeruginosa strains: Practical overview. Microbiol. Immunol. 2020, 64, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Gaviard, C.; Jouenne, T.; Hardouin, J. Proteomics of Pseudomonas aeruginosa: The increasing role of post-translational modifications. Expert Rev. Proteom. 2018, 15, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Darch, S.E.; McNally, A.; Harrison, F.; Corander, J.; Barr, H.L.; Paszkiewicz, K.; Holden, S.; Fogarty, A.; Crusz, S.A.; Diggle, S.P. Recombination is a key driver of genomic and phenotypic diversity in a Pseudomonas aeruginosa population during cystic fibrosis infection. Sci. Rep. 2015, 5, 7649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodman, A.L.; Merighi, M.; Hyodo, M.; Ventre, I.; Filloux, A.; Lory, S.J.G. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009, 23, 249–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desvaux, M.; Hébraud, M.; Talon, R.; Henderson, I.R. Secretion and subcellular localizations of bacterial proteins: A semantic awareness issue. Trends Microbiol. 2009, 17, 139–145. [Google Scholar] [CrossRef]
- Dettman, J.R.; Rodrigue, N.; Aaron, S.D.; Kassen, R. Evolutionary genomics of epidemic and nonepidemic strains of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2013, 110, 21065–21070. [Google Scholar] [CrossRef] [Green Version]
- Sousa, A.M.; Pereira, M.O. Pseudomonas aeruginosa Diversification during Infection Development in Cystic Fibrosis Lungs-A Review. Pathogens 2014, 3, 680–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Q.; Zhai, Y.; Schneider, J.C.; Ramseier, T.M.; Saier, M.H., Jr. Protein secretion systems of Pseudomonas aeruginosa and P fluorescens. Biochim. Biophys. Acta 2003, 1611, 223–233. [Google Scholar] [CrossRef] [Green Version]
- Bleves, S.; Viarre, V.; Salacha, R.; Michel, G.P.; Filloux, A.; Voulhoux, R. Protein secretion systems in Pseudomonas aeruginosa: A wealth of pathogenic weapons. Int. J. Med. Microbiol. 2010, 300, 534–543. [Google Scholar] [CrossRef]
- Delepelaire, P. Type I secretion in gram-negative bacteria. Biochim. Biophys. Acta 2004, 1694, 149–161. [Google Scholar] [CrossRef] [Green Version]
- Guzzo, J.; Pages, J.; Duong, F.; Lazdunski, A.; Murgier, M. Pseudomonas aeruginosa alkaline protease: Evidence for secretion genes and study of secretion mechanism. J. Bacteriol. 1991, 173, 5290–5297. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, K. Role of bacterial proteases in pseudomonal and serratial keratitis. Biol. Chem. 2004, 385, 1007–1016. [Google Scholar] [CrossRef]
- Costa, T.R.; Felisberto-Rodrigues, C.; Meir, A.; Prevost, M.S.; Redzej, A.; Trokter, M.; Waksman, G. Secretion systems in Gram-negative bacteria: Structural and mechanistic insights. Nat. Rev. Microbiol. 2015, 13, 343–359. [Google Scholar] [CrossRef] [PubMed]
- Cianciotto, N.P. Type II secretion: A protein secretion system for all seasons. Trends Microbiol. 2005, 13, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Ball, G.; Durand, É.; Lazdunski, A.; Filloux, A. A novel type II secretion system in Pseudomonas aeruginosa. Mol. Microbiol. 2002, 43, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Rybtke, M.; Berthelsen, J.; Yang, L.; Høiby, N.; Givskov, M.; Tolker-Nielsen, T. The LapG protein plays a role in Pseudomonas aeruginosa biofilm formation by controlling the presence of the CdrA adhesin on the cell surface. Microbiologyopen 2015, 4, 917–930. [Google Scholar] [CrossRef] [Green Version]
- Casilag, F.; Lorenz, A.; Krueger, J.; Klawonn, F.; Weiss, S.; Häussler, S. The LasB Elastase of Pseudomonas aeruginosa Acts in Concert with Alkaline Protease AprA to Prevent Flagellin-Mediated Immune Recognition. Infect. Immun. 2016, 84, 162–171. [Google Scholar] [CrossRef] [Green Version]
- Korotkov, K.V.; Sandkvist, M.; Hol, W.G.J. The type II secretion system: Biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 2012, 10, 336–351. [Google Scholar] [CrossRef] [Green Version]
- Ostroff, R.M.; I Vasil, A.; Vasil, M.L. Molecular comparison of a nonhemolytic and a hemolytic phospholipase C from Pseudomonas aeruginosa. J. Bacteriol. 1990, 172, 5915–5923. [Google Scholar] [CrossRef] [Green Version]
- Allured, V.S.; Collier, R.J.; Carroll, S.F.; McKay, D.B. Structure of exotoxin A of Pseudomonas aeruginosa at 3.0-Angstrom resolution. Proc. Natl. Acad. Sci. USA 1986, 83, 1320–1324. [Google Scholar] [CrossRef] [Green Version]
- Mapipa, Q.; Digban, T.O.; Nnolim, N.E.; Nwodo, U.U. Antibiogram profile and virulence signatures of Pseudomonas aeruginosa isolates recovered from selected agrestic hospital effluents. Sci. Rep. 2021, 11, 11800. [Google Scholar] [CrossRef] [PubMed]
- Sawa, T.; Shimizu, M.; Moriyama, K.; Wiener-Kronish, J.P. Association between Pseudomonas aeruginosa type III secretion, antibiotic resistance, and clinical outcome: A review. Crit. Care 2014, 18, 668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, J.; Alaoui-El-Azher, M.; Chow, M.; Chambers, T.C.; Baker, H.; Jin, S. c-Jun NH 2 -Terminal Kinase-Mediated Signaling Is Essential for Pseudomonas aeruginosa ExoS-Induced Apoptosis. Infect. Immun. 2003, 71, 3361–3370. [Google Scholar] [CrossRef] [Green Version]
- Rangel, S.M.; Logan, L.K.; Hauser, A.R. The ADP-Ribosyltransferase Domain of the Effector Protein ExoS Inhibits Phagocytosis of Pseudomonas aeruginosa during Pneumonia. mBio 2014, 5, e01080-14. [Google Scholar] [CrossRef] [Green Version]
- Rangel, S.M.; Diaz, M.H.; Knoten, C.A.; Zhang, A.; Hauser, A.R. The Role of ExoS in Dissemination of Pseudomonas aeruginosa during Pneumonia. PLoS Pathog. 2015, 11, e1004945. [Google Scholar] [CrossRef] [Green Version]
- Engel, J.; Balachandran, P. Role of Pseudomonas aeruginosa type III effectors in disease. Curr. Opin. Microbiol. 2009, 12, 61–66. [Google Scholar] [CrossRef]
- Urbanowski, M.L.; Lykken, G.L.; Yahr, T. A secreted regulatory protein couples transcription to the secretory activity of the Pseudomonas aeruginosa type III secretion system. Proc. Natl. Acad. Sci. USA 2005, 102, 9930–9935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCaw, M.L.; Lykken, G.L.; Singh, P.K.; Yahr, T.L. ExsD is a negative regulator of the Pseudomonas aeruginosa type III secretion regulon. Mol. Microbiol. 2002, 46, 1123–1133. [Google Scholar] [CrossRef] [Green Version]
- Brutinel, E.D.; Vakulskas, C.A.; Brady, K.M.; Yahr, T.L. Characterization of ExsA and of ExsA-dependent promoters required for expression of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 2008, 68, 657–671. [Google Scholar] [CrossRef]
- Ahator, S.D.; Zhang, L. Small Is Mighty—Chemical Communication Systems in Pseudomonas aeruginosa. Annu. Rev. Microbiol. 2019, 73, 559–578. [Google Scholar] [CrossRef]
- Vakulskas, C.A.; Brady, K.M.; Yahr, T.L. Mechanism of Transcriptional Activation by Pseudomonas aeruginosa ExsA. J. Bacteriol. 2009, 191, 6654–6664. [Google Scholar] [CrossRef] [Green Version]
- Dasgupta, N.; Lykken, G.L.; Wolfgang, M.C.; Yahr, T.L. A novel anti-anti-activator mechanism regulates expression of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 2004, 53, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Rietsch, A.; Vallet-Gely, I.; Dove, S.L.; Mekalanos, J.J. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2005, 102, 8006–8011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yahr, T.L.; Wolfgang, M.C. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 2006, 62, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, J.; Zhang, L.-H. Modulation of Bacterial Type III Secretion System by a Spermidine Transporter Dependent Signaling Pathway. PLoS ONE 2007, 2, e1291. [Google Scholar] [CrossRef]
- Iyer, R.; Williams, C.; Miller, C. Arginine-Agmatine Antiporter in Extreme Acid Resistance in Escherichia coli. J. Bacteriol. 2003, 185, 6556–6561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, D.H.; Lu, C.-D. Polyamines Induce Resistance to Cationic Peptide, Aminoglycoside, and Quinolone Antibiotics in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 2006, 50, 1615–1622. [Google Scholar] [CrossRef] [Green Version]
- El-Halfawy, O.; Valvano, M.A. Antimicrobial Heteroresistance: An Emerging Field in Need of Clarity. Clin. Microbiol. Rev. 2015, 28, 191–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, D.H.; Lu, C.-D. Polyamines Increase Antibiotic Susceptibility in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2006, 50, 1623–1627. [Google Scholar] [CrossRef] [Green Version]
- Filloux, A. Protein Secretion Systems in Pseudomonas aeruginosa: An Essay on Diversity, Evolution, and Function. Front. Microbiol. 2011, 2, 155. [Google Scholar] [CrossRef] [Green Version]
- Wilhelm, S.; Tommassen, J.; Jaeger, K.-E. A Novel Lipolytic Enzyme Located in the Outer Membrane of Pseudomonas aeruginosa. J. Bacteriol. 1999, 181, 6977–6986. [Google Scholar] [CrossRef] [Green Version]
- Wilhelm, S.; Gdynia, A.; Tielen, P.; Rosenau, F.; Jaeger, K.-E. The Autotransporter Esterase EstA of Pseudomonas aeruginosa Is Required for Rhamnolipid Production, Cell Motility, and Biofilm Formation. J. Bacteriol. 2007, 189, 6695–6703. [Google Scholar] [CrossRef] [Green Version]
- Hodak, H.; Jacob-Dubuisson, F. Current challenges in autotransport and two-partner protein secretion pathways. Res. Microbiol. 2007, 158, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Salacha, R.; Kovačić, F.; Brochier-Armanet, C.; Wilhelm, S.; Tommassen, J.; Filloux, A.; Voulhoux, R.; Bleves, S. The Pseudomonas aeruginosa patatin-like protein PlpD is the archetype of a novel Type V secretion system. Environ. Microbiol. 2010, 12, 1498–1512. [Google Scholar] [PubMed]
- Borlee, B.R.; Goldman, A.D.; Murakami, K.; Samudrala, R.; Wozniak, D.J.; Parsek, M.R. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 2010, 75, 827–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allsopp, L.P.; Wood, T.E.; Howard, S.A.; Maggiorelli, F.; Nolan, L.M.; Wettstadt, S.; Filloux, A. RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2017, 114, 7707–7712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pukatzki, S.; Ma, A.; Sturtevant, D.; Krastins, B.; Sarracino, D.; Nelson, W.; Heidelberg, J.; Mekalanos, J.J. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 2006, 103, 1528–1533. [Google Scholar] [CrossRef] [Green Version]
- Kanamaru, S. Structural similarity of tailed phages and pathogenic bacterial secretion systems. Proc. Natl. Acad. Sci. USA 2009, 106, 4067–4068. [Google Scholar] [CrossRef] [Green Version]
- Bingle, L.E.; Bailey, C.M.; Pallen, M. Type VI secretion: A beginner’s guide. Curr. Opin. Microbiol. 2008, 11, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Whitney, J.C.; Beck, C.M.; Goo, Y.A.; Russell, A.; Harding, B.N.; De Leon, J.A.; Cunningham, D.; Tran, B.Q.; Low, D.A.; Goodlett, D.R.; et al. Genetically distinct pathways guide effector export through the type VI secretion system. Mol. Microbiol. 2014, 92, 529–542. [Google Scholar] [CrossRef] [Green Version]
- Russell, A.; Hood, R.D.; Bui, N.K.; LeRoux, M.; Vollmer, W.; Mougous, J.D. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 2011, 475, 343–347. [Google Scholar] [CrossRef] [Green Version]
- Basler, M.; Pilhofer, M.; Henderson, G.P.; Jensen, G.J.; Mekalanos, J.J. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 2012, 483, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Lesic, B.; Starkey, M.; He, J.; Hazan, R.; Rahme, L.G. Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology 2009, 155, 2845–2855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maurice, N.M.; Bedi, B.; Sadikot, R.T. Pseudomonas aeruginosa Biofilms: Host Response and Clinical Implications in Lung Infections. Am. J. Respir. Cell Mol. Biol. 2018, 58, 428–439. [Google Scholar] [CrossRef] [PubMed]
- O’May, C.; Tufenkji, N. The Swarming Motility of Pseudomonas aeruginosa Is Blocked by Cranberry Proanthocyanidins and Other Tannin-Containing Materials. Appl. Environ. Microbiol. 2011, 77, 3061–3067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harshey, R.M. Bacterial Motility on a Surface: Many Ways to a Common Goal. Annu. Rev. Microbiol. 2003, 57, 249–273. [Google Scholar] [CrossRef]
- Partridge, J.; Harshey, R.M. Swarming: Flexible Roaming Plans. J. Bacteriol. 2012, 195, 909–918. [Google Scholar] [CrossRef] [Green Version]
- Guttenplan, S.B.; Kearns, D.B. Regulation of flagellar motility during biofilm formation. FEMS Microbiol. Rev. 2013, 37, 849–871. [Google Scholar] [CrossRef] [Green Version]
- Burrows, L.L. Pseudomonas aeruginosa Twitching Motility: Type IV Pili in Action. Annu. Rev. Microbiol. 2012, 66, 493–520. [Google Scholar] [CrossRef] [Green Version]
- Mattick, J.S. Type IV Pili and Twitching Motility. Annu. Rev. Microbiol. 2002, 56, 289–314. [Google Scholar] [CrossRef]
- Badal, D.; Jayarani, A.V.; Kollaran, M.A.; Kumar, A.; Singh, V. Pseudomonas aeruginosa biofilm formation on endotracheal tubes requires multiple two-component systems. J. Med. Microbiol. 2020, 69, 906–919. [Google Scholar] [CrossRef] [PubMed]
- Strateva, T.; Mitov, I. Contribution of an arsenal of virulence factors to pathogenesis of Pseudomonas aeruginosa infections. Ann. Microbiol. 2011, 61, 717–732. [Google Scholar] [CrossRef]
- Nadal-Jimenez, P.; Koch, G.; Thompson, J.; Xavier, K.B.; Cool, R.H.; Quax, W.J. The Multiple Signaling Systems Regulating Virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2012, 76, 46–65. [Google Scholar] [CrossRef] [Green Version]
- Managò, A.; Becker, K.A.; Carpinteiro, A.; Wilker, B.; Soddemann, M.; Seitz, A.P.; Edwards, M.J.; Grassmé, H.; Szabo, I.; Gulbins, E. Pseudomonas aeruginosa Pyocyanin Induces Neutrophil DeathviaMitochondrial Reactive Oxygen Species and Mitochondrial Acid Sphingomyelinase. Antioxid. Redox Signal. 2015, 22, 1097–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassett, D.J.; Charniga, L.; Bean, K.; E Ohman, D.; Cohen, M.S. Response of Pseudomonas aeruginosa to pyocyanin: Mechanisms of resistance, antioxidant defenses, and demonstration of a manganese-cofactored superoxide dismutase. Infect. Immun. 1992, 60, 328–336. [Google Scholar] [CrossRef] [Green Version]
- Rada, B.; Leto, T.L. Redox warfare between airway epithelial cells and Pseudomonas: Dual oxidase versus pyocyanin. Immunol. Res. 2009, 43, 198–209. [Google Scholar] [CrossRef] [Green Version]
- Mavrodi, D.V.; Bonsall, R.F.; Delaney, S.M.; Soule, M.J.; Phillips, G.; Thomashow, L.S. Functional Analysis of Genes for Biosynthesis of Pyocyanin and Phenazine-1-Carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 2001, 183, 6454–6465. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Huang, Y.; Lou, Y.; Qian, H.; Xu, D.; Ma, L.; Jiang, C.; Zhang, D. Pyocyanin-modifying genes phzM and phzS regulated the extracellular electron transfer in microbiologically-influenced corrosion of X80 carbon steel by Pseudomonas aeruginosa. Corros. Sci. 2019, 164, 108355. [Google Scholar] [CrossRef]
- Lenney, W.; Gilchrist, F.J. Pseudomonas aeruginosa and cyanide production. Eur. Respir. J. 2011, 37, 482–483. [Google Scholar] [CrossRef] [Green Version]
- Pessi, G.; Haas, D. Transcriptional Control of the Hydrogen Cyanide Biosynthetic Genes hcnABC by the Anaerobic Regulator ANR and the Quorum-Sensing Regulators LasR and RhlR in Pseudomonas aeruginosa. J. Bacteriol. 2000, 182, 6940–6949. [Google Scholar] [CrossRef] [Green Version]
- Cooper, C.E.; Brown, G.C. The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: Chemical mechanism and physiological significance. J. Bioenerg. Biomembr. 2008, 40, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Rijavec, T.; Lapanje, A. Hydrogen Cyanide in the Rhizosphere: Not Suppressing Plant Pathogens, but Rather Regulating Availability of Phosphate. Front. Microbiol. 2016, 7, 1785. [Google Scholar] [CrossRef] [Green Version]
- Venturi, V. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol. Rev. 2006, 30, 274–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Li, T.; Cheng, X.-J.; Peng, C.-T.; Li, C.-C.; He, L.-H.; Ju, S.-M.; Wang, N.-Y.; Ye, T.-H.; Lian, M.; et al. Structural and functional studies on Pseudomonas aeruginosa DspI: Implications for its role in DSF biosynthesis. Sci. Rep. 2018, 8, 3928. [Google Scholar] [CrossRef] [Green Version]
- Gallagher, L.A.; McKnight, S.L.; Kuznetsova, M.S.; Pesci, E.C.; Manoil, C.; Couture-Tosi, E.; Delacroix, H.; Mignot, T.; Mesnage, S.; Chami, M.; et al. Functions Required for Extracellular Quinolone Signaling by Pseudomonas aeruginosa. J. Bacteriol. 2002, 184, 6448–6456. [Google Scholar] [CrossRef] [Green Version]
- Kviatkovski, I.; Chernin, L.; Yarnitzky, T.; Frumin, I.; Sobel, N.; Helman, Y. Pseudomonas aeruginosa activates the quorum sensing LuxR response regulator through secretion of 2-aminoacetophenone. Chem. Commun. 2015, 51, 3258–3261. [Google Scholar] [CrossRef]
- Erickson, D.L.; Endersby, R.; Kirkham, A.; Stuber, K.; Vollman, D.D.; Rabin, H.R.; Mitchell, I.; Storey, D.G. Pseudomonas aeruginosa Quorum-Sensing Systems May Control Virulence Factor Expression in the Lungs of Patients with Cystic Fibrosis. Infect. Immun. 2002, 70, 1783–1790. [Google Scholar] [CrossRef] [Green Version]
- Bjarnsholt, T.; Jensen, P.; Jakobsen, T.H.; Phipps, R.; Nielsen, A.K.; Rybtke, M.T.; Tolker-Nielsen, T.; Givskov, M.; Høiby, N.; Ciofu, O. Quorum Sensing and Virulence of Pseudomonas aeruginosa during Lung Infection of Cystic Fibrosis Patients. PLoS ONE 2010, 5, e10115. [Google Scholar] [CrossRef] [Green Version]
- Pearson, J.P.; Pesci, E.; Iglewski, B.H. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 1997, 179, 5756–5767. [Google Scholar] [CrossRef] [Green Version]
- Bredenbruch, F.; Nimtz, M.; Wray, V.; Morr, M.; Müller, R.; Häussler, S. Biosynthetic Pathway of Pseudomonas aeruginosa 4-Hydroxy-2-Alkylquinolines. J. Bacteriol. 2005, 187, 3630–3635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Déziel, E.; Lépine, F.; Milot, S.; He, J.; Mindrinos, M.N.; Tompkins, R.G.; Rahme, L.G. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. USA 2004, 101, 1339–1344. [Google Scholar] [CrossRef] [Green Version]
- De Kievit, T.R.; Gillis, R.; Marx, S.; Brown, C.; Iglewski, B.H. Quorum-Sensing Genes in Pseudomonas aeruginosa Biofilms: Their Role and Expression Patterns. Appl. Environ. Microbiol. 2001, 67, 1865–1873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2014, 6, 26–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulcahy, H.; O’Callaghan, J.; O’Grady, E.P.; Maciá, M.D.; Borrell, N.; Gómez, C.; Casey, P.G.; Hill, C.; Adams, C.; Gahan, C.G.M.; et al. Pseudomonas aeruginosa RsmA Plays an Important Role during Murine Infection by Influencing Colonization, Virulence, Persistence, and Pulmonary Inflammation. Infect. Immun. 2008, 76, 632–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodman, A.L.; Kulasekara, B.; Rietsch, A.; Boyd, D.; Smith, R.S.; Lory, S. A Signaling Network Reciprocally Regulates Genes Associated with Acute Infection and Chronic Persistence in Pseudomonas aeruginosa. Dev. Cell 2004, 7, 745–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broder, U.N.; Jaeger, T.; Jenal, U. LadS is a calcium-responsive kinase that induces acute-to-chronic virulence switch in Pseudomonas aeruginosa. Nat. Microbiol. 2016, 2, 16184. [Google Scholar] [CrossRef]
- Francis, V.I.; Waters, E.; Finton-James, S.E.; Gori, A.; Kadioglu, A.; Brown, A.R.; Porter, S.L. Multiple communication mechanisms between sensor kinases are crucial for virulence in Pseudomonas aeruginosa. Nat. Commun. 2018, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Bhagirath, A.Y.; Pydi, S.P.; Li, Y.; Lin, C.; Kong, W.; Chelikani, P.; Duan, K. Characterization of the Direct Interaction between Hybrid Sensor Kinases PA1611 and RetS That Controls Biofilm Formation and the Type III Secretion System in Pseudomonas aeruginosa. ACS Infect. Dis. 2016, 3, 162–175. [Google Scholar] [CrossRef]
- Inclan, Y.F.; Huseby, M.J.; Engel, J.N. FimL Regulates cAMP Synthesis in Pseudomonas aeruginosa. PLoS ONE 2011, 6, e15867. [Google Scholar] [CrossRef] [Green Version]
- Rietsch, A.; Mekalanos, J.J. Metabolic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Mol. Microbiol. 2005, 59, 807–820. [Google Scholar] [CrossRef]
- Smith, R.S.; Wolfgang, M.C.; Lory, S. An Adenylate Cyclase-Controlled Signaling Network Regulates Pseudomonas aeruginosa Virulence in a Mouse Model of Acute Pneumonia. Infect. Immun. 2004, 72, 1677–1684. [Google Scholar] [CrossRef] [Green Version]
- Almblad, H.; Rybtke, M.; Hendiani, S.; Andersen, J.B.; Givskov, M.; Tolker-Nielsen, T. High levels of cAMP inhibit Pseudomonas aeruginosa biofilm formation through reduction of the c-di-GMP content. Microbiology 2019, 165, 324–333. [Google Scholar] [CrossRef]
- Beatson, S.A.; Whitchurch, C.; Sargent, J.L.; Levesque, R.C.; Mattick, J.S. Differential Regulation of Twitching Motility and Elastase Production by Vfr in Pseudomonas aeruginosa. J. Bacteriol. 2002, 184, 3605–3613. [Google Scholar] [CrossRef] [Green Version]
- Wolfgang, M.C.; Lee, V.T.; Gilmore, M.E.; Lory, S. Coordinate Regulation of Bacterial Virulence Genes by a Novel Adenylate Cyclase-Dependent Signaling Pathway. Dev. Cell 2003, 4, 253–263. [Google Scholar] [CrossRef] [Green Version]
- Potrykus, K.; Cashel, M. (p)ppGpp: Still Magical? Annu. Rev. Microbiol. 2008, 62, 35–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalebroux, Z.D.; Svensson, S.L.; Gaynor, E.C.; Swanson, M.S. ppGpp Conjures Bacterial Virulence. Microbiol. Mol. Biol. Rev. 2010, 74, 171–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amato, S.M.; Fazen, C.H.; Henry, T.C.; Mok, W.W.K.; Orman, M.A.; Sandvik, E.L.; Volzing, K.G.; Brynildsen, M.P. The role of metabolism in bacterial persistence. Front. Microbiol. 2014, 5, 70. [Google Scholar] [CrossRef] [Green Version]
- Dalebroux, Z.D.; Swanson, M. ppGpp: Magic beyond RNA polymerase. Nat. Rev. Microbiol. 2012, 10, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Potvin, E.; Sanschagrin, F.; Levesque, R.C. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 2008, 32, 38–55. [Google Scholar] [CrossRef] [Green Version]
- Zenkin, N.; Severinov, K. The role of RNA polymerase subunit in promoter-independent initiation of transcription. Proc. Natl. Acad. Sci. USA 2004, 101, 4396–4400. [Google Scholar] [CrossRef] [Green Version]
- Murakami, K.; Ono, T.; Viducic, D.; Kayama, S.; Mori, M.; Hirota, K.; Nemoto, K.; Miyake, Y. Role forrpoSgene of Pseudomonas aeruginosa in antibiotic tolerance. FEMS Microbiol. Lett. 2005, 242, 161–167. [Google Scholar] [CrossRef] [Green Version]
- Valentini, M.; Filloux, A. Biofilms and Cyclic di-GMP (c-di-GMP) Signaling: Lessons from Pseudomonas aeruginosa and Other Bacteria. J. Biol. Chem. 2016, 291, 12547–12555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, D.-G.; O’Toole, G.A. c-di-GMP and its Effects on Biofilm Formation and Dispersion: A Pseudomonas aeruginosa Review. Microbiol. Spectr. 2015, 3, 2. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Hu, Y.; Liu, Y.; Zhang, J.; Ulstrup, J.; Molin, S. Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development. Environ. Microbiol. 2011, 13, 1705–1717. [Google Scholar] [CrossRef]
- Høiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.; Moser, C.; Jensen, P.Ø.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Bjarnsholt, T. The clinical impact of bacterial biofilms. Int. J. Oral Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, P.K.; Yeung, A.T.; Hancock, R.E. Antibiotic resistance in Pseudomonas aeruginosa biofilms: Towards the development of novel anti-biofilm therapies. J. Biotechnol. 2014, 191, 121–130. [Google Scholar] [CrossRef]
- Rasamiravaka, T.; Labtani, Q.; Duez, P.; El Jaziri, M. The Formation of Biofilms by Pseudomonas aeruginosa: A Review of the Natural and Synthetic Compounds Interfering with Control Mechanisms. BioMed Res. Int. 2015, 2015, 759348. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Jackson, K.D.; Landry, R.M.; Parsek, M.R.; Wozniak, D.J. Analysis of Pseudomonas aeruginosa Conditional Psl Variants Reveals Roles for the Psl Polysaccharide in Adhesion and Maintaining Biofilm Structure Postattachment. J. Bacteriol. 2006, 188, 8213–8221. [Google Scholar] [CrossRef] [Green Version]
- Franklin, M.J.; Nivens, D.E.; Weadge, J.T.; Howell, P.L. Biosynthesis of the Pseudomonas aeruginosa Extracellular Polysaccharides, Alginate, Pel, and Psl. Front. Microbiol. 2011, 2, 167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, L.; Kolter, R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 2004, 51, 675–690. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, K.B.; Kim, T.H.; Gupta, R.; Greenberg, E.P.; Schuster, M. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol. Microbiol. 2009, 73, 1072–1085. [Google Scholar] [CrossRef] [Green Version]
- Sakuragi, Y.; Kolter, R. Quorum-Sensing Regulation of the Biofilm Matrix Genes (pel) of Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 5383–5386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, G.D. Antibiotic resistance in the environment: A link to the clinic? Curr. Opin. Microbiol. 2010, 13, 589–594. [Google Scholar] [CrossRef]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.-J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2018, 37, 177–192. [Google Scholar] [CrossRef]
- Galdino, A.C.M.; Viganor, L.; de Castro, A.A.; da Cunha, E.F.F.; Mello, T.P.; Mattos, L.M.; Pereira, M.D.; Hunt, M.C.; O’Shaughnessy, M.; Howe, O.; et al. Disarming Pseudomonas aeruginosa Virulence by the Inhibitory Action of 1,10-Phenanthroline-5,6-Dione-Based Compounds: Elastase B (LasB) as a Chemotherapeutic Target. Front. Microbiol. 2019, 10, 1701. [Google Scholar] [CrossRef] [Green Version]
- Evans, A.; Kavanagh, K.A.J.J.o.M.M. Evaluation of metal-based antimicrobial compounds for the treatment of bacterial pathogens. J. Med. Microbiol. 2021, 70, 001363. [Google Scholar] [CrossRef] [PubMed]
- Frei, A. Metal complexes, an untapped source of antibiotic potential? Antibiotics 2020, 9, 90. [Google Scholar] [CrossRef] [Green Version]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.; Silva, V.; Igrejas, G.; Poeta, P. Carbapenems and Pseudomonas aeruginosa: Mechanisms and epidemiology. In Antibiotics and Antimicrobial Resistance Genes in the Environment; Elsevier: Amsterdam, The Netherlands, 2020; pp. 253–268. [Google Scholar]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Hancock, R.E.W. Resistance Mechanisms in Pseudomonas aeruginosa and Other Nonfermentative Gram-Negative Bacteria. Clin. Infect. Dis. 1998, 27 (Suppl. S1), S93–S99. [Google Scholar] [CrossRef] [Green Version]
- Delcour, A.H. Proteomics. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta Proteins Proteom. 2009, 1794, 808–816. [Google Scholar] [CrossRef] [Green Version]
- Hancock, R.E.W.; Brinkman, F.S.L. Function of Pseudomonas Porins in Uptake and Efflux. Annu. Rev. Microbiol. 2002, 56, 17–38. [Google Scholar] [CrossRef] [Green Version]
- Tamber, S.; Hancock, R.E. On the mechanism of solute uptake in Pseudomonas. Front Biosci. 2003, 8, s472–s483. [Google Scholar]
- E El Zowalaty, M.; A Al Thani, A.; Webster, T.J.; Schweizer, H.P.; Nasrallah, G.K.; E Marei, H.; Ashour, H.M. Pseudomonas aeruginosa: Arsenal of resistance mechanisms, decades of changing resistance profiles, and future antimicrobial therapies. Futur. Microbiol. 2015, 10, 1683–1706. [Google Scholar] [CrossRef]
- Marquez, B. Bacterial efflux systems and efflux pumps inhibitors. Biochimie 2005, 87, 1137–1147. [Google Scholar] [CrossRef]
- Venter, H.; Mowla, R.; Ohene-Agyei, T.; Ma, S. RND-type drug efflux pumps from Gram-negative bacteria: Molecular mechanism and inhibition. Front. Microbiol. 2015, 6, 377. [Google Scholar] [CrossRef]
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-Resistant Pseudomonas aeruginosa: Clinical Impact and Complex Regulation of Chromosomally Encoded Resistance Mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef] [Green Version]
- Aendekerk, S.; Diggle, S.P.; Song, Z.; Høiby, N.; Cornelis, P.; Williams, P.; Cámara, M. The MexGHI-OpmD multidrug efflux pump controls growth, antibiotic susceptibility and virulence in Pseudomonas aeruginosa via 4-quinolone-dependent cell-to-cell communication. Microbiology 2005, 151, 1113–1125. [Google Scholar] [CrossRef] [Green Version]
- Wright, G.D. Bacterial resistance to antibiotics: Enzymatic degradation and modification. Adv. Drug Deliv. Rev. 2005, 57, 1451–1470. [Google Scholar] [CrossRef]
- Bush, K.; Jacoby, G.A. Updated Functional Classification of β-Lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef] [Green Version]
- Juan, C.; Torrens, G.; González-Nicolau, M.; Oliver, A. Diversity and regulation of intrinsic β-lactamases from non-fermenting and other Gram-negative opportunistic pathogens. FEMS Microbiol. Rev. 2017, 41, 781–815. [Google Scholar] [CrossRef]
- Rawat, D.; Nair, D. Extended-spectrum β-lactamases in Gram Negative Bacteria. J. Glob. Infect. Dis. 2010, 2, 263. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Zhang, L.; He, Y.; Liu, K.; Zhang, F.; Zhang, H.; Lu, Y.; Yang, C.; Wang, Z.; Fareed, M.S.; et al. An optimized analog of antimicrobial peptide Jelleine-1 shows enhanced antimicrobial activity against multidrug resistant P. aeruginosa and negligible toxicity in vitro and in vivo. Eur. J. Med. Chem. 2021, 219, 113433. [Google Scholar] [CrossRef]
- Grassi, L.; Batoni, G.; Ostyn, L.; Rigole, P.; Bossche, S.V.D.; Rinaldi, A.; Maisetta, G.; Esin, S.; Coenye, T.; Crabbe, A. The Antimicrobial Peptide lin-SB056-1 and Its Dendrimeric Derivative Prevent Pseudomonas aeruginosa Biofilm Formation in Physiologically Relevant Models of Chronic Infections. Front. Microbiol. 2019, 10, 198. [Google Scholar] [CrossRef] [Green Version]
- Parducho, K.R.; Beadell, B.; Ybarra, T.K.; Bush, M.; Escalera, E.; Trejos, A.T.; Chieng, A.; Mendez, M.; Anderson, C.; Park, H.; et al. The Antimicrobial Peptide Human Beta-Defensin 2 Inhibits Biofilm Production of Pseudomonas aeruginosa without Compromising Metabolic Activity. Front. Immunol. 2020, 11, 805. [Google Scholar] [CrossRef]
- Jones, R.A.; Shropshire, H.; Zhao, C.; Murphy, A.; Lidbury, I.; Wei, T.; Scanlan, D.J.; Chen, Y. Phosphorus stress induces the synthesis of novel glycolipids in Pseudomonas aeruginosa that confer protection against a last-resort antibiotic. ISME J. 2021, 15, 3303–3314. [Google Scholar] [CrossRef]
- Breidenstein, E.B.; de la Fuente-Núñez, C.; Hancock, R.E. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef]
- Rodriguez-Valera, F.; Martin-Cuadrado, A.-B.; López-Pérez, M. Flexible genomic islands as drivers of genome evolution. Curr. Opin. Microbiol. 2016, 31, 154–160. [Google Scholar] [CrossRef]
- Botelho, J.; Grosso, F.; Peixe, L. Antibiotic resistance in Pseudomonas aeruginosa—Mechanisms, epidemiology and evolution. Drug Resist. Updat. 2019, 44, 100640. [Google Scholar] [CrossRef]
- Umadevi, S.; Joseph, N.M.; Kumari, K.; Easow, J.M.; Kumar, S.; Stephen, S.; Srirangaraj, S.; Raj, S. Detection of extended spectrum beta lactamases, ampc beta lactamases and metallobetalactamases in clinical isolates of ceftazidime resistant Pseudomonas aeruginosa. Braz. J. Microbiol. 2011, 42, 1284–1288. [Google Scholar] [CrossRef] [Green Version]
- Lopatkin, A.J.; Meredith, H.R.; Srimani, J.K.; Pfeiffer, C.; Durrett, R.; You, L. Persistence and reversal of plasmid-mediated antibiotic resistance. Nat. Commun. 2017, 8, 1689. [Google Scholar] [CrossRef]
- Bennett, P.M. Plasmid encoded antibiotic resistance: Acquisition and transfer of antibiotic resistance genes in bacteria. Br. J. Pharmacol. 2008, 153, S347–S357. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2015, 16, 161–168. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016, 4, 481–511. [Google Scholar] [CrossRef] [Green Version]
- Kao, C.-Y.; Chen, S.-S.; Hung, K.-H.; Wu, H.-M.; Hsueh, P.-R.; Yan, J.-J.; Wu, J.-J. Overproduction of active efflux pump and variations of OprD dominate in imipenem-resistant Pseudomonas aeruginosa isolated from patients with bloodstream infections in Taiwan. BMC Microbiol. 2016, 16, 107. [Google Scholar] [CrossRef] [Green Version]
- Köhler, T.; Epp, S.F.; Curty, L.K.; Pechère, J.-C. Characterization of MexT, the Regulator of the MexE-MexF-OprN Multidrug Efflux System of Pseudomonas aeruginosa. J. Bacteriol. 1999, 181, 6300–6305. [Google Scholar] [CrossRef] [Green Version]
- Berrazeg, M.; Jeannot, K.; Enguéné, V.Y.N.; Broutin, I.; Loeffert, S.; Fournier, D.; Plésiat, P. Mutations in β-Lactamase AmpC Increase Resistance of Pseudomonas aeruginosa Isolates to Antipseudomonal Cephalosporins. Antimicrob. Agents Chemother. 2015, 59, 6248–6255. [Google Scholar] [CrossRef] [Green Version]
- Fernández, L.; Breidenstein, E.B.; Hancock, R.E. Creeping baselines and adaptive resistance to antibiotics. Drug Resist. Updat. 2011, 14, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.J.I.j.o.m.m. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med Microbiol. 2002, 292, 107–113. [Google Scholar] [CrossRef]
- Wilton, M.; Charron-Mazenod, L.; Moore, R.; Lewenza, S. Extracellular DNA Acidifies Biofilms and Induces Aminoglycoside Resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016, 60, 544–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goli, H.R.; Nahaei, M.R.; Rezaee, M.A.; Hasani, A.; Kafil, H.S.; Aghazadeh, M. Emergence of colistin resistant Pseudomonas aeruginosa at Tabriz hospitals, Iran. Iran. J. Microbiol. 2016, 8, 62–69. [Google Scholar] [PubMed]
- Azimi, L.; Lari, A.R. Colistin-resistant Pseudomonas aeruginosa clinical strains with defective biofilm formation. GMS Hyg. Infect. Control. 2019, 14, Doc12. [Google Scholar] [PubMed]
- Haenni, M.; Bour, M.; Châtre, P.; Madec, J.-Y.; Plésiat, P.; Jeannot, K. Resistance of Animal Strains of Pseudomonas aeruginosa to Carbapenems. Front. Microbiol. 2017, 8, 1847. [Google Scholar] [CrossRef]
- Castanon, J.I.R. History of the Use of Antibiotic as Growth Promoters in European Poultry Feeds. Poult. Sci. 2007, 86, 2466–2471. [Google Scholar] [CrossRef]
- Wanty, C.; Anandan, A.; Piek, S.; Walshe, J.; Ganguly, J.; Carlson, R.W.; Stubbs, K.A.; Kahler, C.M.; Vrielink, A. The Structure of the Neisserial Lipooligosaccharide Phosphoethanolamine Transferase A (LptA) Required for Resistance to Polymyxin. J. Mol. Biol. 2013, 425, 3389–3402. [Google Scholar] [CrossRef]
- Andrade, F.F.; Silva, D.; Rodrigues, A.; Pina-Vaz, C. Colistin Update on Its Mechanism of Action and Resistance, Present and Future Challenges. Microorganisms 2020, 8, 1716. [Google Scholar] [CrossRef]
- Dößelmann, B.; Willmann, M.; Steglich, M.; Bunk, B.; Nübel, U.; Peter, S.; Neher, R.A. Rapid and Consistent Evolution of Colistin Resistance in Extensively Drug-Resistant Pseudomonas aeruginosa during Morbidostat Culture. Antimicrob. Agents Chemother. 2017, 61, e00043-17. [Google Scholar] [CrossRef] [Green Version]
- Arcilla, M.S.; van Hattem, J.M.; Matamoros, S.; Melles, D.C.; Penders, J.; de Jong, M.D.; Schultsz, C. Dissemination of the mcr-1 colistin resistance gene. Lancet 2016, 16, 147–149. [Google Scholar] [CrossRef] [Green Version]
- Lima, T.; Domingues, S.; Da Silva, G.J. Plasmid-Mediated Colistin Resistance in Salmonella enterica: A Review. Microorganisms 2019, 7, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pesingi, P.V.; Singh, B.R.; Pesingi, P.K.; Bhardwaj, M.; Singh, S.V.; Kumawat, M.; Sinha, D.K.; Gandham, R.K. MexAB-OprM Efflux Pump of Pseudomonas aeruginosa Offers Resistance to Carvacrol: A Herbal Antimicrobial Agent. Front. Microbiol. 2019, 10, 2664. [Google Scholar] [CrossRef] [PubMed]
- Moskowitz, S.M.; Ernst, R.; Miller, S.I. PmrAB, a Two-Component Regulatory System of Pseudomonas aeruginosa That Modulates Resistance to Cationic Antimicrobial Peptides and Addition of Aminoarabinose to Lipid A. J. Bacteriol. 2004, 186, 575–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macfarlane, E.L.A.; Kwasnicka, A.; Hancock, R.E.W. Role of Pseudomonas aeruginosa PhoP-PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 2000, 146, 2543–2554. [Google Scholar] [CrossRef] [Green Version]
- Ezadi, F.; Ardebili, A.; Mirnejad, R. Antimicrobial Susceptibility Testing for Polymyxins: Challenges, Issues, and Recommendations. J. Clin. Microbiol. 2019, 57, e01390-18. [Google Scholar] [CrossRef] [Green Version]
- McPhee, J.; Bains, M.; Winsor, G.; Lewenza, S.; Kwasnicka, A.; Brazas, M.; Brinkman, F.S.L.; Hancock, R.E.W. Contribution of the PhoP-PhoQ and PmrA-PmrB Two-Component Regulatory Systems to Mg2+ -Induced Gene Regulation in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 3995–4006. [Google Scholar] [CrossRef] [Green Version]
- Fernández, L.; Gooderham, W.J.; Bains, M.; McPhee, J.; Wiegand, I.; Hancock, R.E.W. Adaptive Resistance to the “Last Hope” Antibiotics Polymyxin B and Colistin in Pseudomonas aeruginosa Is Mediated by the Novel Two-Component Regulatory System ParR-ParS. Antimicrob. Agents Chemother. 2010, 54, 3372–3382. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Sousa, T.; Hébraud, M.; Dapkevicius, M.L.N.E.; Maltez, L.; Pereira, J.E.; Capita, R.; Alonso-Calleja, C.; Igrejas, G.; Poeta, P. Genomic and Metabolic Characteristics of the Pathogenicity in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2021, 22, 12892. https://doi.org/10.3390/ijms222312892
de Sousa T, Hébraud M, Dapkevicius MLNE, Maltez L, Pereira JE, Capita R, Alonso-Calleja C, Igrejas G, Poeta P. Genomic and Metabolic Characteristics of the Pathogenicity in Pseudomonas aeruginosa. International Journal of Molecular Sciences. 2021; 22(23):12892. https://doi.org/10.3390/ijms222312892
Chicago/Turabian Stylede Sousa, Telma, Michel Hébraud, Maria L. N. Enes Dapkevicius, Luís Maltez, José Eduardo Pereira, Rosa Capita, Carlos Alonso-Calleja, Gilberto Igrejas, and Patricia Poeta. 2021. "Genomic and Metabolic Characteristics of the Pathogenicity in Pseudomonas aeruginosa" International Journal of Molecular Sciences 22, no. 23: 12892. https://doi.org/10.3390/ijms222312892
APA Stylede Sousa, T., Hébraud, M., Dapkevicius, M. L. N. E., Maltez, L., Pereira, J. E., Capita, R., Alonso-Calleja, C., Igrejas, G., & Poeta, P. (2021). Genomic and Metabolic Characteristics of the Pathogenicity in Pseudomonas aeruginosa. International Journal of Molecular Sciences, 22(23), 12892. https://doi.org/10.3390/ijms222312892