Patterns of Maternal Neutrophil Gene Expression at 30 Weeks of Gestation, but Not DNA Methylation, Distinguish Mild from Severe Preeclampsia
Abstract
:1. Introduction
2. Results
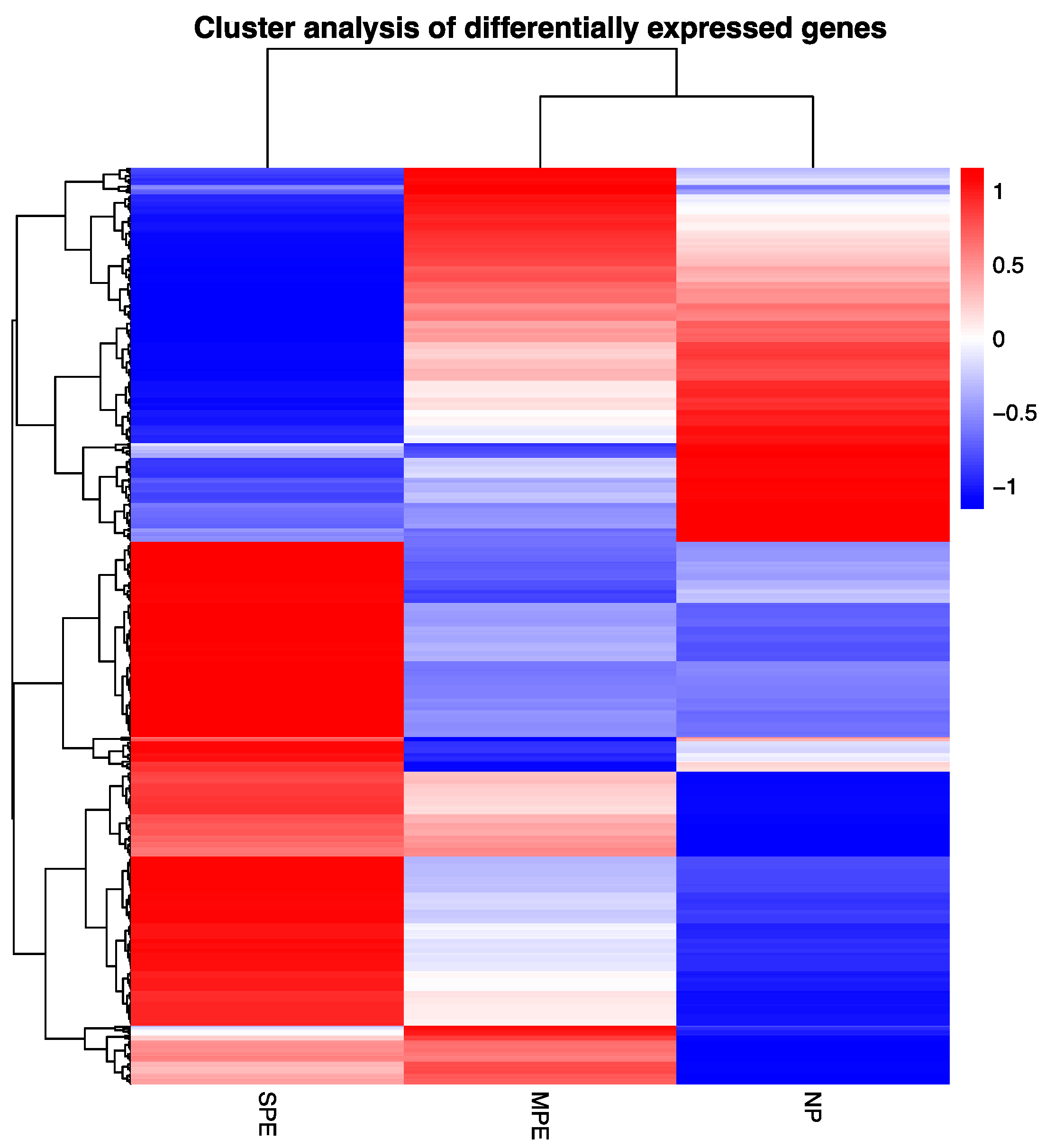
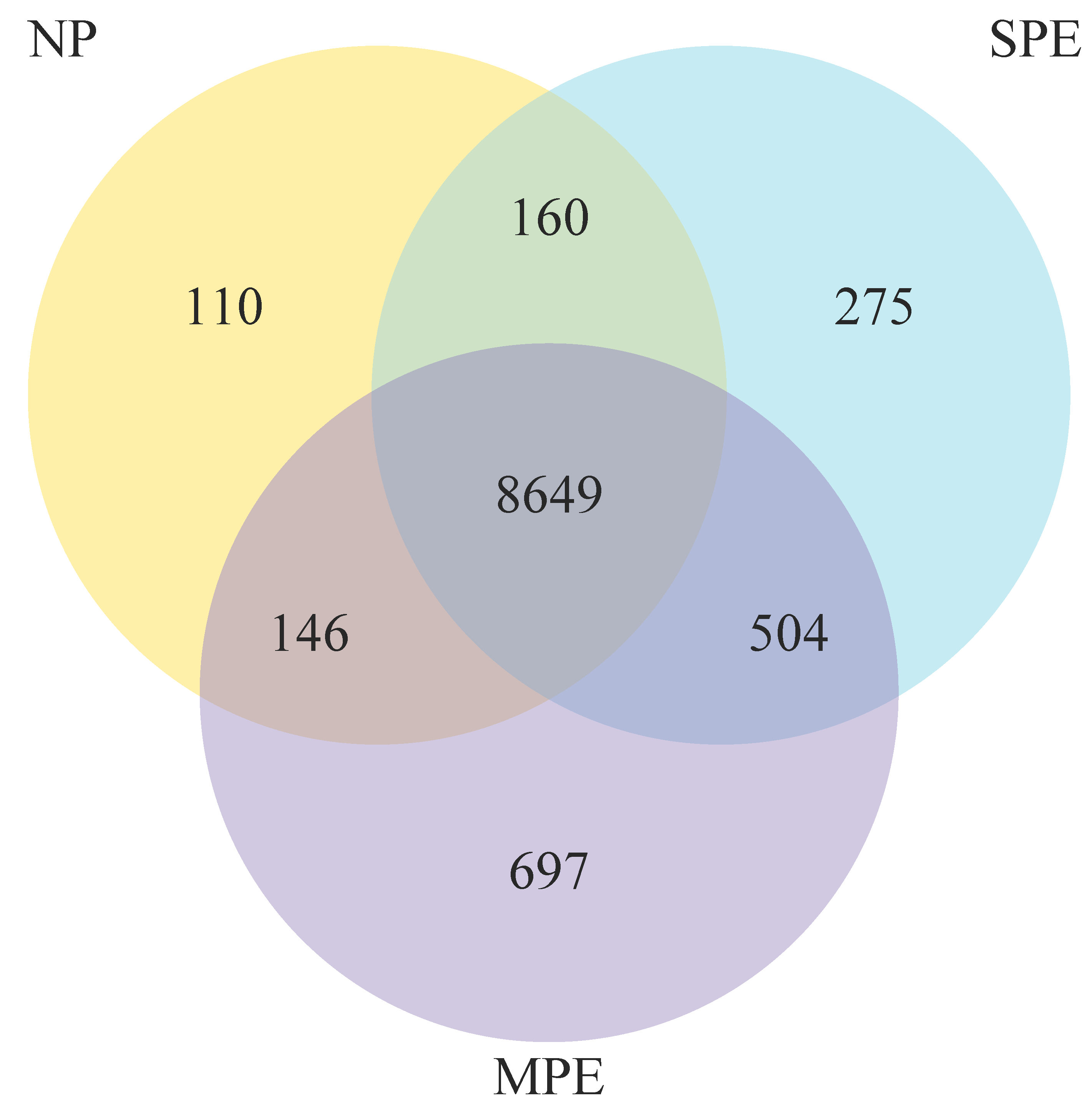
2.1. RNA Sequencing of Pregnancy Neutrophils
2.2. hMeDIP (5-hmC) DNA Sequencing of Pregnancy Neutrophils
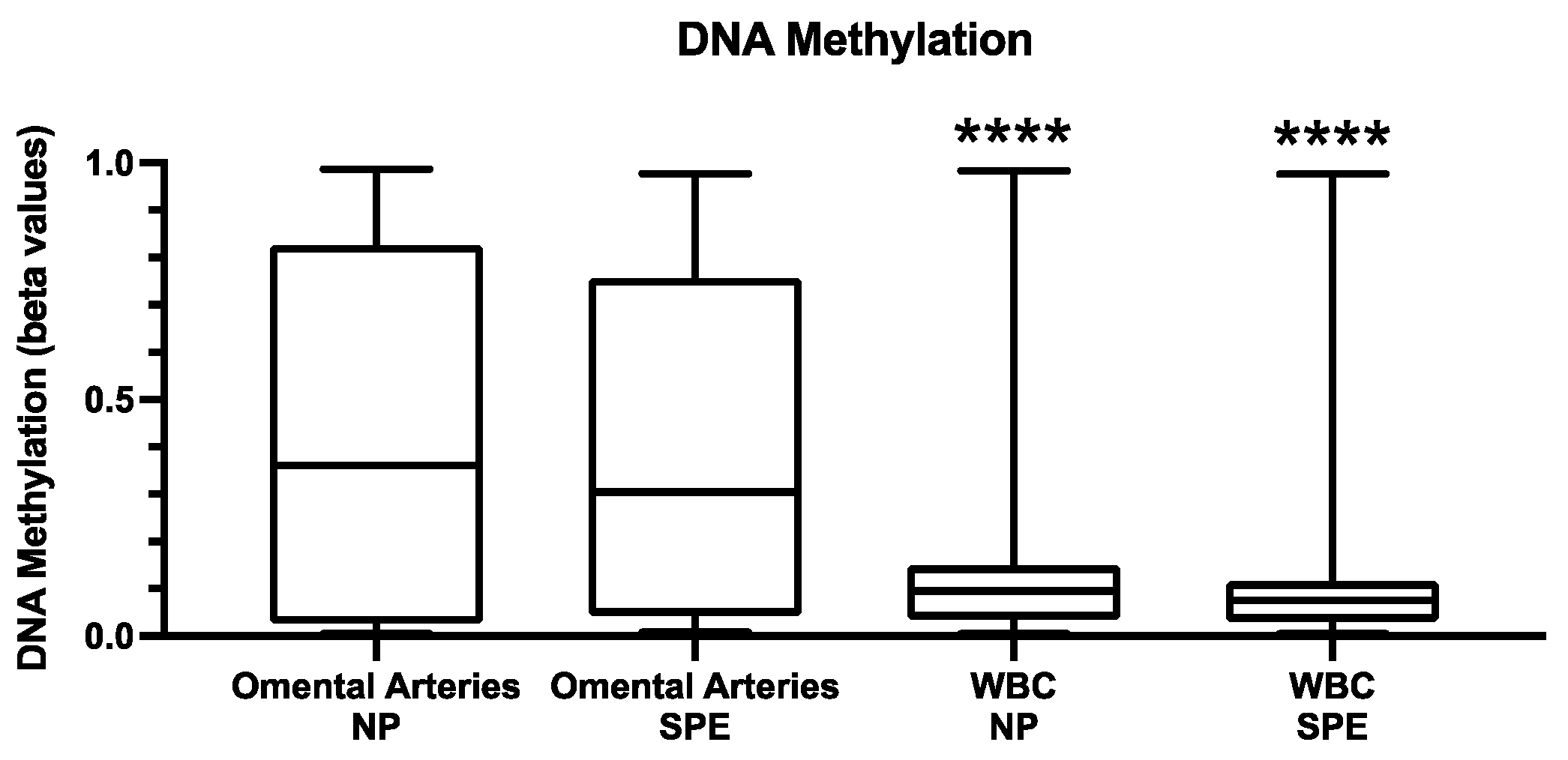
2.3. DNA Methylation in Leukocytes
3. Discussion
4. Materials and Methods
4.1. Study Subjects
4.2. RNA Sequencing and hMeDIP (5-hmC) DNA Sequencing of Neutrophils
4.3. DNA Methylation in Leukocytes
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cunningham, F.G.; Gant, N.F.; Leveno, K.J.; Gilstrap, L.C., III; Hauth, J.C.; Wenstrom, K.D. Williams Obstetrics, 21st ed.; McGraw-Hill: New York, NY, USA, 2001. [Google Scholar]
- Leik, C.E.; Walsh, S.W. Neutrophils infiltrate resistance-sized vessels of subcutaneous fat in women with preeclampsia. Hypertension 2004, 44, 72–77. [Google Scholar] [CrossRef] [Green Version]
- Mishra, N.; Nugent, W.H.; Mahavadi, S.; Walsh, S.W. Mechanisms of enhanced vascular reactivity in preeclampsia. Hypertension 2011, 58, 867–873. [Google Scholar] [CrossRef]
- Shah, T.J.; Walsh, S.W. Activation of NF-kappaB and expression of COX-2 in association with neutrophil infiltration in systemic vascular tissue of women with preeclampsia. Am. J. Obstet. Gynecol. 2007, 196, 48.e1–48.e8. [Google Scholar] [CrossRef]
- Mousa, A.A.; Archer, K.J.; Cappello, R.; Estrada-Gutierrez, G.; Isaacs, C.R.; Strauss, J.F., 3rd; Walsh, S.W. DNA Methylation is Altered in Maternal Blood Vessels of Women With Preeclampsia. Reprod. Sci. 2012, 19, 1332–1342. [Google Scholar] [CrossRef] [Green Version]
- Mousa, A.A.; Cappello, R.E.; Estrada-Gutierrez, G.; Shukla, J.; Romero, R.; Strauss, J.F., 3rd; Walsh, S.W. Preeclampsia is associated with alterations in DNA methylation of genes involved in collagen metabolism. Am. J. Pathol. 2012, 181, 1455–1463. [Google Scholar] [CrossRef] [Green Version]
- Mousa, A.A.; Strauss, J.F., 3rd; Walsh, S.W. Reduced methylation of the thromboxane synthase gene is correlated with its increased vascular expression in preeclampsia. Hypertension 2012, 59, 1249–1255. [Google Scholar] [CrossRef] [Green Version]
- He, Y.F.; Li, B.Z.; Li, Z.; Liu, P.; Wang, Y.; Tang, Q.; Ding, J.; Jia, Y.; Chen, Z.; Li, L.; et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 2011, 333, 1303–1307. [Google Scholar] [CrossRef] [Green Version]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef] [Green Version]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef] [Green Version]
- Yin, R.; Mao, S.Q.; Zhao, B.; Chong, Z.; Yang, Y.; Zhao, C.; Zhang, D.; Huang, H.; Gao, J.; Li, Z.; et al. Ascorbic Acid enhances tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J. Am. Chem. Soc. 2013, 135, 10396–10403. [Google Scholar] [CrossRef]
- Nabel, C.S.; Kohli, R.M. Molecular biology. Demystifying DNA demethylation. Science 2011, 333, 1229–1230. [Google Scholar] [CrossRef]
- Klug, M.; Schmidhofer, S.; Gebhard, C.; Andreesen, R.; Rehli, M. 5-Hydroxymethylcytosine is an essential intermediate of active DNA demethylation processes in primary human monocytes. Genome Biol. 2013, 14, R46. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Zhang, Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011, 25, 2436–2452. [Google Scholar] [CrossRef] [Green Version]
- Klug, M.; Heinz, S.; Gebhard, C.; Schwarzfischer, L.; Krause, S.W.; Andreesen, R.; Rehli, M. Active DNA demethylation in human postmitotic cells correlates with activating histone modifications, but not transcription levels. Genome Biol. 2010, 11, R63. [Google Scholar] [CrossRef] [Green Version]
- Walsh, S.W.; Nugent, W.H.; Al Dulaimi, M.; Washington, S.L.; Dacha, P.; Strauss, J.F., 3rd. Proteases Activate Pregnancy Neutrophils by a Protease-Activated Receptor 1 Pathway: Epigenetic Implications for Preeclampsia. Reprod. Sci. 2020, 27, 2115–2127. [Google Scholar] [CrossRef]
- Munchel, S.; Rohrback, S.; Randise-Hinchliff, C.; Kinnings, S.; Deshmukh, S.; Alla, N.; Tan, C.; Kia, A.; Greene, G.; Leety, L.; et al. Circulating transcripts in maternal blood reflect a molecular signature of early-onset preeclampsia. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef]
- Walsh, S.W.; Reep, D.T.; Alam, S.M.K.; Washington, S.L.; Al Dulaimi, M.; Lee, S.M.; Springel, E.H.; Strauss, J.F.; Stephenson, D.J.; Chalfant, C.E. Placental Production of Eicosanoids and Sphingolipids in Women Who Developed Preeclampsia on Low-Dose Aspirin. Reprod. Sci. 2020, 27, 2158–2169. [Google Scholar] [CrossRef]
- Zhang, X.; Kluger, Y.; Nakayama, Y.; Poddar, R.; Whitney, C.; DeTora, A.; Weissman, S.M.; Newburger, P. Gene expression in mature neutrophils: Early responses to inflammatory stimuli. J. Leukoc. Biol. 2004, 75, 358–372. [Google Scholar] [CrossRef] [Green Version]
- Naegelen, I.; Beaume, N.; Plançon, S.; Schenten, V.; Tschirhart, E.J.; Bréchard, S. Regulation of Neutrophil Degranulation and Cytokine Secretion: A Novel Model Approach Based on Linear Fitting. J. Immunol. Res. 2015, 2015, 817038. [Google Scholar] [CrossRef] [Green Version]
- Coughlin, S.R. Protease-activated receptors in hemostasis, thrombosis and vascular biology. J. Thromb. Haemost. 2005, 3, 1800–1814. [Google Scholar] [CrossRef]
- Estrada-Gutierrez, G.; Cappello, R.E.; Mishra, N.; Romero, R.; Strauss, J.F., 3rd; Walsh, S.W. Increased expression of matrix metalloproteinase-1 in systemic vessels of preeclamptic women a critical mediator of vascular dysfunction. Am. J. Pathol. 2011, 178, 451–460. [Google Scholar] [CrossRef]
- Vaughan, J.E.; Walsh, S.W.; Ford, G.D. Thromboxane mediates neutrophil superoxide production in pregnancy. Am. J. Obstet. Gynecol. 2006, 195, 1415–1420. [Google Scholar] [CrossRef] [PubMed]
- Dunning, M.J.; Smith, M.L.; Ritchie, M.E.; Tavare, S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics 2007, 23, 2183–2184. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [Green Version]

| • Regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis |
| • Blood vessel endothelial cell proliferation involved in sprouting angiogenesis |
| • Regulation of endothelial cell apoptotic process |
| • Endothelial cell apoptotic process |
| • Regulation of cell migration involved in sprouting angiogenesis |
| • Multicellular organismal response to stress |
| • Cell migration involved in sprouting angiogenesis |
| • Positive regulation of blood vessel endothelial cell migration |
| • Regulation of megakaryocyte differentiation |
| • Megakaryocyte differentiation |
| • Phagocytosis, engulfment |
| • Positive regulation of endothelial cell migration |
| • Regulation of blood vessel endothelial cell migration |
| • Sprouting angiogenesis |
| • Blood vessel endothelial cell migration |
| • Regulation of endothelial cell proliferation |
| • Endothelial cell proliferation |
| • Regulation of endothelial cell migration |
| • Positive regulation of angiogenesis |
| • Positive regulation of vasculature development |
| • Endothelial cell migration |
| • Regulation of myeloid cell differentiation |
| • Positive regulation of MAP kinase activity |
| • Tissue migration |
| • Regulation of angiogenesis |
| • Blood coagulation |
| • Hemostasis |
| • Coagulation |
| • Activation of protein kinase activity |
| • Positive regulation of protein serine/threonine kinase activity |
| • Regulation of MAP kinase activity |
| • Regulation of cGMP-mediated signaling |
| • Regulation of transforming growth factor beta1 production |
| • Regulation of vasculature development |
| • Phagocytosis |
| • Transforming growth factor beta1 production |
| • Regulation of fibrinolysis |
| • Positive regulation of leukocyte activation |
| • Programmed cell death involved in cell development |
| • Positive regulation of fibroblast migration |
| • Epithelial cell proliferation |
| • Positive regulation of cell activation |
| • Chemosensory behavior |
| • Regulation of plasminogen activation |
| • Positive regulation of phagocytosis, engulfment |
| • Myeloid cell differentiation |
| • Regulation of phagocytosis, engulfment |
| • Fatty acid transmembrane transport |
| • Regulation of endothelial cell chemotaxis |
| • Chronic inflammatory response |
| • Plasminogen activation |
| • Regulation of hemopoiesis |
| • Positive regulation of tumor necrosis factor biosynthetic process |
| • Positive regulation of MAPK cascade |
| • Regulation of protein serine/threonine kinase activity |
| • Positive regulation of blood coagulation |
| • Regulation of macrophage differentiation |
| • Positive regulation of hemostasis |
| • Positive regulation of cell migration |
| • Leukocyte differentiation |
| • Angiogenesis |
| • Positive regulation of protein kinase activity |
| • Nitric-oxide-mediated signal transduction |
| • Positive regulation of leukocyte degranulation |
| • Positive regulation of coagulation |
| • cGMP-mediated signaling |
| • Positive regulation of transforming growth factor beta receptor signaling pathway |
| • Positive regulation of cellular response to transforming growth factor beta stimulus |
| • Regulation of fatty acid transport |
| • Positive regulation of macrophage activation |
| • Tumor necrosis factor biosynthetic process |
| • Regulation of tumor necrosis factor biosynthetic process |
| • Granulocyte differentiation |
| • Regulation of granulocyte chemotaxis |
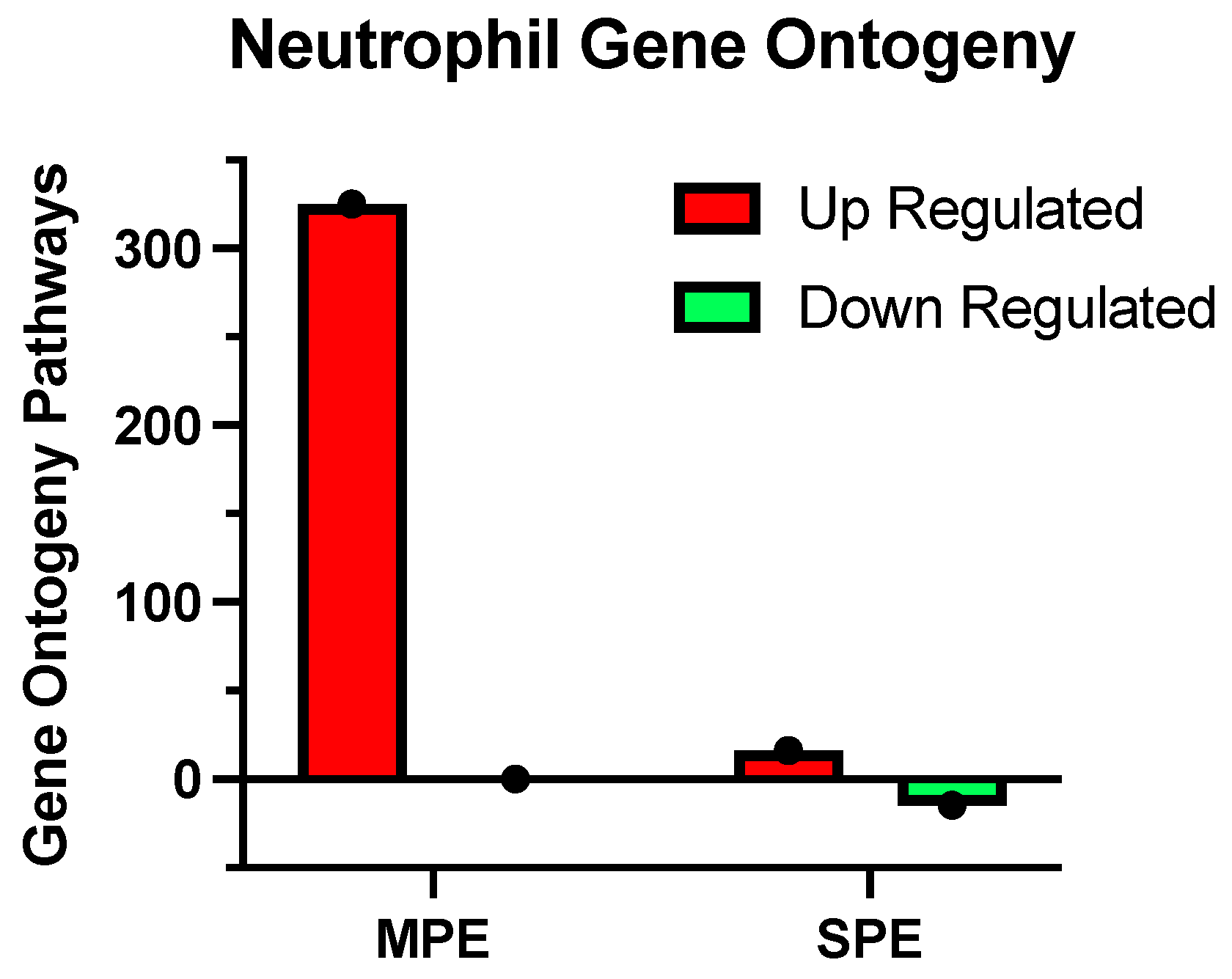
| A. Upregulated Pathways |
| • Regulation of leukocyte apoptotic process |
| • Leukocyte apoptotic process |
| The adjusted p-value (padj) for these pathways was < 0.05 |
| B. Downregulated Pathways |
| • Neutrophil degranulation |
| • Neutrophil activation involved in immune response |
| • Neutrophil-mediated immunity |
| • Neutrophil activation |
| • Granulocyte activation |
| • Tumor-necrosis-factor-mediated signaling pathway |
| Differentially expressed genes identified by RNA-seq were subjected to gene ontology analysis by Novogene. The adjusted p-value (padj) for these pathways was < 0.0001. |
| Variable | NP n = 11 | MPE n = 3 | SPE n = 10 |
|---|---|---|---|
| Maternal age (years) | 29.8 ± 5.6 | 31.3 ± 7.0 | 28.4 ± 2.2 |
| Pre-pregnancy BMI (kg/m2) | 27.6 ± 5.8 | 27.5 ± 5.5 | 32.8 ± 10.0 |
| BMI at sample collection (kg/m2) | 32.2 ± 6.1 | 33.7 ± 7.6 | 38.0 ± 10.6 |
| Systolic blood pressure at 30 weeks (mmHg) | 112 ± 9 | 116 ± 12 | 179 ± 16 **** |
| Diastolic blood pressure at 30 weeks (mmHg) | 69 ± 8 | 77 ± 13 | 109 ± 12 **** |
| Protein/creatinine ratio | ND | 0.4 ± 0.1 | 1.2 ± 1.4 |
| Primiparous | 5 | 1 | 6 |
| Multiparous | 6 | 2 | 4 |
| Race | |||
| White | 6 | 2 | 3 |
| Black | 2 | 4 | |
| Hispanic | 1 | 1 | 3 |
| Asian | 2 | ||
| Type of delivery C-section Vaginal | 2 9 | 2 1 | 4 6 |
| Gestational age at sample collection (weeks) | 32.6 ± 2.3 | 30.5 ± 2.1 | 31.8 ± 4.2 |
| Gestational age at delivery (weeks) | 38.6 ± 1.3 | 38.9 ± 3.2 | 32.8 ± 4.2 *** |
| Infant birth weight (grams) | 2986 ± 476 | 2818 ± 836 | 1700 ± 670 *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walsh, S.W.; Al Dulaimi, M.; Archer, K.J.; Strauss, J.F., III. Patterns of Maternal Neutrophil Gene Expression at 30 Weeks of Gestation, but Not DNA Methylation, Distinguish Mild from Severe Preeclampsia. Int. J. Mol. Sci. 2021, 22, 12876. https://doi.org/10.3390/ijms222312876
Walsh SW, Al Dulaimi M, Archer KJ, Strauss JF III. Patterns of Maternal Neutrophil Gene Expression at 30 Weeks of Gestation, but Not DNA Methylation, Distinguish Mild from Severe Preeclampsia. International Journal of Molecular Sciences. 2021; 22(23):12876. https://doi.org/10.3390/ijms222312876
Chicago/Turabian StyleWalsh, Scott W., Marwah Al Dulaimi, Kellie J. Archer, and Jerome F. Strauss, III. 2021. "Patterns of Maternal Neutrophil Gene Expression at 30 Weeks of Gestation, but Not DNA Methylation, Distinguish Mild from Severe Preeclampsia" International Journal of Molecular Sciences 22, no. 23: 12876. https://doi.org/10.3390/ijms222312876






