Reduced Zeb1 Expression in Prostate Cancer Cells Leads to an Aggressive Partial-EMT Phenotype Associated with Altered Global Methylation Patterns
Abstract
1. Introduction
2. Results
2.1. Inducible Knockdown of Zeb1 in PC-3 Human Prostate Cancer Cells Results in Enhanced Expression of Epithelial Proteins
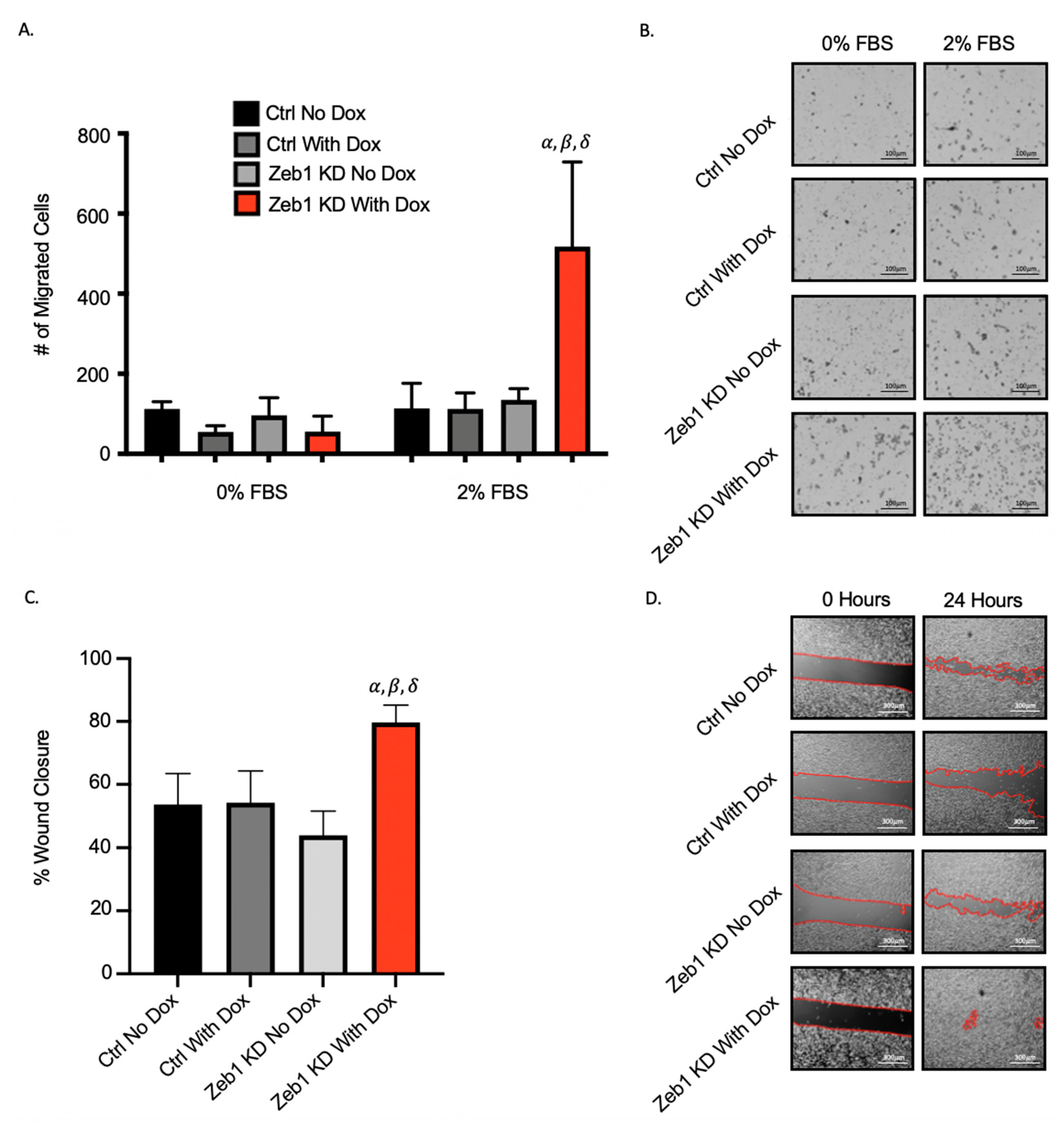
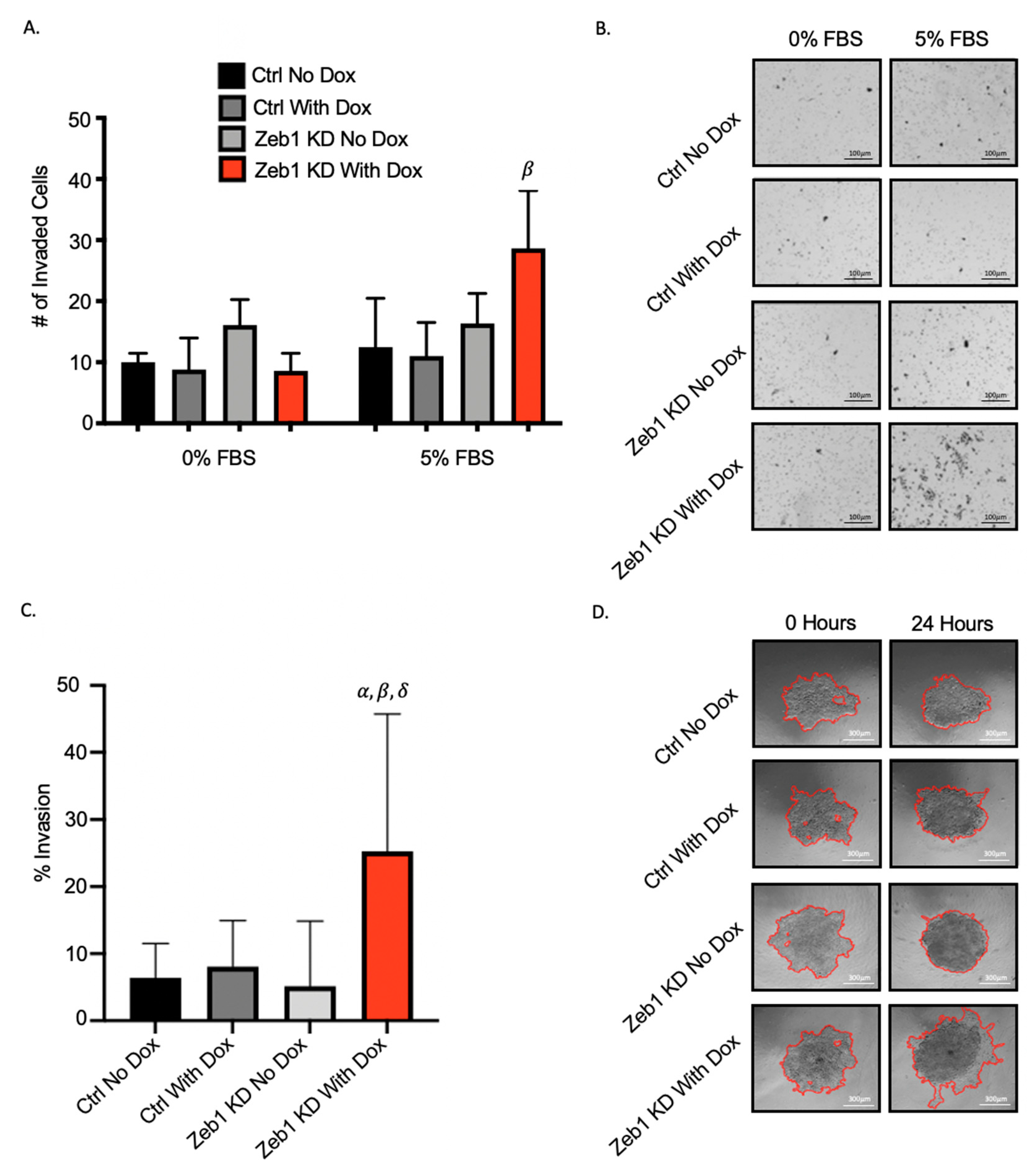
2.2. Knockdown of Zeb1 in PC-3 Prostate Cancer Cells Increases Migration and Invasion but Does Not Alter Proliferation
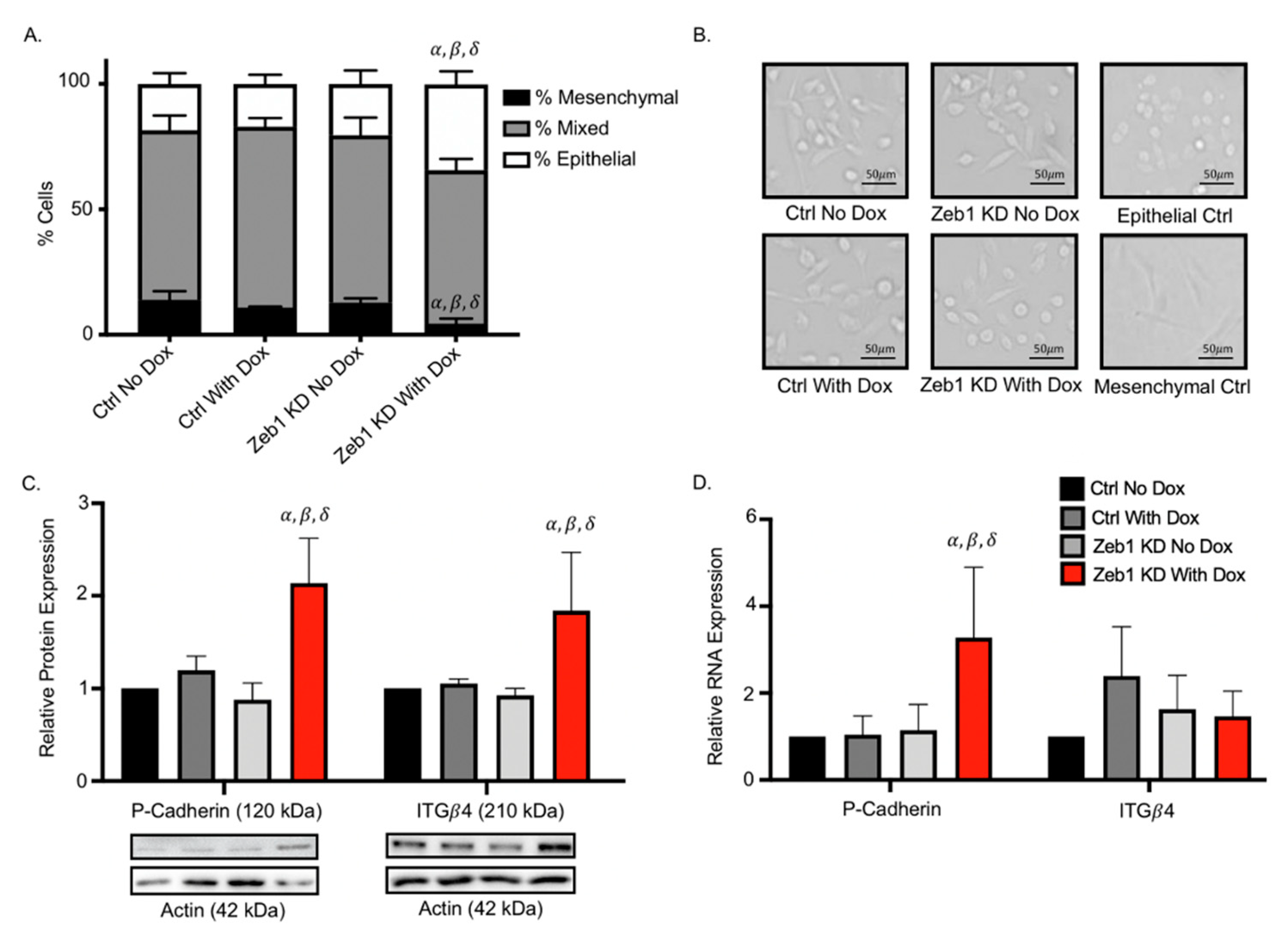
2.3. Knockdown of Zeb1 in PC-3 Prostate Cancer Cells Leads to a Partial EMT Phenotype at the Cellular and Molecular Level
2.4. Treatment of PC-3 Zeb1KD Prostate Cancer Cells with the Global Demethylating Agent 5-Azacitadine Results in Decreased DNA Methylation, Migration, and Invasion
2.5. Methylation Chip Analysis of Zeb1KD PC-3 Prostate Cancer Cells Identified 10 Genes Associated with a p-EMT Phenotype
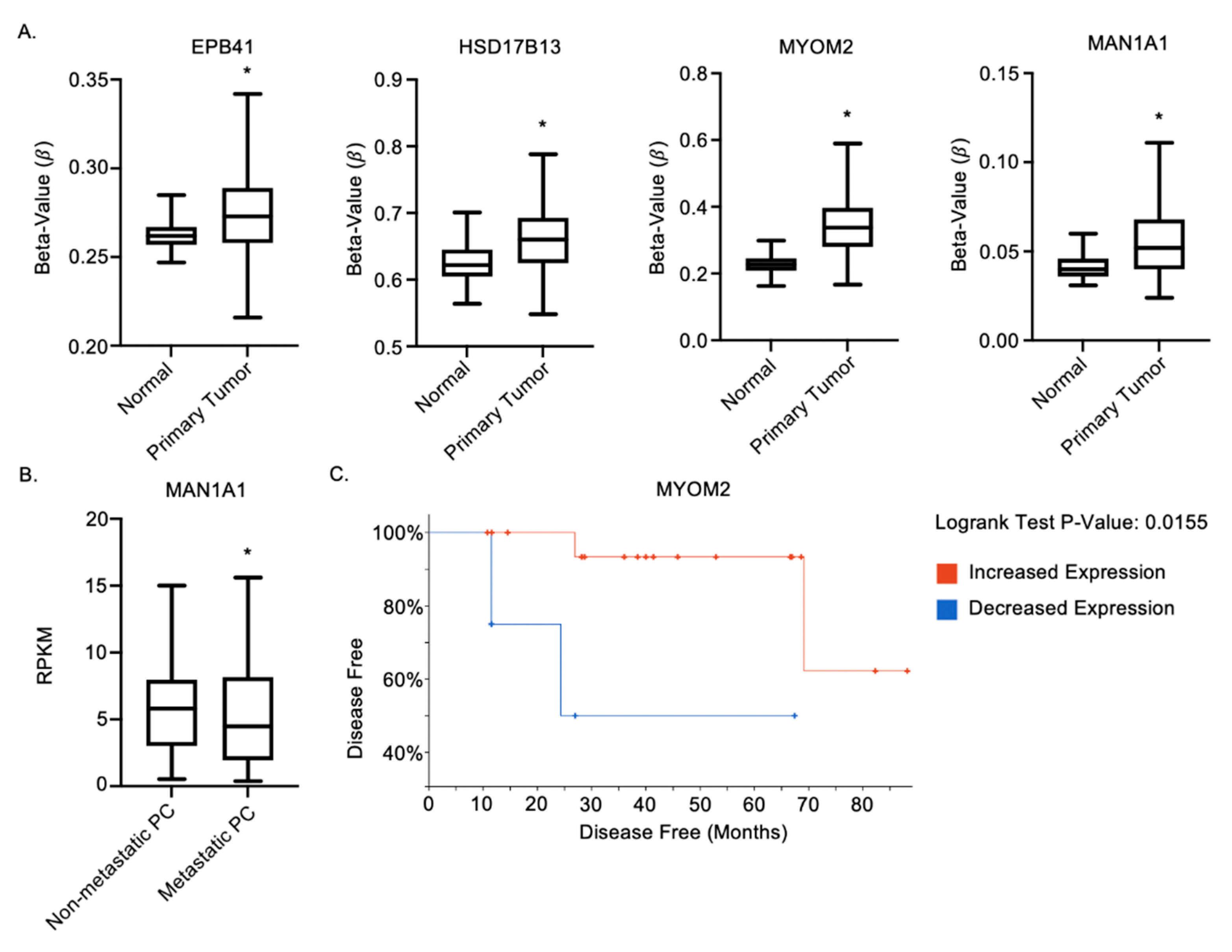
2.6. MAN1A1, EPB41, HSD17B13 and MYOM2 Are Altered in Prostate Cancer Patients
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Transductions
4.3. Immunoblotting
4.4. Quantitative Real-Time PCR
4.5. Transwell Migration and Invasion Assays
4.6. Physical Barrier Wound Healing Assay
4.7. Spheroid Invasion Assay
4.8. BrdU Proliferation Assay
4.9. Cell Morphology Assay
4.10. DNA Extraction and Dot-Blot DNA Analysis
4.11. DNA Methylation Chip Analysis
4.12. Patient Sample Analysis
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Pantel, K.; Brakenhoff, R.H.; Brandt, B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat. Rev. Cancer 2008, 8, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Fidler, I.J. The pathogenesis of cancer metastasis: The “seed and soil” hypothesis revisited. Nat. Rev. Cancer 2003, 3, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Kano, A. Tumor cell secretion of soluble factor(s) for specific immunosuppression. Sci. Rep. 2015, 5, srep08913. [Google Scholar] [CrossRef]
- Weidner, N.; Semple, J.P.; Welch, W.R.; Folkman, J. Tumor angiogenesis and metastasis–Correlation in invasive breast carcinoma. N. Engl. J. Med. 1991, 324, 1–8. [Google Scholar] [CrossRef]
- Scott, J.; Kuhn, P.; Anderson, A.R.A. Unifying metastasis-integrating intravasation, circulation and end-organ colonization. Nat. Rev. Cancer 2012, 12, 445–446. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hou, J.M.; Krebs, M.; Ward, T.; Sloane, R.; Priest, L.; Hughes, A.; Clack, G.; Ranson, M.; Blackhall, F.; Dive, C. Circulating tumor cells as a window on metastasis biology in lung cancer. Am. J. Pathol. 2011, 178, 989–996. [Google Scholar] [CrossRef] [PubMed]
- Palstra, R.J.; Grosveld, F. Transcription factor binding at enhancers: Shaping a genomic regulatory landscape in flux. Front. Genet. 2012, 3, 195. [Google Scholar] [CrossRef] [PubMed]
- Takeyama, Y.; Sato, M.; Horio, M.; Hase, T.; Yoshida, K.; Yokoyama, T.; Nakashima, H.; Hashimoto, N.; Sekido, Y.; Gazdar, A.F.; et al. Knockdown of ZEB1, a master epithelial-to-mesenchymal transition (EMT) gene, suppresses anchorage-independent cell growth of lung cancer cells. Cancer Lett. 2010, 296, 216–224. [Google Scholar] [CrossRef]
- Graham, T.R.; Yacoub, R.; Taliaferro-Smith, L.; Osunkoya, A.O.; Odero-Marah, V.A.; Liu, T.; Kimbro, K.S.; Sharma, D.; O’Regan, R.M. Reciprocal regulation of ZEB1 and AR in triple negative breast cancer cells. Breast Cancer Res. Treat. 2010, 123, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yan, L.; Xia, L.; Lu, X.; Zhu, W.; Ding, D.; Du, M.; Zhang, D.; Wang, H.; Hu, B. Zinc finger E-box– binding homeobox 1 (ZEB1) is required for neural differentiation of human embryonic stem cells. J. Biol. Chem. 2018, 293, 19317–19329. [Google Scholar] [CrossRef] [PubMed]
- Liao, T.T.; Yang, M.H. Revisiting epithelial-mesenchymal transition in cancer metastasis: The connection between epithelial plasticity and stemness. Mol. Oncol. 2017, 11, 792–804. [Google Scholar] [CrossRef]
- Jolly, M.K.; Somarelli, J.A.; Sheth, M.; Biddle, A.; Tripathi, S.C.; Armstrong, A.J.; Hanash, S.M.; Bapat, S.A.; Rangarajan, A.; Levine, H. Hybrid epithelial/mesenchymal phenotypes promote metastasis and therapy resistance across carcinomas. Pharmacol. Ther. 2019, 194, 161–184. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef] [PubMed]
- DeAngelis, J.T.; Farrington, W.J.; Tollefsbol, T.O. An overview of epigenetic assays. Mol. Biotechnol. 2008, 38, 179–183. [Google Scholar] [CrossRef]
- Kitagawa, Y.; Kyo, S.; Takakura, M.; Kanaya, T.; Koshida, K.; Namiki, M.; Inoue, M. Demethylating reagent 5-azacytidine inhibits telomerase activity in human prostate cancer cells through transcriptional repression of hTERT. Clin. Cancer Res. 2000, 6, 2868–2875. [Google Scholar]
- Ribeiro, A.S.; Paredes, J. P-Cadherin Linking Breast Cancer Stem Cells and Invasion: A Promising Marker to Identify an “Intermediate/Metastable” EMT State. Front. Oncol. 2015, 4, 371. [Google Scholar] [CrossRef] [PubMed]
- Bierie, B.; Pierce, S.E.; Kroeger, C.; Stover, D.G.; Pattabiraman, D.R.; Thiru, P.; Liu Donaher, J.; Reinhardt, F.; Chaffer, C.L.; Keckesova, Z.; et al. Integrin-β4 identifies cancer stem cell-enriched populations of partially mesenchymal carcinoma cells. Proc. Natl. Acad. Sci. USA 2017, 114, E2337–E2346. [Google Scholar] [CrossRef] [PubMed]
- Czibere, A.; Bruns, I.; Kröger, N.; Platzbecker, U.; Lind, J.; Zohren, F.; Fenk, R.; Germing, U.; Schröder, T.; Gräf, T.; et al. 5-Azacytidine for the treatment of patients with acute myeloid leukemia or myelodysplastic syndrome who relapse after allo-SCT: A retrospective analysis. Bone Marrow Transplant. 2010, 45, 872–876. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Wang, L.; Ren, X.; Zhang, Y.; Zhang, H. LRPPRC: A multifunctional protein involved in energy metabolism and human disease. Front. Physiol. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, R.; Mori, Y.; Cheng, Y.; Jin, Z.; Olaru, A.V.; Hamilton, J.P.; David, S.; Selaru, F.M.; Yang, J.; Abraham, J.M.; et al. Silencing of claudin-11 is associated with increased invasiveness of gastric cancer cells. PLoS ONE 2009, 4, e8002. [Google Scholar] [CrossRef] [PubMed]
- Kohn, E.C.; Liotta, L.A. Molecular insights into cancer invasion: Strategies for prevention and intervention. Cancer Res. 1995, 55, 1856–1862. [Google Scholar] [PubMed]
- Liu, S.; Liu, J.; Yu, X.; Shen, T.; Fu, Q. Identification of a Two-Gene (PML-EPB41) Signature With Independent Prognostic Value in Osteosarcoma. Front. Oncol. 2020, 9, 1–11. [Google Scholar] [CrossRef]
- Yuan, W.; Ji, J.; Shu, Y.; Chen, J.; Liu, S.; Wu, L.; Zhou, Z.; Liu, Z.; Tang, Q.; Zhang, X.; et al. Downregulation of DAPK1 promotes the stemness of cancer stem cells and EMT process by activating ZEB1 in colorectal cancer. J. Mol. Med. 2019, 97, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, A.; Kulkarni, D.; Grochola, L.F.; Bond, G.L.; Barnard, N.; Toppmeyer, D.; Levine, A.J.; Hirshfield, K.M. A genetic variant in a PP2A regulatory subunit encoded by the PPP2R2B gene associates with altered breast cancer risk and recurrence. Int. J. Cancer 2011, 128, 2335–2343. [Google Scholar] [CrossRef]
- Peng, C.; Zhang, Z.; Wu, J.; Lv, Z.; Tang, J.; Xie, H.; Zhou, L.; Zheng, S. A critical role for ZDHHC2 in metastasis and recurrence in human hepatocellular carcinoma. BioMed Res. Int. 2014, 2014, 832712. [Google Scholar] [CrossRef]
- Chen, J.; Zhuo, J.Y.; Yang, F.; Liu, Z.K.; Zhou, L.; Xie, H.Y.; Xu, X.; Zheng, S. Sen 17-Beta-Hydroxysteroid Dehydrogenase 13 Inhibits the Progression and Recurrence of Hepatocellular Carcinoma. Hepatobiliary Pancreat. Dis. Int. 2018, 17, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, F.; Yamamoto, M. Identification of genes that exhibit changes in expression on the 8p chromosomal arm by the Systematic Multiplex RT-PCR (SM RT-PCR) and DNA microarray hybridization methods. Gene Expr. 2008, 14, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Legler, K.; Rosprim, R.; Karius, T.; Eylmann, K.; Rossberg, M.; Wirtz, R.M.; Müller, V.; Witzel, I.; Schmalfeldt, B.; Milde-Langosch, K.; et al. Reduced mannosidase MAN1A1 expression leads to aberrant N-glycosylation and impaired survival in breast cancer. Br. J. Cancer 2018, 118, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Tam, W.L.; Weinberg, R.A. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med. 2013, 19, 1438–1449. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Kang, Y. Epithelial-Mesenchymal Plasticity in Cancer Progression and Metastasis. Dev. Cell 2019, 49, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Point, S.S. Infinium HD Methylation Assay Manual Workflow Checklist Convert DNA Create the BCD Plate Infinium HD Methylation Assay Manual Workflow Checklist Incubate DNA Fragment DNA; Illumina: San Diego, CA, USA, 2019; pp. 1–5. [Google Scholar]
- Hansen, K.D.; Aryee, M.; Irizarry, R.A.; Jaffe, A.E.; Maksimovic, J.; Houseman, E.A.; Fortin, J.-P.; Triche, T.; Andrews, S.V.; Hickey, P.F. Available online: https://bioconductor.org/packages/release/bioc/html/minfi.html (accessed on 10 September 2021).
- Tian, Y.; Morris, T.J.; Webster, A.P.; Yang, Z.; Beck, S.; Feber, A.; Teschendorff, A.E. The Chip Analysis Methylation Pipeline. Available online: https://www.bioconductor.org/packages/devel/bioc/vignettes/ChAMP/inst/doc/ChAMP.html (accessed on 10 September 2021).
- Teschendorff, A.E.; Marabita, F.; Lechner, M.; Bartlett, T.; Tegner, J.; Gomez-Cabrero, D.; Beck, S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 2013, 29, 189–196. [Google Scholar] [CrossRef] [PubMed]


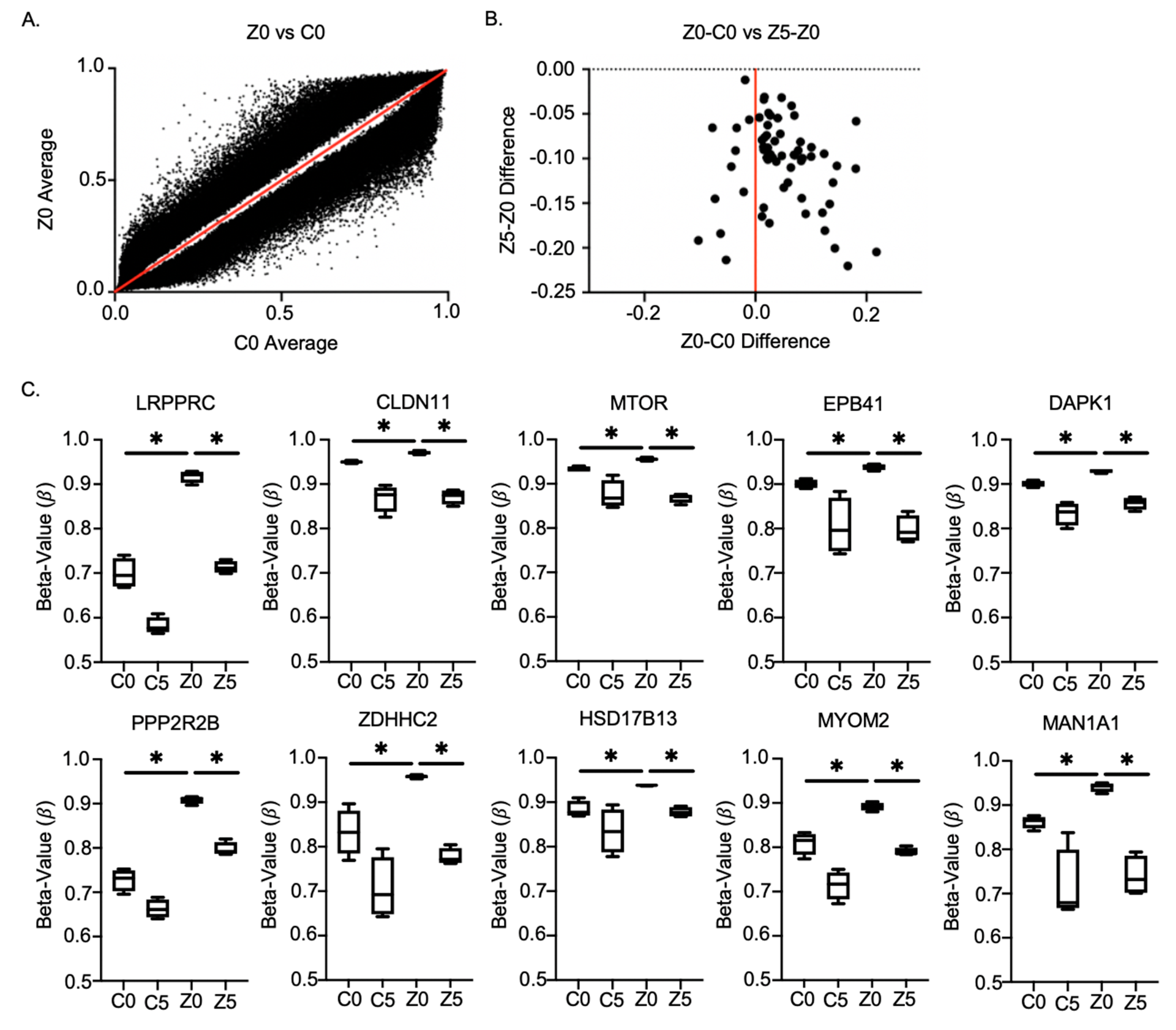
| Gene | Function Relative to Cancer Aggressiveness |
|---|---|
| LRPPRC | Dysregulation is related to various diseases ranging from tumors to viral infections [21]. |
| Claudin-11 | Plays an important role in cellular proliferation and migration [22]. |
| mTOR | Regulates cell growth, proliferation, motility, survival, protein synthesis, autophagy, and transcription [23]. |
| EPB41 | Expression is significantly decreased in HCC tissue specimens, especially in portal vein metastasis or intrahepatic metastasis, compared to normal tissues [24]. |
| DAPK1 | Downregulation promotes the stemness of cancer stem cells and EMT process by activating ZEB1 in colorectal cancer [25]. |
| PPP2RR2B | Negative control of cell growth and division [26]. |
| ZDHHC2 | Tumor suppressor in metastasis and recurrence of HCC [27]. |
| HSD17B13 | Downregulated in hepatocellular carcinoma [28]. |
| MYOM2 | Downregulation was observed in a clinical assessment of breast cancer patients [29]. |
| MAN1A1 | Reduced expression leads to impaired survival in breast cancer [30]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kitz, J.; Lefebvre, C.; Carlos, J.; Lowes, L.E.; Allan, A.L. Reduced Zeb1 Expression in Prostate Cancer Cells Leads to an Aggressive Partial-EMT Phenotype Associated with Altered Global Methylation Patterns. Int. J. Mol. Sci. 2021, 22, 12840. https://doi.org/10.3390/ijms222312840
Kitz J, Lefebvre C, Carlos J, Lowes LE, Allan AL. Reduced Zeb1 Expression in Prostate Cancer Cells Leads to an Aggressive Partial-EMT Phenotype Associated with Altered Global Methylation Patterns. International Journal of Molecular Sciences. 2021; 22(23):12840. https://doi.org/10.3390/ijms222312840
Chicago/Turabian StyleKitz, Jenna, Cory Lefebvre, Joselia Carlos, Lori E. Lowes, and Alison L. Allan. 2021. "Reduced Zeb1 Expression in Prostate Cancer Cells Leads to an Aggressive Partial-EMT Phenotype Associated with Altered Global Methylation Patterns" International Journal of Molecular Sciences 22, no. 23: 12840. https://doi.org/10.3390/ijms222312840
APA StyleKitz, J., Lefebvre, C., Carlos, J., Lowes, L. E., & Allan, A. L. (2021). Reduced Zeb1 Expression in Prostate Cancer Cells Leads to an Aggressive Partial-EMT Phenotype Associated with Altered Global Methylation Patterns. International Journal of Molecular Sciences, 22(23), 12840. https://doi.org/10.3390/ijms222312840






