Identification of Impacted Pathways and Transcriptomic Markers as Potential Mediators of Pulmonary Fibrosis in Transgenic Mice Expressing Human IGFBP5
Abstract
:1. Introduction
2. Results
2.1. Transcriptomic Signature and Functional Enrichment of hIGFBP5 pFBs
2.2. Hub Gene Analysis
2.3. Sex Affects the Impact of hIGFBP5 on the mRNA Levels of Genes of Interest in pFBs
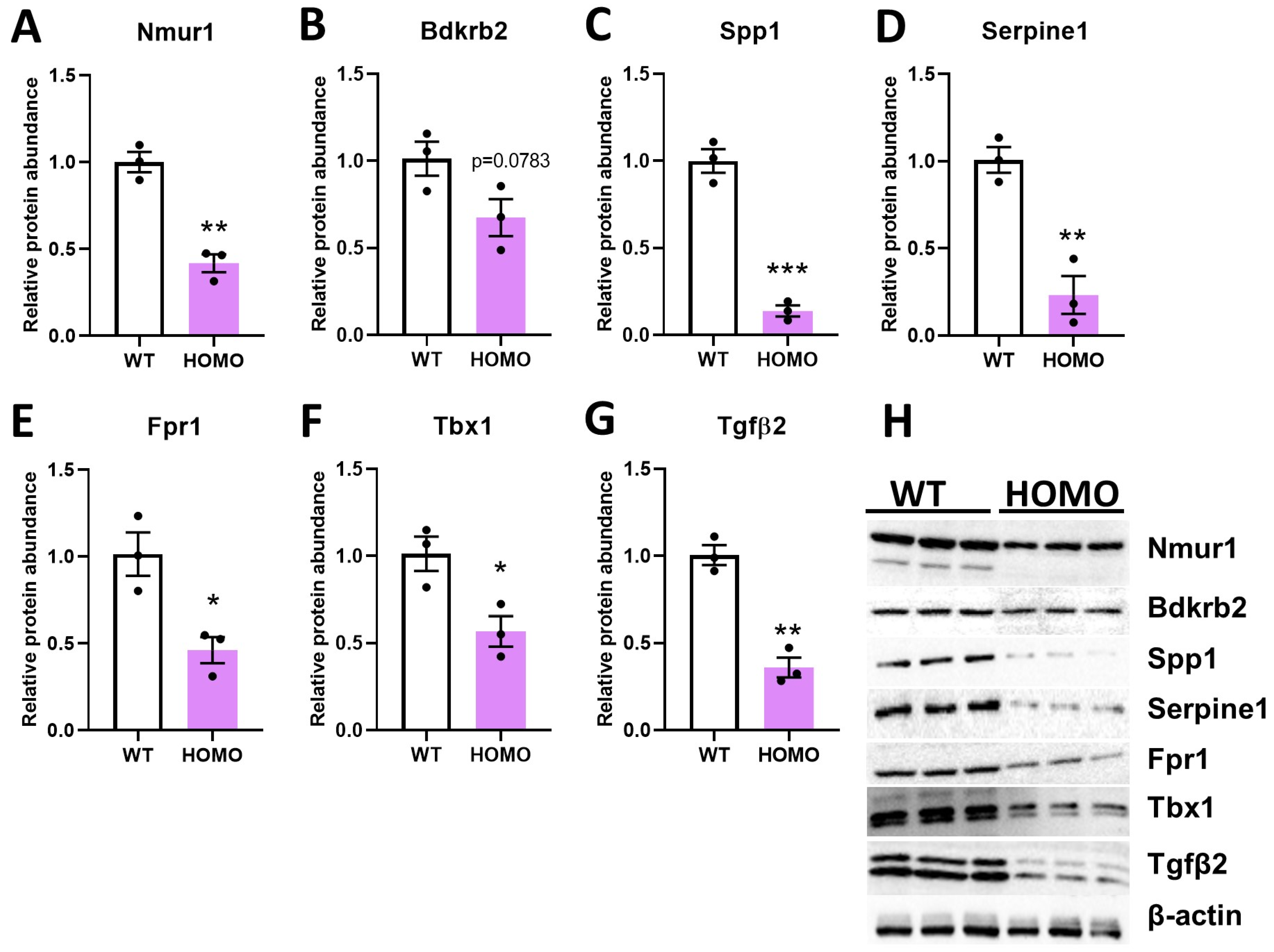
2.4. Effect of hIGFBP5 on Intracellular Protein Abundance
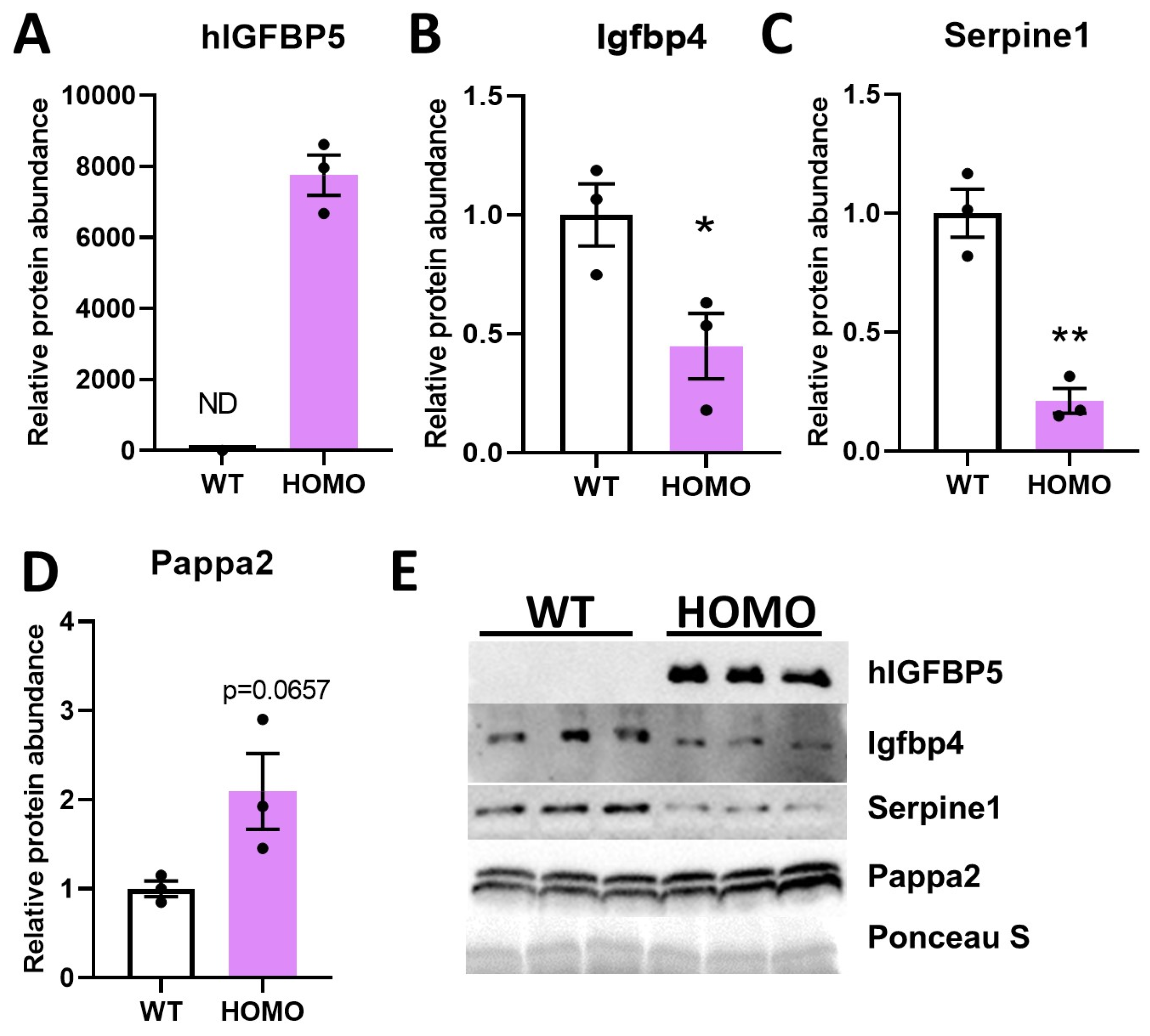
2.5. Effect of hIGFBP5 on the Secretion of Igfbp4, Serpine1 and Pappa2
3. Discussion
3.1. Effect of hIGFBP5 on ECM Macromolecules and ECM-Receptor Interaction
3.2. Effect of hIGFBP5 on AGE-RAGE and Egr1-MAPK Dependent Signaling Pathways
3.3. Effect of hIGFBP5 on Focal Adhesion, Calcium Signaling and Actin Polymerization
3.4. Effect of hIGFBP5 on PI3K-Akt, Cell Survival, and Angiogenesis
3.5. hIGFBP5 Downregulated Igfbp3, Igfbp4 and Igfbp7
3.6. Effect of hIGFBP5 on Neuroactive Ligand-Receptor Interaction
3.7. Overall Impact on Receptors and Integrins
3.8. Downregulation of Bdkrb2, one of the Most Relevant Hub Genes
3.9. Effect of hIGFBP5 on Chemokine and Cytokine Signaling
3.10. Endogenous vs. Exogenous IGFBP5
3.11. Sex Differences in the Response of Fibroblasts
3.12. Conclusions
4. Materials and Methods
4.1. Ethics Statement
4.2. Culture of Primary Mouse Lung Fibroblasts
4.3. RNA Extraction and Preparation
4.4. RNA Sequencing & Differential Expression Analysis
4.5. Functional Enrichment & Hub Gene Network Analysis
4.6. Quantitative Reverse Transcription (qRT-PCR)
4.7. Immunoblotting
4.8. Ponceau S Staining
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Varga, J.; Abraham, D. Systemic sclerosis: A prototypic multisystem fibrotic disorder. J Clin. Investig. 2007, 117, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.H.; Feghali-Bostwick, C.A.; Silver, R.M. Update on scleroderma-associated interstitial lung disease. Curr. Opin. Rheumatol. 2014, 26, 630–636. [Google Scholar] [CrossRef]
- Steen, V.D.; Medsger, T.A. Changes in causes of death in systemic sclerosis, 1972–2002. Ann. Rheum. Dis. 2007, 66, 940–944. [Google Scholar] [CrossRef] [Green Version]
- Tyndall, A.J.; Bannert, B.; Vonk, M.; Airò, P.; Cozzi, F.; Carreira, P.E.; Bancel, D.F.; Allanore, Y.; Müller-Ladner, U.; Distler, O. Causes and risk factors for death in systemic sclerosis: A study from the EULAR Scleroderma Trials and Research (EUSTAR) database. Ann. Rheum. Dis. 2010, 69, 1809–1815. [Google Scholar] [CrossRef] [Green Version]
- Herzog, E.L.; Mathur, A.; Tager, A.M.; Feghali-Bostwick, C.; Schneider, F.; Varga, J. Interstitial lung disease associated with systemic sclerosis and idiopathic pulmonary fibrosis: How similar and distinct? Arthritis Rheumatol. 2014, 66, 1967. [Google Scholar] [CrossRef] [PubMed]
- James, P.L.; Jones, S.; Busby, W., Jr.; Clemmons, D.; Rotwein, P. A highly conserved insulin-like growth factor-binding protein (IGFBP-5) is expressed during myoblast differentiation. J. Biol. Chem. 1993, 268, 22305–22312. [Google Scholar] [CrossRef]
- Shimasaki, S.; Ling, N. Identification and molecular characterization of insulin-like growth factor binding proteins (IGFBP-1,-2,-3,-4,-5 and -6). Prog. Growth Factor Res. 1991, 3, 243–266. [Google Scholar] [CrossRef]
- Xu, Q.; Li, S.; Zhao, Y.; Maures, T.J.; Yin, P.; Duan, C. Evidence that IGF binding protein-5 functions as a ligand-independent transcriptional regulator in vascular smooth muscle cells. Circ. Res. 2004, 94, E46–E54. [Google Scholar] [CrossRef]
- Su, Y.; Nishimoto, T.; Feghali-Bostwick, C. IGFBP-5 promotes fibrosis independently of its translocation to the nucleus and its interaction with nucleolin and IGF. PLoS ONE 2015, 10, e0130546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feghali, C.A.; Wright, T.M. Identification of multiple, differentially expressed messenger RNAs in dermal fibroblasts from patients with systemic sclerosis. Arthritis Rheum. 1999, 42, 1451–1457. [Google Scholar] [CrossRef]
- Nguyen, X.X.; Muhammad, L.; Nietert, P.J.; Feghali-Bostwick, C. IGFBP-5 Promotes Fibrosis via Increasing Its Own Expression and That of Other Pro-fibrotic Mediators. Front. Endocrinol. 2018, 9, 601. [Google Scholar] [CrossRef] [PubMed]
- Pilewski, J.M.; Liu, L.; Henry, A.C.; Knauer, A.V.; Feghali-Bostwick, C.A. Insulin-like growth factor binding proteins 3 and 5 are overexpressed in idiopathic pulmonary fibrosis and contribute to extracellular matrix deposition. Am. J. Pathol. 2005, 166, 399–407. [Google Scholar] [CrossRef] [Green Version]
- Yasuoka, H.; Jukic, D.M.; Zhou, Z.; Choi, A.M.; Feghali-Bostwick, C.A. Insulin-like growth factor binding protein 5 induces skin fibrosis: A novel murine model for dermal fibrosis. Arthritis Rheum. 2006, 54, 3001–3010. [Google Scholar] [CrossRef]
- Yasuoka, H.; Zhou, Z.; Pilewski, J.M.; Oury, T.D.; Choi, A.M.; Feghali-Bostwick, C.A. Insulin-like growth factor-binding protein-5 induces pulmonary fibrosis and triggers mononuclear cellular infiltration. Am. J. Pathol. 2006, 169, 1633–1642. [Google Scholar] [CrossRef] [Green Version]
- Yasuoka, H.; Larregina, A.T.; Yamaguchi, Y.; Feghali-Bostwick, C.A. Human skin culture as an ex vivo model for assessing the fibrotic effects of insulin-like growth factor binding proteins. Open Rheumatol. J. 2008, 2, 17–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, X.X.; Sanderson, M.; Helke, K.; Feghali-Bostwick, C. Phenotypic Characterization of Transgenic Mice Expressing Human IGFBP-5. Int. J. Mol. Sci. 2020, 22, 335. [Google Scholar] [CrossRef] [PubMed]
- Yasuoka, H.; Hsu, E.; Ruiz, X.D.; Steinman, R.A.; Choi, A.M.; Feghali-Bostwick, C.A. The fibrotic phenotype induced by IGFBP-5 is regulated by MAPK activation and egr-1-dependent and-independent mechanisms. Am. J. Pathol. 2009, 175, 605–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasuoka, H.; Yamaguchi, Y.; Feghali-Bostwick, C.A. The membrane-associated adaptor protein DOK5 is upregulated in systemic sclerosis and associated with IGFBP-5-induced fibrosis. PLoS ONE 2014, 9, e87754. [Google Scholar] [CrossRef] [Green Version]
- Overgaard, M.T.; Boldt, H.B.; Laursen, L.S.; Sottrup-Jensen, L.; Conover, C.A.; Oxvig, C. Pregnancy-associated plasma protein-A2 (PAPP-A2): A novel insulin-like growth factor-binding protein-5 proteinase. J. Biol. Chem. 2001, 276, 21849–21853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil-Cayuela, C.; Rivera, M.; Ortega, A.; Tarazón, E.; Triviño, J.C.; Lago, F.; González-Juanatey, J.R.; Almenar, L.; Martínez-Dolz, L.; Portolés, M. RNA sequencing analysis identifies new human collagen genes involved in cardiac remodeling. J. Am. Coll. Cardiol. 2015, 65, 1265–1267. [Google Scholar] [CrossRef] [Green Version]
- Matson, S.; Lee, J.; Ren, W.; Collard, H.; Matthay, M.; Achtar-Zadeh, N.; Wolters, P.; Hansen, K.; Eickelberg, O. Common and Distinct Transcriptome and Proteome Expression Patterns from Lungs in Idiopathic Pulmonary Fibrosis (IPF) and Rheumatoid Arthritis-Associated Interstitial Lung Disease (RA-ILD). In Proceedings of the C58. Mechanisms of pulmonary fibrosis: American Thoracic Society 2019 International Conference, Dallas, TX, USA, 17–22 May 2019; p. A5258. [Google Scholar]
- Pace, J.M.; Corrado, M.; Missero, C.; Byers, P.H. Identification, characterization and expression analysis of a new fibrillar collagen gene, COL27A1. Matrix Biol. 2003, 22, 3–14. [Google Scholar] [CrossRef]
- Deng, G.; Le Li, Y.O. Modeling paraquat-induced lung fibrosis in C. elegans reveals KRIT1 as a key regulator of collagen gene transcription. Aging 2021, 13, 4452. [Google Scholar] [CrossRef]
- Bornstein, P. Diversity of function is inherent in matricellular proteins: An appraisal of thrombospondin 1. J. Cell Biol. 1995, 130, 503–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Xiong, Z.; Lechner, E.J.; Klenotic, P.A.; Hamburg, B.J.; Hulver, M.; Khare, A.; Oriss, T.; Mangalmurti, N.; Chan, Y. Thrombospondin-1 triggers macrophage IL-10 production and promotes resolution of experimental lung injury. Mucosal Immunol. 2014, 7, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Latoche, J.D.; Ufelle, A.C.; Fazzi, F.; Ganguly, K.; Leikauf, G.D.; Fattman, C.L. Secreted phosphoprotein 1 and sex-specific differences in silica-induced pulmonary fibrosis in mice. Environ. Health Perspect. 2016, 124, 1199–1207. [Google Scholar] [CrossRef] [Green Version]
- Sato, S.; Nagaoka, T.; Hasegawa, M.; Tamatani, T.; Nakanishi, T.; Takigawa, M.; Takehara, K. Serum levels of connective tissue growth factor are elevated in patients with systemic sclerosis: Association with extent of skin sclerosis and severity of pulmonary fibrosis. J. Rheumatol. 2000, 27, 149–154. [Google Scholar]
- Cui, Y.; Ji, J.; Hou, J.; Tan, Y.; Han, X. Identification of Key Candidate Genes Involved in the Progression of Idiopathic Pulmonary Fibrosis. Molecules 2021, 26, 1123. [Google Scholar] [CrossRef]
- Garner, I.M.; Evans, I.C.; Barnes, J.L.; Maher, T.M.; Renzoni, E.A.; Denton, C.P.; Scotton, C.J.; Abraham, D.J.; McAnulty, R.J. Hypomethylation of the TNXB gene contributes to increased expression and deposition of tenascin-X in idiopathic pulmonary fibrosis. In Proceedings of the B59: Putting the Genie Back in The Bottle: Regulating Gene Expression—American Thoracic Society 2014 International Conference, San Diego, CA, USA, 16–21 May 2014; p. A3378. [Google Scholar]
- Estany, S.; Vicens-Zygmunt, V.; Llatjós, R.; Montes, A.; Penín, R.; Escobar, I.; Xaubet, A.; Santos, S.; Manresa, F.; Dorca, J. Lung fibrotic tenascin-C upregulation is associated with other extracellular matrix proteins and induced by TGFβ1. BMC Pulm. Med. 2014, 14, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sureshbabu, A.; Tonner, E.; Allan, G.; Flint, D. Relative roles of TGF-β and IGFBP-5 in idiopathic pulmonary fibrosis. Pulm. Med. 2011, 2011, 517687. [Google Scholar] [CrossRef] [Green Version]
- Steiglitz, B.M.; Keene, D.R.; Greenspan, D.S. PCOLCE2 encodes a functional procollagen C-proteinase enhancer (PCPE2) that is a collagen-binding protein differing in distribution of expression and post-translational modification from the previously described PCPE1. J. Biol. Chem. 2002, 277, 49820–49830. [Google Scholar] [CrossRef] [Green Version]
- Baicu, C.F.; Zhang, Y.; Van Laer, A.O.; Renaud, L.; Zile, M.R.; Bradshaw, A.D. Effects of the absence of procollagen C-endopeptidase enhancer-2 on myocardial collagen accumulation in chronic pressure overload. Am. J. Physiol. Heart Circ. Physiol. 2012, 303, H234–H240. [Google Scholar] [CrossRef] [Green Version]
- Ramasamy, R.; Yan, S.F.; Schmidt, A.M. Receptor for AGE (RAGE): Signaling mechanisms in the pathogenesis of diabetes and its complications. Ann. N. Y. Acad. Sci. 2011, 1243, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machahua, C.; Montes-Worboys, A.; Llatjos, R.; Escobar, I.; Dorca, J.; Molina-Molina, M.; Vicens-Zygmunt, V. Increased AGE-RAGE ratio in idiopathic pulmonary fibrosis. Respir. Res. 2016, 17, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Nedić, O.; Rattan, S.; Grune, T.; Trougakos, I. Molecular effects of advanced glycation end products on cell signalling pathways, ageing and pathophysiology. Free Radic. Res. 2013, 47, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Kliment, C.R.; Oury, T.D. Oxidative stress, extracellular matrix targets, and idiopathic pulmonary fibrosis. Free Radic. Biol. Med. 2010, 49, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, I.; Xu, S.; Denton, C.P.; Abraham, D.J.; Ponticos, M. STAT3 controls COL1A2 enhancer activation cooperatively with JunB, regulates type I collagen synthesis posttranscriptionally, and is essential for lung myofibroblast differentiation. Mol. Biol. Cell 2018, 29, 84–95. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Lu, W.; Zhang, X.; Lu, J.; Xu, S.; Chen, S.; Zhong, Z.; Zhou, T.; Wang, Q.; Chen, J. Cryptotanshinone protects against pulmonary fibrosis through inhibiting Smad and STAT3 signaling pathways. Pharmacol. Res. 2019, 147, 104307. [Google Scholar] [CrossRef]
- Kinoshita, K.; Aono, Y.; Azuma, M.; Kishi, J.; Takezaki, A.; Kishi, M.; Makino, H.; Okazaki, H.; Uehara, H.; Izumi, K. Antifibrotic effects of focal adhesion kinase inhibitor in bleomycin-induced pulmonary fibrosis in mice. Am. J. Respir. Cell Mol. Biol. 2013, 49, 536–543. [Google Scholar] [CrossRef] [PubMed]
- Kuragano, M.; Uyeda, T.Q.; Kamijo, K.; Murakami, Y.; Takahashi, M. Different contributions of nonmuscle myosin IIA and IIB to the organization of stress fiber subtypes in fibroblasts. Mol. Biol. Cell 2018, 29, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Webb, D.J.; Donais, K.; Whitmore, L.A.; Thomas, S.M.; Turner, C.E.; Parsons, J.T.; Horwitz, A.F. FAK–Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 2004, 6, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Levinson, H.; Moyer, K.E.; Saggers, G.C.; Paul Ehrlich, H. Calmodulin-myosin light chain kinase inhibition changes fibroblast-populated collagen lattice contraction, cell migration, focal adhesion formation, and wound contraction. Wound Repair Regen. 2004, 12, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Lembong, J.; Sabass, B.; Stone, H.A. Calcium oscillations in wounded fibroblast monolayers are spatially regulated through substrate mechanics. Phys. Biol. 2017, 14, 045006. [Google Scholar] [CrossRef] [PubMed]
- Janssen, L.J.; Mukherjee, S.; Ask, K. Calcium Homeostasis and Ionic Mechanisms in Pulmonary Fibroblasts. Am. J. Respir. Cell Mol. Biol. 2015, 53, 135–148. [Google Scholar] [CrossRef]
- Mukherjee, S.; Ayaub, E.A.; Murphy, J.; Lu, C.; Kolb, M.; Ask, K.; Janssen, L.J. Disruption of Calcium Signaling in Fibroblasts and Attenuation of Bleomycin-Induced Fibrosis by Nifedipine. Am. J. Respir. Cell Mol. Biol. 2015, 53, 450–458. [Google Scholar] [CrossRef] [Green Version]
- Jaffar, J.; Yang, S.-H.; Kim, S.Y.; Kim, H.-W.; Faiz, A.; Chrzanowski, W.; Burgess, J.K. Greater cellular stiffness in fibroblasts from patients with idiopathic pulmonary fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2018, 315, L59–L65. [Google Scholar] [CrossRef] [Green Version]
- Chaigne, B.; Clary, G.; Le Gall, M.; Dumoitier, N.; Fernandez, C.; Lofek, S.; Chafey, P.; Moinzadeh, P.; Krieg, T.; Denton, C.P. Proteomic Analysis of Human Scleroderma Fibroblasts Response to Transforming Growth Factor-β. Proteom. Clin. Appl. 2019, 13, 1800069. [Google Scholar] [CrossRef] [PubMed]
- Berfield, A.K.; Andress, D.L.; Abrass, C.K. IGFBP-5201-218 stimulates Cdc42GAP aggregation and filopodia formation in migrating mesangial cells. Kidney Int. 2000, 57, 1991–2003. [Google Scholar] [CrossRef] [Green Version]
- Kojima, H.; Kunimoto, H.; Inoue, T.; Nakajima, K. The STAT3-IGFBP5 axis is critical for IL-6/gp130-induced premature senescence in human fibroblasts. Cell Cycle 2012, 11, 730–739. [Google Scholar] [CrossRef]
- Cayrol, C.; Girard, J.-P. IL-33: An alarmin cytokine with crucial roles in innate immunity, inflammation and allergy. Curr. Opin. Immunol. 2014, 31, 31–37. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, V.N.; Zhou, H.; Ghandhi, S.A.; Karasic, T.B.; Yaghoubian, B.; Amundson, S.A.; Hei, T.K. Radiation-induced bystander signaling pathways in human fibroblasts: A role for interleukin-33 in the signal transmission. Cell. Signal. 2010, 22, 1076–1087. [Google Scholar] [CrossRef] [Green Version]
- Luzina, I.G.; Kopach, P.; Lockatell, V.; Kang, P.H.; Nagarsekar, A.; Burke, A.P.; Hasday, J.D.; Todd, N.W.; Atamas, S.P. Interleukin-33 potentiates bleomycin-induced lung injury. Am. J. Respir. Cell Mol. Biol. 2013, 49, 999–1008. [Google Scholar] [CrossRef]
- Ebina, M.; Shimizukawa, M.; Shibata, N.; Kimura, Y.; Suzuki, T.; Endo, M.; Sasano, H.; Kondo, T.; Nukiwa, T. Heterogeneous increase in CD34-positive alveolar capillaries in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2004, 169, 1203–1208. [Google Scholar] [CrossRef]
- Hsu, E.; Shi, H.; Jordan, R.M.; Lyons-Weiler, J.; Pilewski, J.M.; Feghali-Bostwick, C.A. Lung tissues in patients with systemic sclerosis have gene expression patterns unique to pulmonary fibrosis and pulmonary hypertension. Arthritis Rheum. 2011, 63, 783–794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, Y.; Nishimoto, T.; Hoffman, S.; Nguyen, X.X.; Pilewski, J.M.; Feghali-Bostwick, C. Insulin-like growth factor binding protein-4 exerts antifibrotic activity by reducing levels of connective tissue growth factor and the C-X-C chemokine receptor 4. FASEB Bioadv. 2019, 1, 167–179. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Liu, Y.; Wang, B. Identification of transcriptomic markers for developing idiopathic pulmonary fibrosis: An integrative analysis of gene expression profiles. Int. J. Clin. Exp. Pathol. 2020, 13, 1698–1706. [Google Scholar]
- Mishra, S.; Shah, M.I.; Udhaya Kumar, S.; Thirumal Kumar, D.; Gopalakrishnan, C.; Al-Subaie, A.M.; Magesh, R.; George Priya Doss, C.; Kamaraj, B. Networsk analysis of transcriptomics data for the prediction and prioritization of membrane-associated biomarkers for idiopathic pulmonary fibrosis (IPF) by bioinformatics approach. Adv. Protein Chem. Struct. Biol. 2021, 123, 241–273. [Google Scholar] [CrossRef] [PubMed]
- Malaab, M.; Renaud, L.; Takamura, N.; Zimmerman, K.D.; da Silveira, W.A.; Ramos, P.S.; Haddad, S.; Peters-Golden, M.; Penke, L.R.; Wolf, B. Antifibrotic factor KLF4 is repressed by the miR-10/TFAP2A/TBX5 axis in dermal fibroblasts: Insights from twins discordant for systemic sclerosis. Ann. Rheum. Dis. 2021, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.Y.; Yu, J. Sensory nerves in lung and airways. Compr. Physiol. 2014, 4, 287–324. [Google Scholar] [CrossRef]
- Lee, Y.S.; Jung, W.Y.; Heo, H.; Park, M.G.; Oh, S.-H.; Park, B.-G.; Kim, S. Exosome-mediated ultra-effective direct conversion of human fibroblasts into neural progenitor-like cells. ACS Nano 2018, 12, 2531–2538. [Google Scholar] [CrossRef]
- Wan, X.Y.; Xu, L.Y.; Li, B.; Sun, Q.H.; Ji, Q.L.; Huang, D.D.; Zhao, L.; Xiao, Y.T. Chemical conversion of human lung fibroblasts into neuronal cells. Int. J. Mol. Med. 2018, 41, 1463–1468. [Google Scholar] [CrossRef] [Green Version]
- Sureshbabu, A.; Okajima, H.; Yamanaka, D.; Tonner, E.; Shastri, S.; Maycock, J.; Szymanowska, M.; Shand, J.; Takahashi, S.-I.; Beattie, J. IGFBP5 induces cell adhesion, increases cell survival and inhibits cell migration in MCF-7 human breast cancer cells. J. Cell Sci. 2012, 125, 1693–1705. [Google Scholar] [PubMed] [Green Version]
- Nam, T.-J.; Busby, W.H., Jr.; Rees, C.; Clemmons, D.R. Thrombospondin and osteopontin bind to insulin-like growth factor (IGF)-binding protein-5 leading to an alteration in IGF-I-stimulated cell growth. Endocrinology 2000, 141, 1100–1106. [Google Scholar] [CrossRef]
- Dagher, O.K.; Jaffa, M.A.; Habib, A.; Ziyadeh, F.N.; Jaffa, A.A. Heteromerization fingerprints between bradykinin B2 and thromboxane TP receptors in native cells. PLoS ONE 2019, 14, e0216908. [Google Scholar] [CrossRef]
- Schanstra, J.P.; Neau, E.; Drogoz, P.; Arevalo Gomez, M.A.; Lopez Novoa, J.M.; Calise, D.; Pecher, C.; Bader, M.; Girolami, J.P.; Bascands, J.L. In vivo bradykinin B2 receptor activation reduces renal fibrosis. J. Clin. Investig. 2002, 110, 371–379. [Google Scholar] [CrossRef]
- De Prins, A.; Martin, C.; Van Wanseele, Y.; Skov, L.J.; Tömböly, C.; Tourwé, D.; Caveliers, V.; Van Eeckhaut, A.; Holst, B.; Rosenkilde, M.M. Development of potent and proteolytically stable human neuromedin U receptor agonists. Eur. J. Med. Chem. 2018, 144, 887–897. [Google Scholar] [CrossRef] [Green Version]
- Takayama, K.; Mori, K.; Tanaka, A.; Nomura, E.; Sohma, Y.; Mori, M.; Taguchi, A.; Taniguchi, A.; Sakane, T.; Yamamoto, A. Discovery of a human neuromedin U receptor 1-selective hexapeptide agonist with enhanced serum stability. J. Med. Chem. 2017, 60, 5228–5234. [Google Scholar] [CrossRef]
- Numajiri, H.; Kuzumi, A.; Fukasawa, T.; Ebata, S.; Yoshizaki-Ogawa, A.; Asano, Y.; Kazoe, Y.; Mawatari, K.; Kitamori, T.; Yoshizaki, A. B cell depletion inhibits fibrosis via suppressing pro-fibrotic macrophage differentiation in a mouse model of systemic sclerosis. Arthritis Rheumatol. 2021, 73, 2086–2095. [Google Scholar] [CrossRef]
- Fraticelli, P.; Fischetti, C.; Salaffi, F.; Carotti, M.; Mattioli, M.; Pomponio, G.; Gabrielli, A. Combination therapy with rituximab and mycophenolate mofetil in systemic sclerosis. A single-centre case series study. Clin. Exp. Rheumatol. 2018, 36, 142–145. [Google Scholar] [PubMed]
- Servettaz, A.; Kavian, N.; Nicco, C.; Deveaux, V.; Chéreau, C.; Wang, A.; Zimmer, A.; Lotersztajn, S.; Weill, B.; Batteux, F. Targeting the cannabinoid pathway limits the development of fibrosis and autoimmunity in a mouse model of systemic sclerosis. Am. J. Pathol. 2010, 177, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Makino, H.; Aono, Y.; Azuma, M.; Kishi, M.; Yokota, Y.; Kinoshita, K.; Takezaki, A.; Kishi, J.; Kawano, H.; Ogawa, H. Antifibrotic effects of CXCR4 antagonist in bleomycin-induced pulmonary fibrosis in mice. J. Med. Investig. 2013, 60, 127–137. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Xu, X.; Geng, J.; Wan, X.; Dai, H. The autocrine CXCR4/CXCL12 axis contributes to lung fibrosis through modulation of lung fibroblast activity. Exp. Ther. Med. 2020, 19, 1844–1854. [Google Scholar] [CrossRef]
- Griffiths, K.; Habiel, D.; Jaffar, J.; Binder, U.; Darby, W.; Hosking, C.; Skerra, A.; Westall, G.; Hogaboam, C.; Foley, M. Anti-fibrotic effects of CXCR4-targeting i-body AD-114 in preclinical models of pulmonary fibrosis. Sci. Rep. 2018, 8, 3212. [Google Scholar] [CrossRef] [Green Version]
- Higashi, A.Y.; Aronow, B.J.; Dressler, G.R. Expression profiling of fibroblasts in chronic and acute disease models reveals novel pathways in kidney fibrosis. J. Am. Soc. Nephrol. 2019, 30, 80–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Black, L.M.; Lever, J.M.; Agarwal, A. Renal inflammation and fibrosis: A double-edged sword. J. Histochem. Cytochem. 2019, 67, 663–681. [Google Scholar] [CrossRef] [Green Version]
- Arauz, J.; Rivera-Espinoza, Y.; Shibayama, M.; Favari, L.; Flores-Beltrán, R.E.; Muriel, P. Nicotinic acid prevents experimental liver fibrosis by attenuating the prooxidant process. Int. Immunopharmacol. 2015, 28, 244–251. [Google Scholar] [CrossRef]
- Strieter, R.M.; Gomperts, B.N.; Keane, M.P. The role of CXC chemokines in pulmonary fibrosis. J. Clin. Investig. 2007, 117, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Das, A.M.; Seideman, J.; Griswold, D.; Afuh, C.N.; Kobayashi, T.; Abe, S.; Fang, Q.; Hashimoto, M.; Kim, H.; et al. The CC chemokine ligand 2 (CCL2) mediates fibroblast survival through IL-6. Am. J. Respir. Cell Mol. Biol. 2007, 37, 121–128. [Google Scholar] [CrossRef] [Green Version]
- Phillips, R.J.; Burdick, M.D.; Hong, K.; Lutz, M.A.; Murray, L.A.; Xue, Y.Y.; Belperio, J.A.; Keane, M.P.; Strieter, R.M. Circulating fibrocytes traffic to the lungs in response to CXCL12 and mediate fibrosis. J. Clin. Investig. 2004, 114, 438–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, B.B.; Murray, L.; Das, A.; Wilke, C.A.; Herrygers, A.B.; Toews, G.B. The role of CCL12 in the recruitment of fibrocytes and lung fibrosis. Am. J. Respir. Cell Mol. Biol. 2006, 35, 175–181. [Google Scholar] [CrossRef] [Green Version]
- Yin, P.; Xu, Q.; Duan, C. Paradoxical actions of endogenous and exogenous insulin-like growth factor-binding protein-5 revealed by RNA interference analysis. J. Biol. Chem. 2004, 279, 32660–32666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voltz, J.W.; Card, J.W.; Carey, M.A.; DeGraff, L.M.; Ferguson, C.D.; Flake, G.P.; Bonner, J.C.; Korach, K.S.; Zeldin, D.C. Male sex hormones exacerbate lung function impairment after bleomycin-induced pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2008, 39, 45–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gharaee-Kermani, M.; Hatano, K.; Nozaki, Y.; Phan, S.H. Gender-based differences in bleomycin-induced pulmonary fibrosis. Am. J. Pathol. 2005, 166, 1593–1606. [Google Scholar] [CrossRef] [Green Version]
- Salih, D.A.; Mohan, S.; Kasukawa, Y.; Tripathi, G.; Lovett, F.A.; Anderson, N.F.; Carter, E.J.; Wergedal, J.E.; Baylink, D.J.; Pell, J.M. Insulin-like growth factor-binding protein-5 induces a gender-related decrease in bone mineral density in transgenic mice. Endocrinology 2005, 146, 931–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adesanya, O.O.; Zhou, J.; Bondy, C.A. Cellular localization and sex steroid regulation of insulin-like growth factor binding protein messenger ribonucleic acids in the primate myometrium. J. Clin. Endocrinol. Metab. 1996, 81, 2495–2501. [Google Scholar]
- Onoda, N.; Li, D.; Mickey, G.; Erickson, G.; Shimasaki, S. Gonadotropin-releasing hormone overcomes follicle-stimulating hormone’s inhibition of insulin-like growth factor-5 synthesis and promotion of its degradation in rat granulosa cells. Mol. Cell. Endocrinol. 1995, 110, 17–25. [Google Scholar] [CrossRef]
- Boonyaratanakornkit, V.; Strong, D.D.; Mohan, S.; Baylink, D.J.; Beck, C.A.; Linkhart, T.A. Progesterone stimulation of human insulin-like growth factor-binding protein-5 gene transcription in human osteoblasts is mediated by a CACCC sequence in the proximal promoter. J. Biol. Chem. 1999, 274, 26431–26438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregory, C.W.; Kim, D.; Ye, P.; D’Ercole, A.J.; Pretlow, T.G.; Mohler, J.L.; French, F.S. Androgen receptor up-regulates insulin-like growth factor binding protein-5 (IGFBP-5) expression in a human prostate cancer xenograft. Endocrinology 1999, 140, 2372–2381. [Google Scholar] [CrossRef]
- Peoples, C.; Medsger, T.A., Jr.; Lucas, M.; Rosario, B.L.; Feghali-Bostwick, C.A. Gender differences in systemic sclerosis: Relationship to clinical features, serologic status and outcomes. J. Scleroderma Relat. Disord. 2016, 1, 204–212. [Google Scholar] [CrossRef] [Green Version]
- Aida-Yasuoka, K.; Peoples, C.; Yasuoka, H.; Hershberger, P.; Thiel, K.; Cauley, J.A.; Medsger, T.A.; Feghali-Bostwick, C.A. Estradiol promotes the development of a fibrotic phenotype and is increased in the serum of patients with systemic sclerosis. Arthritis Res. Ther. 2013, 15, R10. [Google Scholar] [CrossRef] [Green Version]
- Lou, Q.; Cao, S.; Xu, W.; Zhang, Y.; Qin, Z.; Wei, W. Molecular characterization and mRNA expression of ribosomal protein L8 in Rana nigromaculata during development and under exposure to hormones. J. Environ. Sci. 2014, 26, 2331–2339. [Google Scholar] [CrossRef]
- Stephens, S.B.; Kauffman, A.S. Estrogen regulation of the molecular phenotype and active translatome of AVPV kisspeptin neurons. Endocrinology 2021, 162, bqab080. [Google Scholar] [CrossRef] [PubMed]
- Oliva, M.; Muñoz-Aguirre, M.; Kim-Hellmuth, S.; Wucher, V.; Gewirtz, A.D.; Cotter, D.J.; Parsana, P.; Kasela, S.; Balliu, B.; Viñuela, A. The impact of sex on gene expression across human tissues. Science 2020, 369, eaba3066. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cahill, T.; da Silveira, W.A.; Renaud, L.; Williamson, T.; Wang, H.; Chung, D.; Overton, I.; Chan, S.S.; Hardiman, G. Induced Torpor as a Countermeasure for Low Dose Radiation Exposure in a Zebrafish Model. Cells 2021, 10, 906. [Google Scholar] [CrossRef]
- Zaman, M.S.; Barman, S.K.; Corley, S.M.; Wilkins, M.R.; Malladi, C.S.; Wu, M.J. Transcriptomic insights into the zinc homeostasis of MCF-7 breast cancer cells via next-generation RNA sequencing. Metallomics 2021, 13, mfab026. [Google Scholar] [CrossRef]
- Draghici, S.; Khatri, P.; Tarca, A.L.; Amin, K.; Done, A.; Voichita, C.; Georgescu, C.; Romero, R. A systems biology approach for pathway level analysis. Genome Res. 2007, 17, 1537–1545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarca, A.L.; Draghici, S.; Khatri, P.; Hassan, S.S.; Mittal, P.; Kim, J.-S.; Kim, C.J.; Kusanovic, J.P.; Romero, R. A novel signaling pathway impact analysis. Bioinformatics 2009, 25, 75–82. [Google Scholar] [CrossRef] [Green Version]
- Khatri, P.; Draghici, S.; Tarca, A.L.; Hassan, S.S.; Romero, R. A system biology approach for the steady-state analysis of gene signaling networks. In Proceedings of the Iberoamerican Congress on Pattern Recognition, Valparaíso, Chile, 13–16 November 2007; pp. 32–41. [Google Scholar]
- Nguyen, T.-M.; Shafi, A.; Nguyen, T.; Draghici, S. Identifying significantly impacted pathways: A comprehensive review and assessment. Genome Biol. 2019, 20, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Nakaya, A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002, 30, 42–46. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Gene Ontology Consortium. Creating the gene ontology resource: Design and implementation. Genome Res. 2001, 11, 1425–1433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koschützki, D.; Schreiber, F. Centrality analysis methods for biological networks and their application to gene regulatory networks. Gene Regul. Syst. Biol. 2008, 2, GRSB-S702. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef] [PubMed]

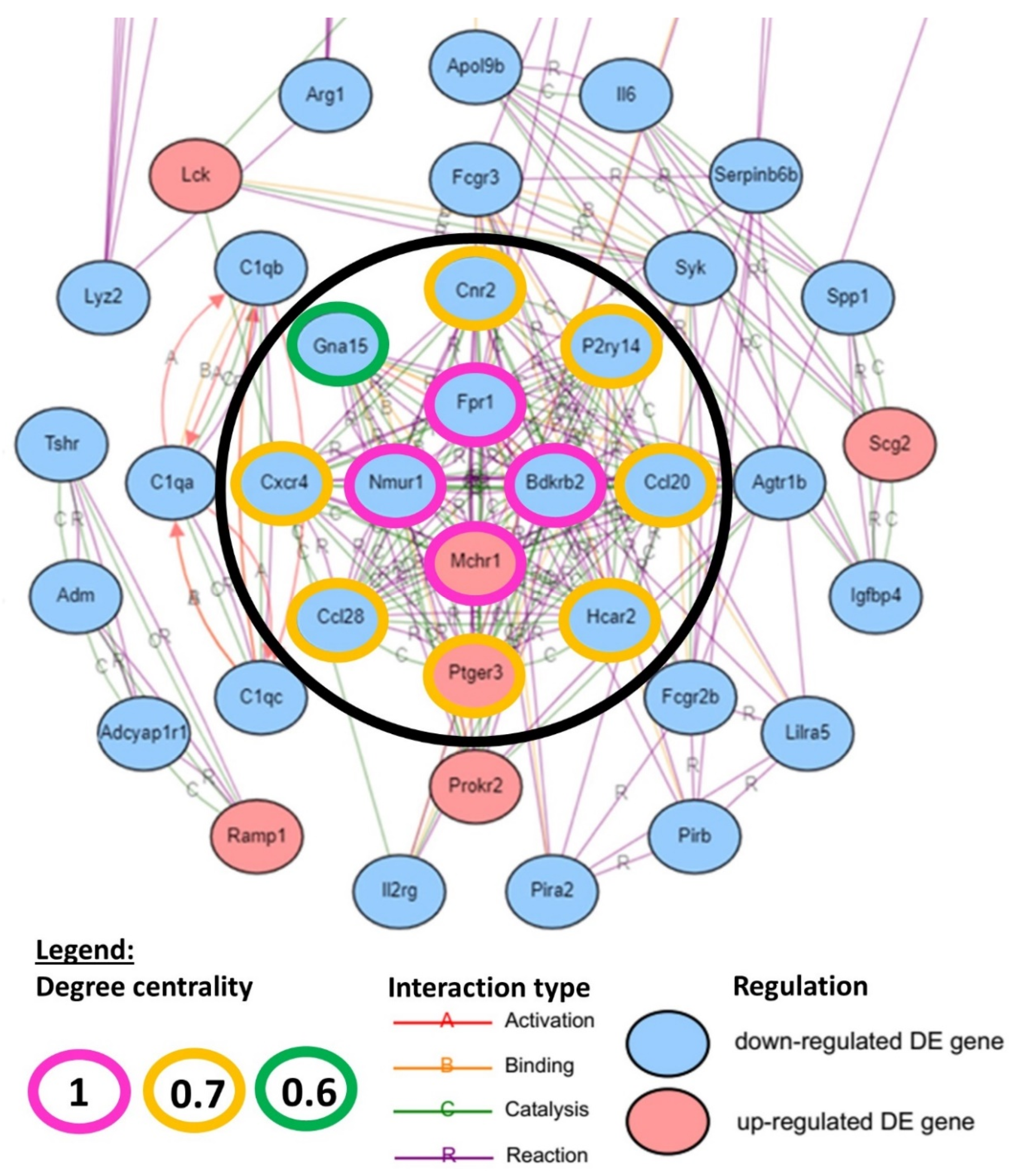

| Category | Name | Count DEG | Count All | p-Value | Correction |
|---|---|---|---|---|---|
| BP | Angiogenesis | 134 | 446 | 3.67E-04 | SCDP |
| BP | positive regulation of MAPK cascade | 117 | 412 | 8.15E-03 | SCDP |
| BP | regulation of blood coagulation | 28 | 62 | 1.32E-02 | SCDP |
| BP | regulation of cell migration | 217 | 798 | 2.55E-02 | SCDP |
| BP | morphogenesis of a branching structure | 70 | 199 | 3.05E-02 | SCDP |
| BP | positive regulation of cell population proliferation | 190 | 764 | 4.07E-02 | SCDP |
| BP | biological adhesion | 287 | 1046 | 4.30E-02 | SCDP |
| BP | Ossification | 88 | 312 | 4.30E-02 | SCDP |
| BP | response to gonadotropin | 8 | 10 | 4.30E-02 | SCDP |
| MF | calcium ion binding | 111 | 411 | 1.71E-02 | highSP |
| MF | cytokine activity | 39 | 110 | 1.71E-02 | highSP |
| MF | growth factor activity | 33 | 91 | 2.96E-02 | highSP |
| MF | receptor ligand activity | 81 | 246 | 5.69E-05 | SCDP |
| MF | calcium ion binding | 111 | 411 | 8.65E-03 | SCDP |
| MF | carbohydrate binding | 58 | 185 | 9.67E-03 | SCDP |
| MF | cell adhesion molecule binding | 60 | 195 | 9.67E-03 | SCDP |
| MF | extracellular matrix structural constituent | 39 | 117 | 3.03E-02 | SCDP |
| MF | glycosaminoglycan binding | 48 | 154 | 3.03E-02 | SCDP |
| CC | extracellular space | 271 | 867 | 9.14E-17 | highSP |
| CC | integral component of plasma membrane | 231 | 819 | 4.75E-08 | highSP |
| CC | cell surface | 189 | 625 | 9.10E-08 | highSP |
| CC | extracellular matrix | 122 | 377 | 1.04E-05 | highSP |
| CC | integral component of membrane | 749 | 3218 | 3.56E-05 | highSP |
| CC | collagen-containing extracellular matrix | 85 | 293 | 2.37E-03 | highSP |
| CC | external side of plasma membrane | 75 | 266 | 2.37E-02 | highSP |
| CC | Z disc | 32 | 95 | 3.71E-02 | highSP |
| CC | extracellular region | 395 | 1362 | 7.12E-18 | SCDP |
| CC | intrinsic component of membrane | 770 | 3318 | 1.96E-12 | SCDP |
| CC | cell surface | 189 | 625 | 4.75E-11 | SCDP |
| CC | apical part of cell | 91 | 318 | 1.25E-03 | SCDP |
| CC | I band | 35 | 103 | 2.61E-02 | SCDP |
| PW | Cytokine–cytokine receptor interaction | 52 | 163 | 6.45E-07 | none |
| PW | Focal adhesion | 47 | 187 | 2.46E-05 | none |
| PW | Staphylococcus aureus infection | 14 | 37 | 3.04E-05 | none |
| PW | Viral protein interaction with cytokine and cytokine receptor | 18 | 50 | 6.95E-05 | none |
| PW | AGE-RAGE signaling pathway in diabetic complications | 34 | 98 | 1.29E-04 | none |
| PW | Systemic lupus erythematosus | 20 | 77 | 1.97E-04 | none |
| PW | Proteoglycans in cancer | 54 | 186 | 2.79E-04 | none |
| PW | Chemokine signaling pathway | 35 | 147 | 7.48E-04 | none |
| PW | Neuroactive ligand-receptor interaction | 38 | 133 | 1.10E-03 | none |
| PW | Calcium signaling pathway | 34 | 150 | 2.06E-03 | none |
| Pathways | Genes | p-Value | Outcomes |
|---|---|---|---|
| Cytokine–cytokine receptor interaction | Cxcr4, Ccl20, Ccl28 | 6.45E-07 | Innate and adaptive inflammatory host defenses, cell growth, differentiation, cell death, angiogenesis, development and repair processes aimed at the restoration of homeostasis |
| Chemokine signaling pathway | Cxcr4, Ccl20, Ccl28 | 7.48E-04 | Inflammatory immune response, cellular activation, differentiation and survival, cellular polarization and actin reorganization |
| Neuroactive ligand-receptor interaction | Fpr1, Bdkrb2, Mchr1, Nmur1, Cnr2, P2ry14, Ptger3 | 1.10E-03 | Environmental information processing |
| Calcium signaling pathway | Gna15, Bdkrb2, Cxcr4, Ptger3 | 2.06E-03 | MAPK signaling, apoptosis, long-term potentiation/depression, phosphatidylinositol signaling pathway, contraction, metabolism, proliferation |
| Axon guidance | Cxcr4 | 2.15E-03 | Formation of neuronal network, cytoskeletal organization |
| Complement and coagulation cascades | Bdkrb2 | 4.48E-03 | Innate immunity, recruitment of inflammatory and immunocompetent cells |
| cGMP-PKG signaling pathway | Bdkrb2 | 1.98E-02 | Physiologic processes, regulation of cytosolic calcium concentration and sensitivity of myofilaments to Ca2+, ROS release from mitochondria |
| Regulation of actin cytoskeleton | Bdkrb2 | 2.92E-02 | Focal adhesion, MAPK signaling, adherens junction, |
| Endocrine and other factor-regulated calcium reabsorption | Bdkrb2 | 4.33E-02 | Intracellular signalling processes, neuronal excitability, muscle contraction and bone formation |
| Rap1 signaling pathway | Fpr1 | 5.01E-03 | Cell adhesion, cell–cell junction formation and cell polarity, control of cellcell and cell-matrix interactions by regulating the function of integrins and other adhesion molecules, MAPK signaling |
| TNF signaling pathway | Ccl20 | 2.71E-02 | Apoptosis and cell survival as well as inflammation and immunity, MAPK cascade, apoptosis, necroptosis, PI3K-dependent NF-kappa B pathway, JNK pathway, survival. |
| cAMP signaling pathway | Hcar2, Ptger3 | 0.487 | Metabolism, secretion, calcium homeostasis, muscle contraction, cell fate, and gene transcription |
| Genes | Full Name | RNAseq (F) | qRT-PCR | Immunoblot Cell Lysates | Immunoblot Cond. Media | |
|---|---|---|---|---|---|---|
| Log2Fc | q-Value | Regulation | Regulation | Regulation | ||
| Col27a1 | collagen, type XXVII, alpha 1 | 3.201 | 1.00E-06 | Up | NA | NA |
| Pcolce2 | procollagen C-endopeptidase enhancer 2 | 3.124 | 1.12E-03 | Up | NA | NA |
| Pappa2 | pappalysin 2 | 1.351 | 2.00E-03 | NA | NA | Up |
| Nmur1 | neuromedin U receptor 1 | −3.449 | 4.61E-02 | Down | Down | NA |
| Il6 | interleukin 6 | −3.021 | 1.00E-06 | Down | NA | NA |
| Bdkrb2 | bradykinin receptor, beta 2 | −5.230 | 5.77E-04 | Down | Down | NA |
| Cav3 | caveolin 3 | −7.164 | 1.00E-06 | Down | NA | NA |
| Spp1 | secreted phosphoprotein 1 | −3.034 | 2.21E-02 | Down | Down | NA |
| Serpine1 | serpin family E member 1 | −2.515 | 1.00E-06 | NA | Down | Down |
| Fpr1 | formyl peptide receptor 1 | −5.713 | 7.96E-03 | NA | Down | NA |
| Tbx1 | T-box 1 | −9.079 | 4.20E-02 | NA | Down | NA |
| Tgfβ2 | transforming growth factor, beta 2 | −1.178 | 1.00E-06 | NA | Down | NA |
| Igfbp4 | insulin-like growth factor binding protein 4 | −3.539 | 1.00E-06 | NA | NA | Down |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, X.-X.; Renaud, L.; Feghali-Bostwick, C. Identification of Impacted Pathways and Transcriptomic Markers as Potential Mediators of Pulmonary Fibrosis in Transgenic Mice Expressing Human IGFBP5. Int. J. Mol. Sci. 2021, 22, 12609. https://doi.org/10.3390/ijms222212609
Nguyen X-X, Renaud L, Feghali-Bostwick C. Identification of Impacted Pathways and Transcriptomic Markers as Potential Mediators of Pulmonary Fibrosis in Transgenic Mice Expressing Human IGFBP5. International Journal of Molecular Sciences. 2021; 22(22):12609. https://doi.org/10.3390/ijms222212609
Chicago/Turabian StyleNguyen, Xinh-Xinh, Ludivine Renaud, and Carol Feghali-Bostwick. 2021. "Identification of Impacted Pathways and Transcriptomic Markers as Potential Mediators of Pulmonary Fibrosis in Transgenic Mice Expressing Human IGFBP5" International Journal of Molecular Sciences 22, no. 22: 12609. https://doi.org/10.3390/ijms222212609
APA StyleNguyen, X.-X., Renaud, L., & Feghali-Bostwick, C. (2021). Identification of Impacted Pathways and Transcriptomic Markers as Potential Mediators of Pulmonary Fibrosis in Transgenic Mice Expressing Human IGFBP5. International Journal of Molecular Sciences, 22(22), 12609. https://doi.org/10.3390/ijms222212609






