Synaptotagmin-13 Is a Neuroendocrine Marker in Brain, Intestine and Pancreas
Abstract
:1. Introduction
2. Results and Discussion
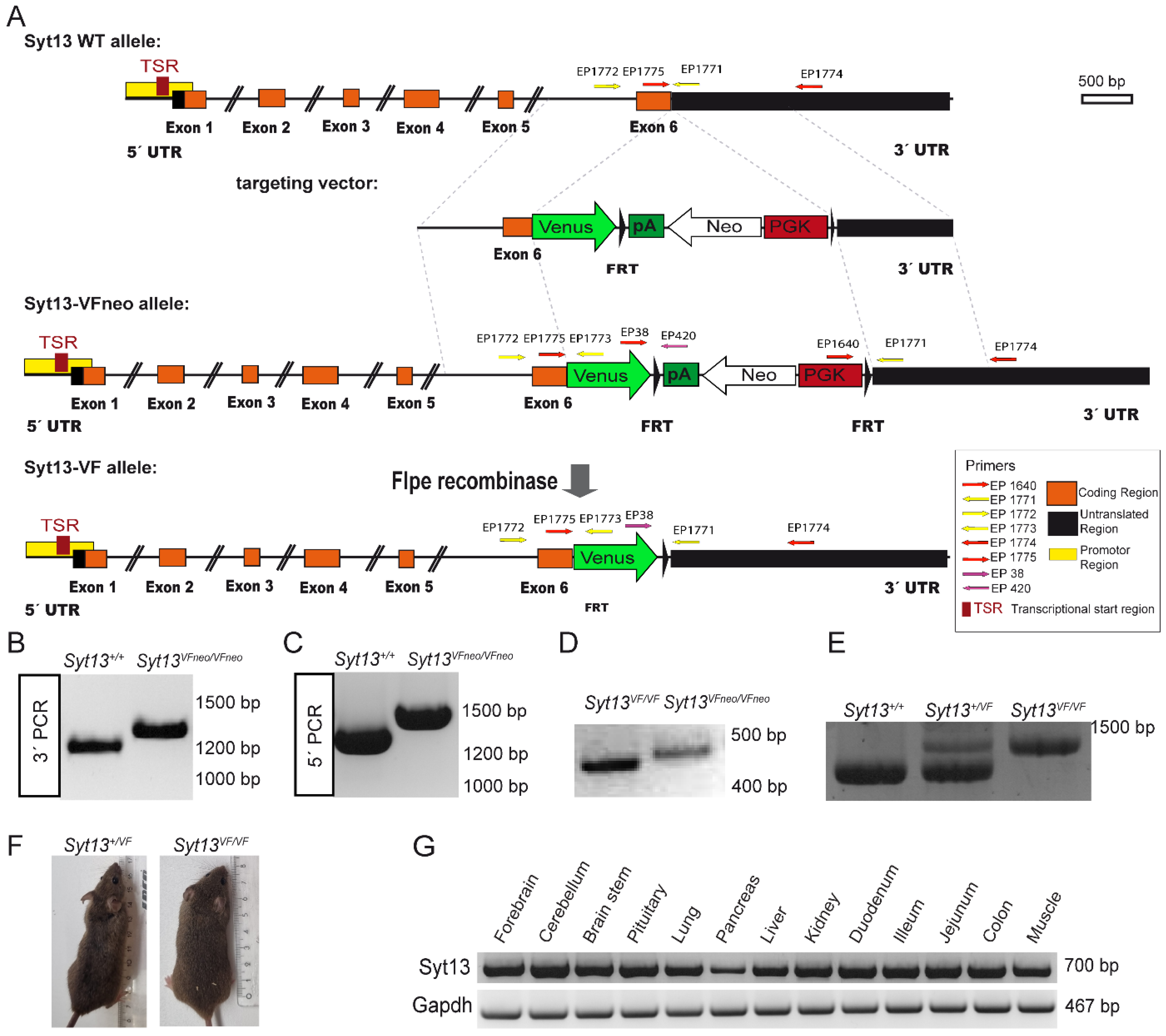
2.1. Generation of the Syt13-Venus Fusion Mouse Line
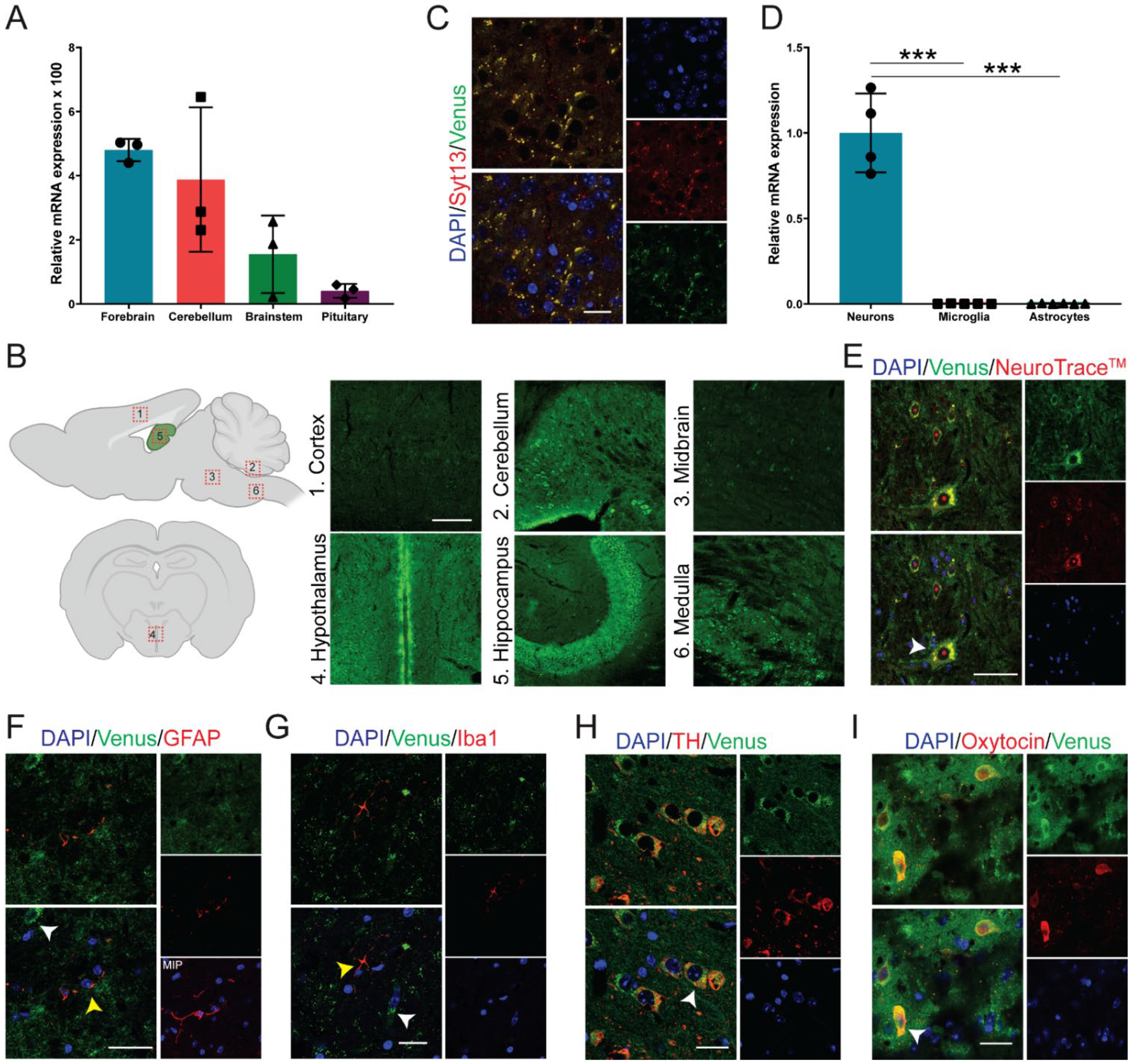
2.2. Syt13 Is Highly Expressed in Neuroendocrine Cells
2.3. Syt13 Expression Is Restricted to Enteroendocrine Cells in the Adult Intestine
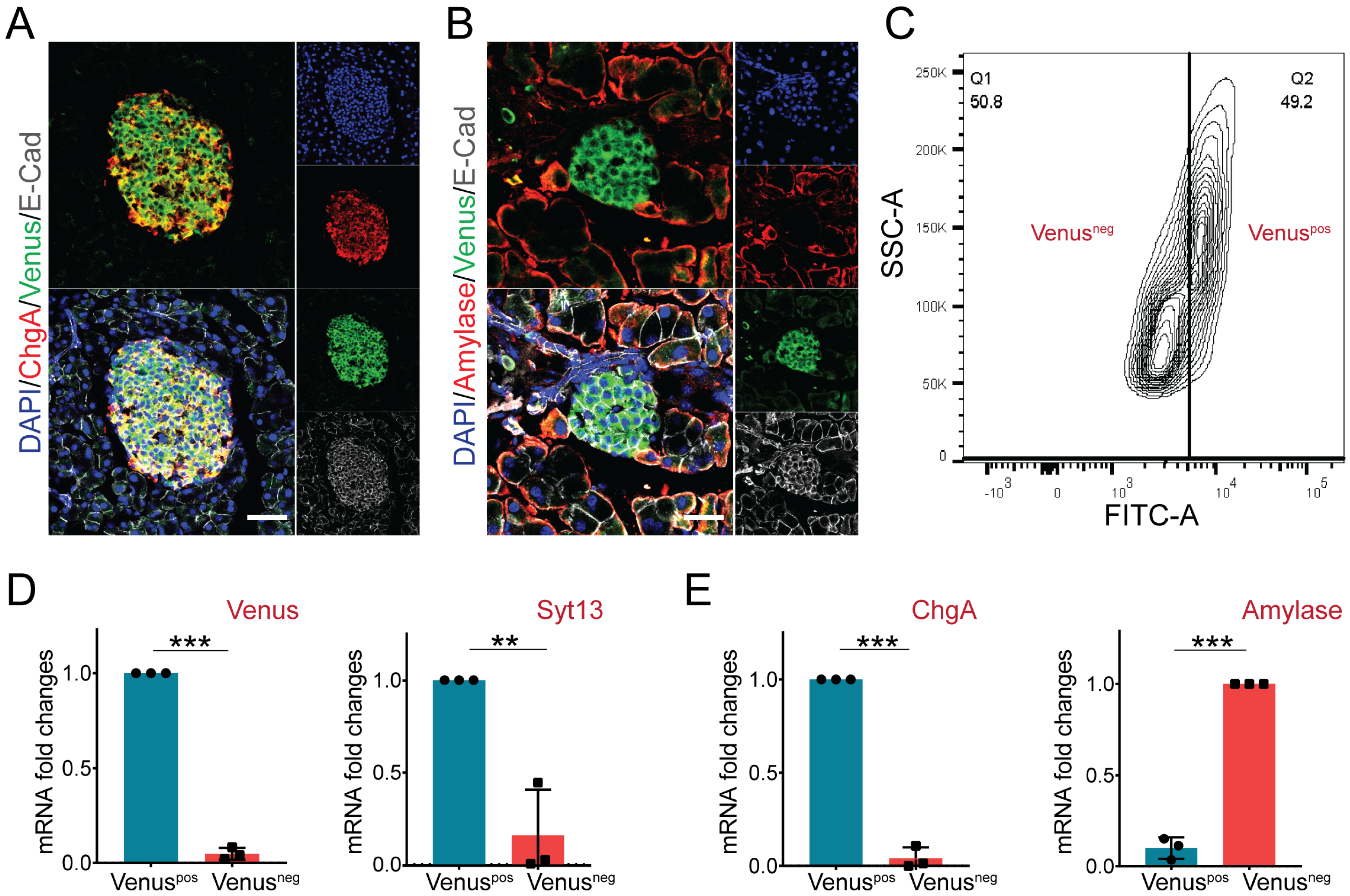
2.4. Pancreatic Endocrine but Not Exocrine Cells Specifically Express Syt13 Protein
3. Materials and Methods
3.1. Generation of the Targeting Vector
3.2. Embryonic Stem Cell (ESC) Homologous Recombination and Generation of Mouse Line
3.3. Genotyping
3.4. Organ Dissection and Immunostaining
3.5. Neuronal and Glia Cell Isolation
3.6. Crypt Isolation, Culturing and Live Imaging
3.7. Pancreatic Islet and Exocrine Tissue Isolation and FACS Sorting
3.8. RNA Isolation, RT-PCR and qPCR
3.9. ScRNA-Seq Data Sources and Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Südhof, T.C. Synaptotagmins: Why so many? J. Biol. Chem. 2002, 277, 7629–7632. [Google Scholar] [CrossRef] [Green Version]
- Rickman, C.; Craxton, M.; Osborne, S.; Davletov, B. Comparative analysis of tandem C2 domains from the mammalian synaptotagmin family. Biochem. J. 2004, 378, 681–686. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Shin, O.H.; Machius, M.; Tomchick, D.R.; Südhof, T.C.; Rizo, J. Structural basis for the evolutionary inactivation of Ca2+ binding to synaptotagmin 4. Nat. Struct. Mol. Biol. 2004, 11, 844–849. [Google Scholar] [CrossRef] [PubMed]
- Sugita, S.; Han, W.; Butz, S.; Lao, Y.; Liu, X.; Ferna, R.; Su, T.C.; Hines, H.; Na, B. Synaptotagmin VII as a plasma membrane Ca(2+) sensor in exocytosis. Neuron 2001, 30, 459–473. [Google Scholar] [CrossRef] [Green Version]
- Bhalla, A.; Tucker, W.C.; Chapman, E.R. Synaptotagmin Isoforms Couple Distinct Ranges of Ca2+, Ba2+ and Sr2+ Concentration to SNARE-mediated Membrane Fusion. Mol. Biol. Cell 2005, 16, 4755–4764. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Ullrich, B.; Zhang, J.Z.; Anderson, R.G.W.; Brose, N.; Südhof, T.C. Ca2+-dependent and -independent activities of neural and non-neural synaptotagmins. Nature 1995, 375, 594–599. [Google Scholar] [CrossRef]
- Wolfes, A.C.; Dean, C. The diversity of synaptotagmin isoforms. Curr. Opin. Neurobiol. 2020, 63, 198–209. [Google Scholar] [CrossRef]
- Yoo, E.S.; Yu, J.; Sohn, J.W. Neuroendocrine control of appetite and metabolism. Exp. Mol. Med. 2021, 53, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Moghadam, P.K.; Jackson, M.B. The Functional Significance of Synaptotagmin Diversity in Neuroendocrine Secretion. Front. Endocrinol. 2013, 4, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Poser, C.; Ichtchenko, K.; Shao, X.; Rizo, J.; Südhof, T.C. The evolutionary pressure to inactivate: A subclass of synaptotagmins with an amino acid substitution that abolishes Ca2+ binding. J. Biol. Chem. 1997, 272, 14314–14319. [Google Scholar] [CrossRef] [Green Version]
- Von Poser, C.; Südhof, T.C. Synaptotagmin 13: Structure and expression of a novel synaptotagmin. Eur. J. Cell Biol. 2001, 80, 41–47. [Google Scholar] [CrossRef]
- Fukuda, M.; Mikoshiba, K. Synaptotagmin-like protein 1-3: A novel family of C-terminal-type tandem C2 proteins. Biochem. Biophys. Res. Commun. 2001, 281, 1226–1233. [Google Scholar] [CrossRef] [PubMed]
- Willmann, S.J.; Mueller, N.S.; Engert, S.; Sterr, M.; Burtscher, I.; Raducanu, A.; Irmler, M.; Beckers, J.; Sass, S.; Theis, F.J.; et al. The global gene expression profile of the secondary transition during pancreatic development. Mech. Dev. 2016, 139, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Hong, S.; Lee, D.; Lee, M.H.; Choi, J.S.; Koh, M.J.; Sun, W.; Kim, H.; Lee, H.W. Altered expression of synaptotagmin 13 mRNA in adult mouse brain after contextual fear conditioning. Biochem. Biophys. Res. Commun. 2012, 425, 880–885. [Google Scholar] [CrossRef] [PubMed]
- Nizzardo, M.; Taiana, M.; Rizzo, F.; Aguila Benitez, J.; Nijssen, J.; Allodi, I.; Melzi, V.; Bresolin, N.; Comi, G.P.; Hedlund, E.; et al. Synaptotagmin 13 is neuroprotective across motor neuron diseases. Acta Neuropathol. 2020, 139, 837–853. [Google Scholar] [CrossRef] [Green Version]
- Kanda, M.; Shimizu, D.; Tanaka, H.; Tanaka, C.; Kobayashi, D.; Hayashi, M.; Takami, H.; Niwa, Y.; Yamada, S.; Fujii, T.; et al. Synaptotagmin XIII expression and peritoneal metastasis in gastric cancer. Br. J. Surg. 2018, 105, 1349–1358. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, S.; Hu, M.; Xu, M.; Jiang, X. Silencing of synaptotagmin 13 inhibits tumor growth through suppressing proliferation and promoting apoptosis of colorectal cancer cells. Int. J. Mol. Med. 2020, 45, 237–244. [Google Scholar] [CrossRef]
- Zhang, L.; Fan, B.; Zheng, Y.; Lou, Y.; Cui, Y.; Wang, K.; Zhang, T.; Tan, X. Identification SYT13 as a novel biomarker in lung adenocarcinoma. J. Cell. Biochem. 2020, 121, 963–973. [Google Scholar] [CrossRef]
- Hitz, C.; Wurst, W.; Kühn, R. Conditional brain-specific knockdown of MAPK using Cre/loxP regulated RNA interference. Nucleic Acids Res. 2007, 35, e90. [Google Scholar] [CrossRef]
- Gronostajski, R.M.; Sadowski, P.D. The FLP recombinase of the Saccharomyces cerevisiae 2 microns plasmid attaches covalently to DNA via a phosphotyrosyl linkage. Mol. Cell. Biol. 1985, 5, 3274–3279. [Google Scholar] [CrossRef] [Green Version]
- Mittelsteadt, T.; Seifert, G.; Alvárez-Barón, E.; Steinhäuser, C.; Becker, A.J.; Schoch, S. Differential mRNA expression patterns of the synaptotagmin gene family in the rodent brain. J. Comp. Neurol. 2009, 512, 514–528. [Google Scholar] [CrossRef]
- Watson, C.; Kirkcaldie, M.; Paxinos, G. Techniques for studying the brain. In The Brain, 1st ed.; Academic Press: San Diego, CA, USA; pp. 153–165. [CrossRef]
- Böttcher, A.; Büttner, M.; Tritschler, S.; Sterr, M.; Aliluev, A.; Oppenländer, L.; Burtscher, I.; Sass, S.; Irmler, M.; Beckers, J.; et al. Non-canonical Wnt/PCP signalling regulates intestinal stem cell lineage priming towards enteroendocrine and Paneth cell fates. Nat. Cell Biol. 2021, 23, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Walker, E.M.; Dadi, P.K.; Hu, R.; Xu, Y.; Zhang, W.; Sanavia, T.; Mun, J.; Liu, J.; Nair, G.G.; et al. Synaptotagmin 4 Regulates Pancreatic β Cell Maturation by Modulating the Ca2+ Sensitivity of Insulin Secretion Vesicles. Dev. Cell 2018, 45, 347–361. [Google Scholar] [CrossRef] [PubMed]
- Dolai, S.; Xie, L.; Zhu, D.; Liang, T.; Qin, T.; Xie, H.; Kang, Y.; Chapman, E.R.; Gaisano, H.Y. Synaptotagmin-7 functions to replenish insulin granules for exocytosis in human islet β-cell. Diabetes 2016, 65, 1962–1976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, S.A.; Olsson, A.H.; Esguerra, J.L.S.; Heimann, E.; Ladenvall, C.; Edlund, A.; Salehi, A.; Taneera, J.; Degerman, E.; Groop, L.; et al. Reduced insulin secretion correlates with decreased expression of exocytotic genes in pancreatic islets from patients with type 2 diabetes. Mol. Cell. Endocrinol. 2012, 364, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Kanda, M.; Kasahara, Y.; Shimizu, D.; Miwa, T.; Umeda, S.; Sawaki, K.; Nakamura, S.; Kodera, Y.; Obika, S. Amido-Bridged Nucleic Acid-Modified Antisense Oligonucleotides Targeting SYT13 to Treat Peritoneal Metastasis of Gastric Cancer. Mol. Ther. Nucl. Acids 2020, 22, 791–802. [Google Scholar] [CrossRef] [PubMed]
- Bastidas-Ponce, A.; Tritschler, S.; Dony, L.; Scheibner, K.; Tarquis-Medina, M.; Salinno, C.; Schirge, S.; Burtscher, I.; Böttcher, A.; Theis, F.J.; et al. Comprehensive single cell mRNA profiling reveals a detailed roadmap for pancreatic endocrinogenesis. Development 2019, 146, dev173849. [Google Scholar] [CrossRef] [Green Version]
- Salinno, C.; Büttner, M.; Cota, P.; Tritschler, S.; Tarquis-Medina, M.; Bastidas-Ponce, A.; Scheibner, K.; Burtscher, I.; Böttcher, A.; Theis, F.J.; et al. CD81 marks immature and dedifferentiated pancreatic β-cells. Mol. Metab. 2021, 49, 101188. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tarquis-Medina, M.; Scheibner, K.; González-García, I.; Bastidas-Ponce, A.; Sterr, M.; Jaki, J.; Schirge, S.; García-Cáceres, C.; Lickert, H.; Bakhti, M. Synaptotagmin-13 Is a Neuroendocrine Marker in Brain, Intestine and Pancreas. Int. J. Mol. Sci. 2021, 22, 12526. https://doi.org/10.3390/ijms222212526
Tarquis-Medina M, Scheibner K, González-García I, Bastidas-Ponce A, Sterr M, Jaki J, Schirge S, García-Cáceres C, Lickert H, Bakhti M. Synaptotagmin-13 Is a Neuroendocrine Marker in Brain, Intestine and Pancreas. International Journal of Molecular Sciences. 2021; 22(22):12526. https://doi.org/10.3390/ijms222212526
Chicago/Turabian StyleTarquis-Medina, Marta, Katharina Scheibner, Ismael González-García, Aimée Bastidas-Ponce, Michael Sterr, Jessica Jaki, Silvia Schirge, Cristina García-Cáceres, Heiko Lickert, and Mostafa Bakhti. 2021. "Synaptotagmin-13 Is a Neuroendocrine Marker in Brain, Intestine and Pancreas" International Journal of Molecular Sciences 22, no. 22: 12526. https://doi.org/10.3390/ijms222212526
APA StyleTarquis-Medina, M., Scheibner, K., González-García, I., Bastidas-Ponce, A., Sterr, M., Jaki, J., Schirge, S., García-Cáceres, C., Lickert, H., & Bakhti, M. (2021). Synaptotagmin-13 Is a Neuroendocrine Marker in Brain, Intestine and Pancreas. International Journal of Molecular Sciences, 22(22), 12526. https://doi.org/10.3390/ijms222212526





