HBV HBx-Downregulated lncRNA LINC01010 Attenuates Cell Proliferation by Interacting with Vimentin
Abstract
:1. Introduction
2. Results
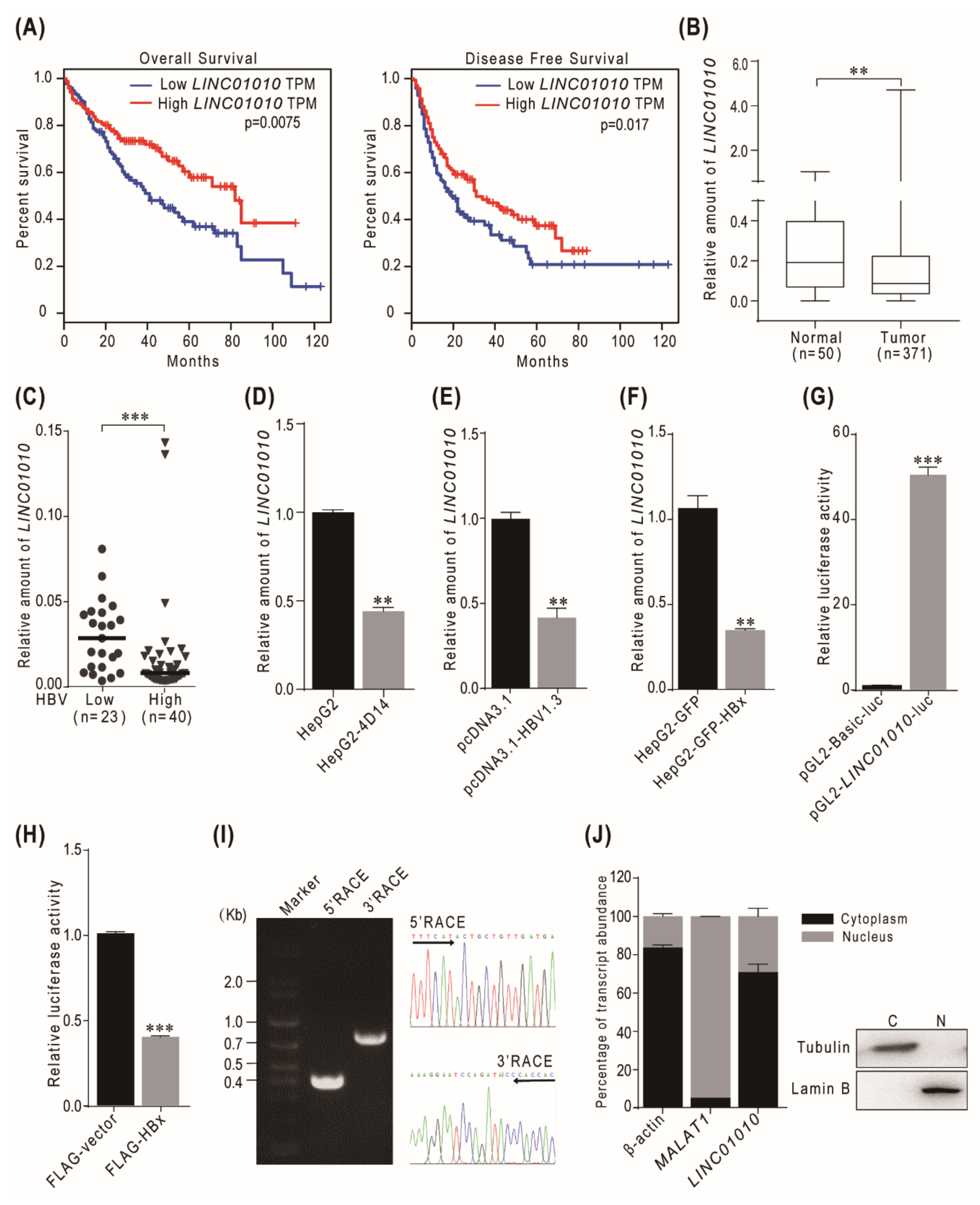
2.1. HBV HBx-Downregulated lncRNA LINC01010 Is Abnormally Expressed in HCC
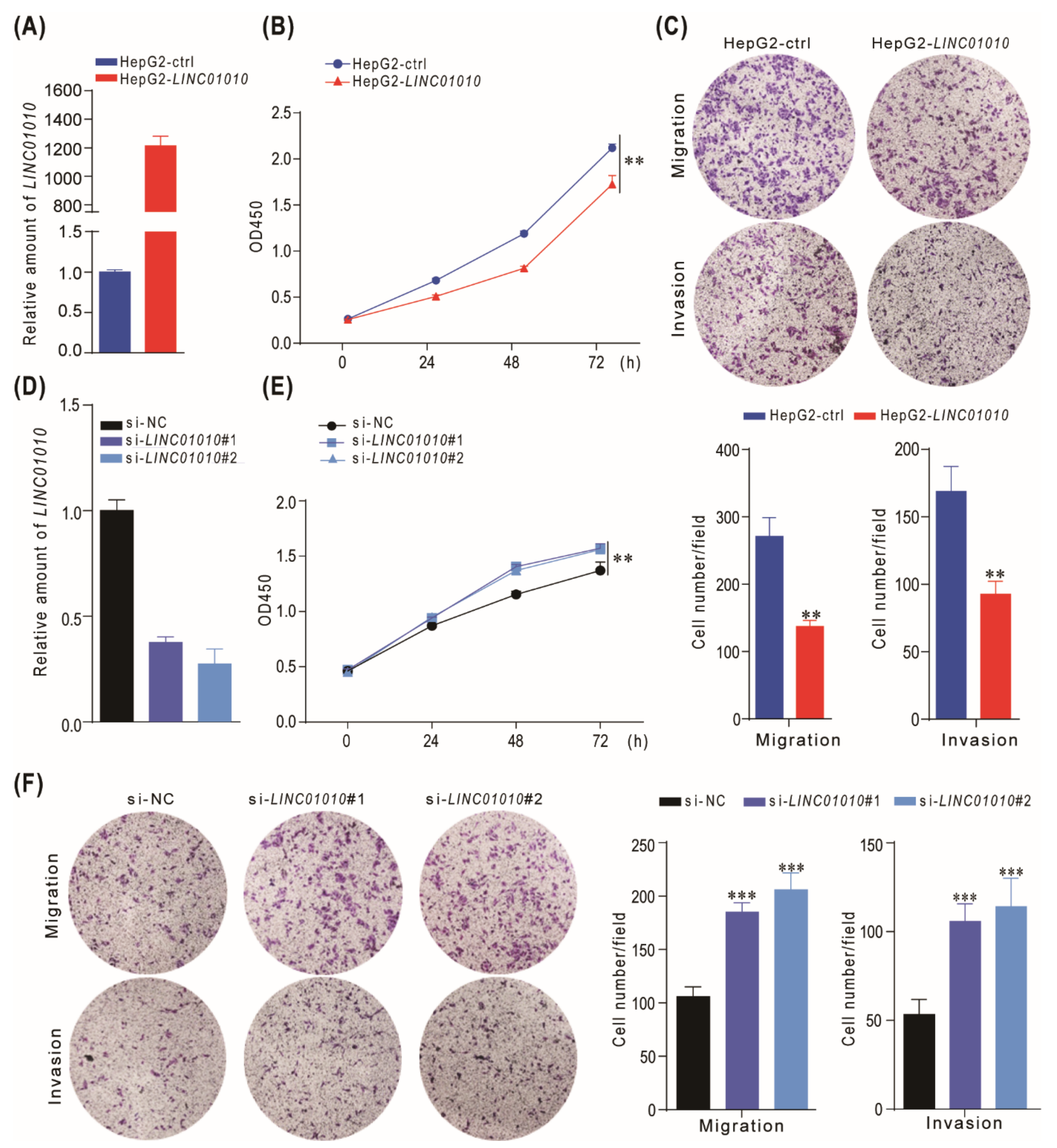
2.2. LINC01010 Attenuates the Cell Proliferation, Migration and Invasion
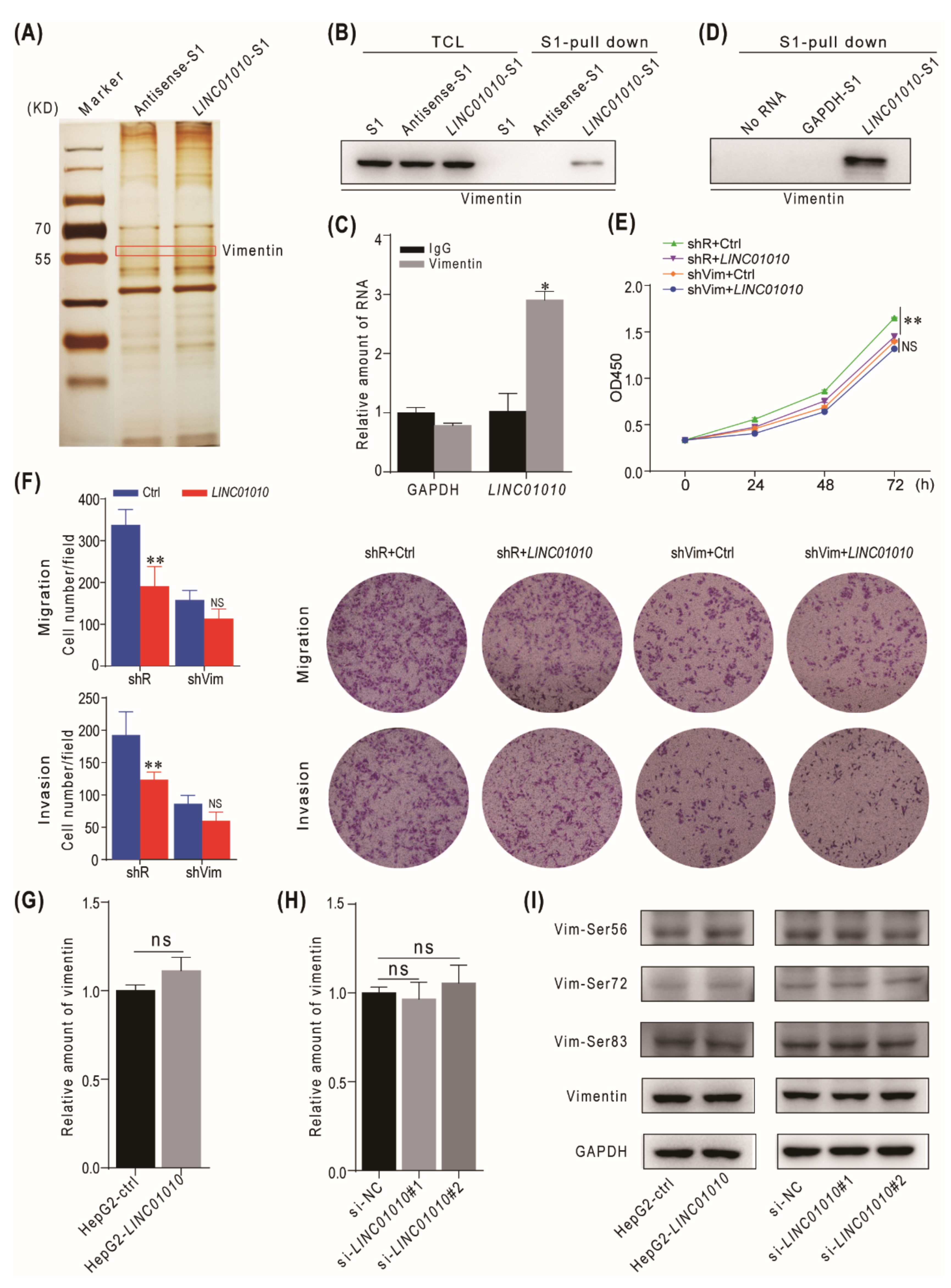
2.3. LINC01010 Interacts with Vimentin
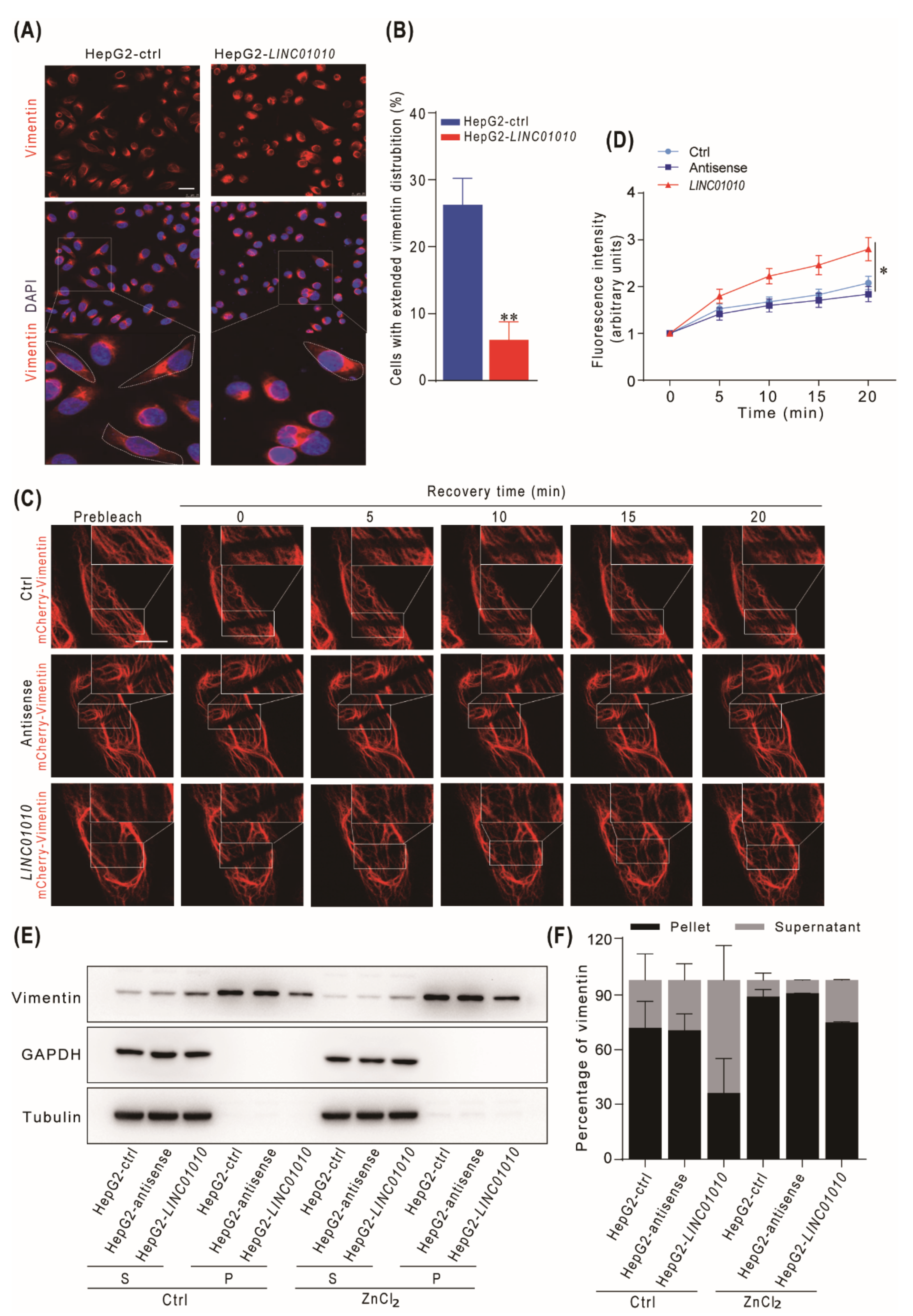
2.4. Overexpression of LINC01010 Inhibits the Network Extension and Assembly of Vimentin
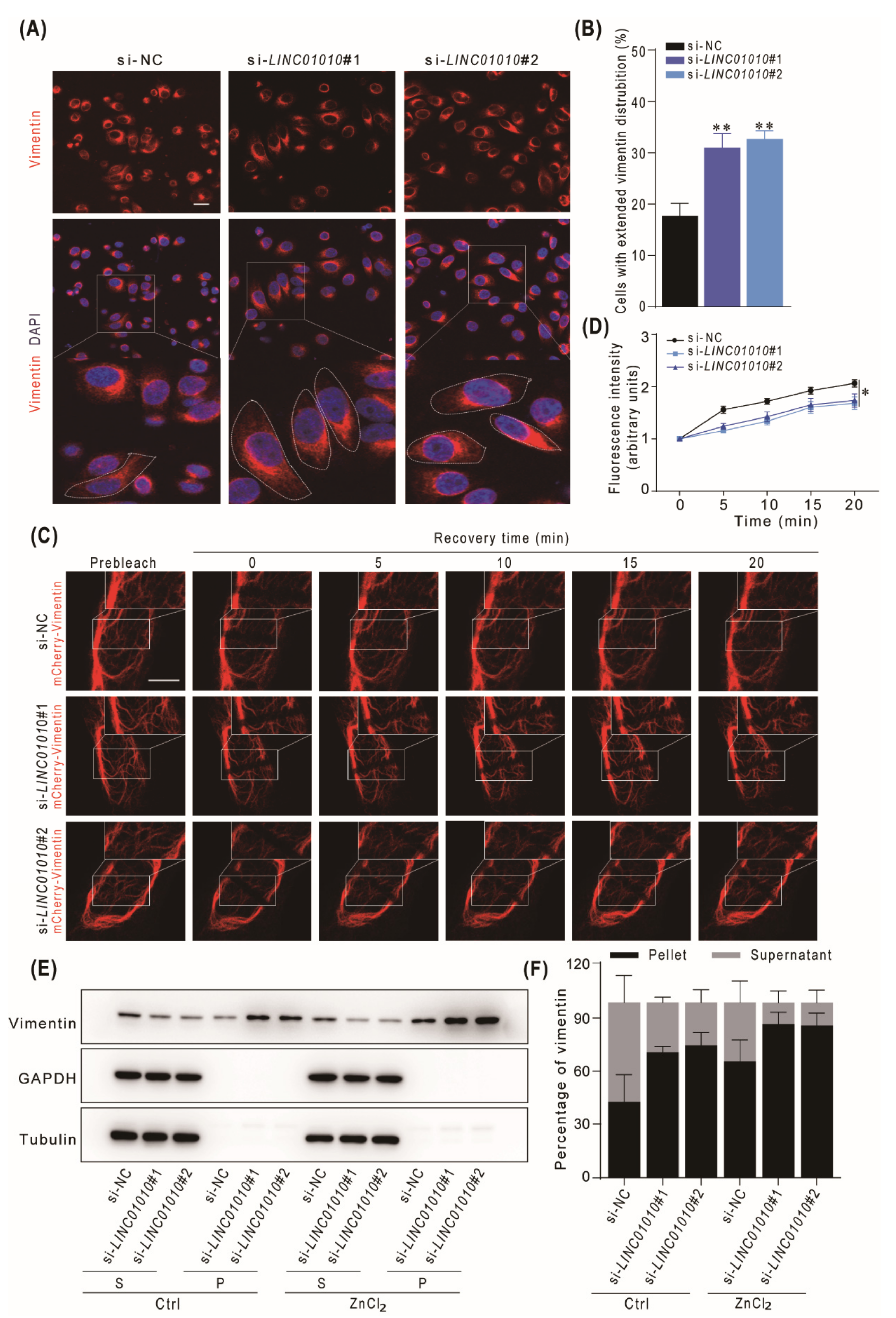
2.5. Knockdown of LINC01010 Promotes the Network Extension and Assembly of Vimentin
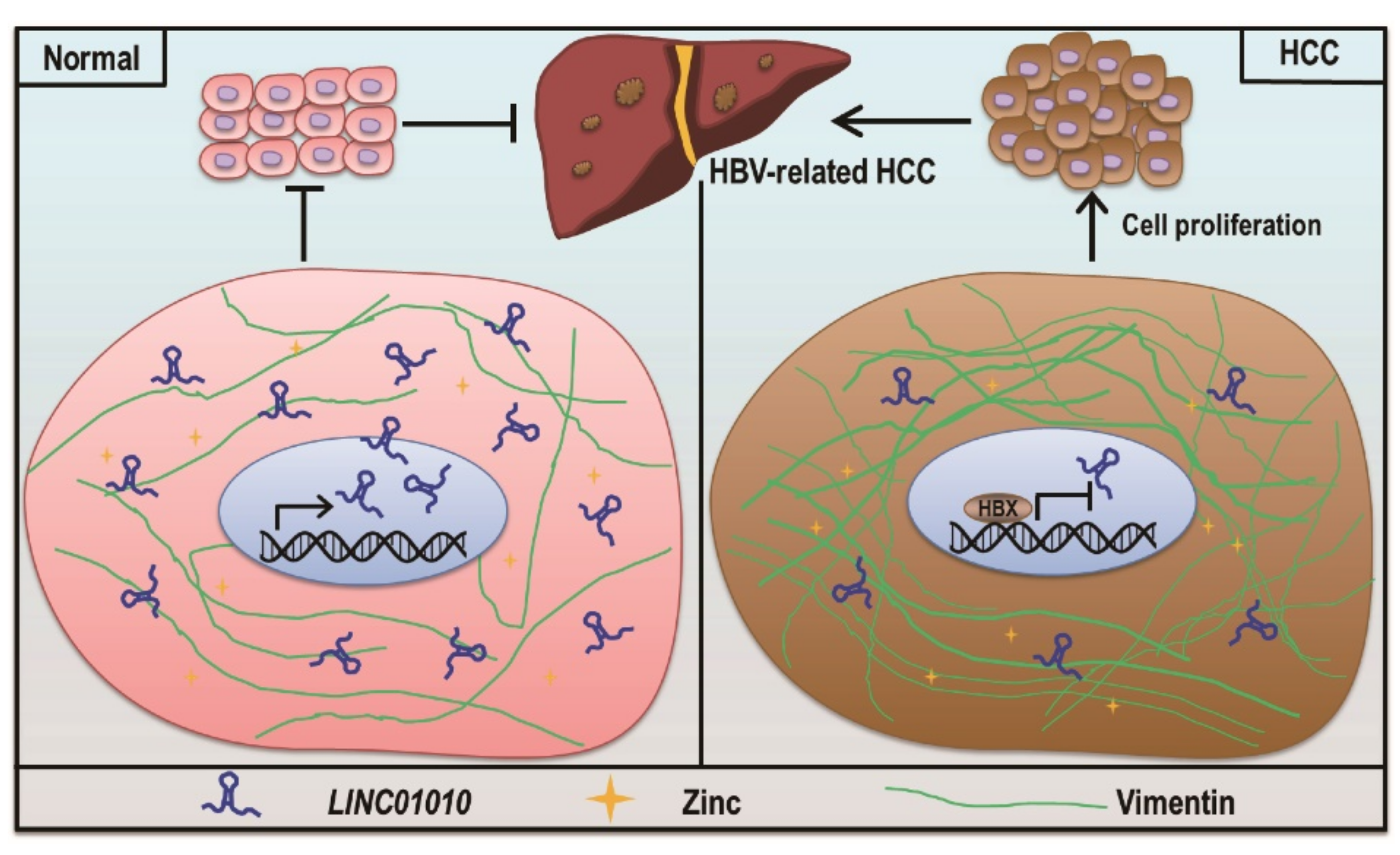
3. Discussion
4. Materials and Methods
4.1. Plasmids and Antibodies
4.2. Cell Lines and Cell Culture
4.3. Tumor Samples
4.4. RNA Interference
4.5. Rapid Amplification of cDNA Ends (RACE)
4.6. Luciferase Reporter Assay
4.7. Quantitative RT-PCR (qRT-PCR)
4.8. Immunoblotting
4.9. Cell Growth Assay
4.10. Migration and Invasion Assays
4.11. S1 Pull-Down
4.12. RNA Immunoprecipitation (RIP)
4.13. In Vitro RNA Pull-Down Assay
4.14. Cell Extension Formation and Immunofluorescence Assays
4.15. Fluorescence Recovery after Photobleaching (FRAP)
4.16. Vimentin Solubility Assay
4.17. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGlynn, K.A.; Petrick, J.L.; London, W.T. Global epidemiology of hepatocellular carcinoma: An emphasis on demographic and regional variability. Clin. Liver Dis. 2015, 19, 223–238. [Google Scholar] [CrossRef] [Green Version]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- El Serag, H.B.; Rudolph, K.L. Hepatocellular Carcinoma: Epidemiology and Molecular Carcinogenesis. Gastroenterology 2007, 132, 2557–2576. [Google Scholar] [CrossRef] [PubMed]
- Venook, A.P.; Papandreou, C.; Furuse, J.; de Guevara, L.L. The incidence and epidemiology of hepatocellular carcinoma: A global and regional perspective. Oncologist 2010, 15 (Suppl. 4), 5–13. [Google Scholar] [CrossRef]
- Yan, H.; Zhong, G.; Xu, G.; He, W.; Jing, Z.; Gao, Z.; Huang, Y.; Qi, Y.; Peng, B.; Wang, H.; et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 2012, 1, e49. [Google Scholar] [CrossRef]
- Kawai, H.; Suda, T.; Aoyagi, Y.; Isokawa, O.; Mita, Y.; Waguri, N.; Kuroiwa, T.; Igarashi, M.; Tsukada, K.; Mori, S.; et al. Quantitative evaluation of genomic instability as a possible predictor for development of hepatocellular carcinoma: Comparison of loss of heterozygosity and replication error. Hepatology 2000, 31, 1246–1250. [Google Scholar] [CrossRef] [PubMed]
- Ferber, M.J.; Montoya, D.P.; Yu, C.; Aderca, I.; McGee, A.; Thorland, E.C.; Nagorney, D.M.; Gostout, B.S.; Burgart, L.J.; Boix, L.; et al. Integrations of the hepatitis B virus (HBV) and human papillomavirus (HPV) into the human telomerase reverse transcriptase (hTERT) gene in liver and cervical cancers. Oncogene 2003, 22, 3813–3820. [Google Scholar] [CrossRef] [Green Version]
- Horikawa, I.; Barrett, J.C. cis-Activation of the human telomerase gene (hTERT) by the hepatitis B virus genome. J. Natl. Cancer Inst. 2001, 93, 1171–1173. [Google Scholar] [CrossRef]
- Wang, J.; Zindy, F.; Chenivesse, X.; Lamas, E.; Henglein, B.; Brechot, C. Modification of cyclin A expression by hepatitis B virus DNA integration in a hepatocellular carcinoma. Oncogene 1992, 7, 1653–1656. [Google Scholar] [PubMed]
- Chaturvedi, V.K.; Singh, A.; Dubey, S.K.; Hetta, H.F.; John, J.; Singh, M.P. Molecular mechanistic insight of hepatitis B virus mediated hepatocellular carcinoma. Microb. Pathog. 2019, 128, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.H.; Chen, M.; Mo, H.H.; Tsai, W.C.; Chang, Y.C.; Chang, C.C.; Chen, K.C.; Wu, H.Y.; Yuan, C.H.; Lee, C.H.; et al. Utilizing Experimental Mouse Model to Identify Effectors of Hepatocellular Carcinoma Induced by HBx Antigen. Cancers 2020, 12, 409. [Google Scholar] [CrossRef] [Green Version]
- Elmore, L.W.; Hancock, A.R.; Chang, S.F.; Wang, X.W.; Chang, S.; Callahan, C.P.; Geller, D.A.; Will, H.; Harris, C.C. Hepatitis B virus X protein and p53 tumor suppressor interactions in the modulation of apoptosis. Proc. Natl. Acad. Sci. USA 1997, 94, 14707–14712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cougot, D.; Wu, Y.; Cairo, S.; Caramel, J.; Renard, C.A.; Levy, L.; Buendia, M.A.; Neuveut, C. The hepatitis B virus X protein functionally interacts with CREB-binding protein/p300 in the regulation of CREB-mediated transcription. J. Biol. Chem. 2007, 282, 4277–4287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, Y.G.; Oh, S.H.; Park, E.S.; Cho, H.; Lee, N.; Park, H.; Kim, D.K.; Yu, D.Y.; Seong, J.K.; Lee, M.O. Hepatitis B virus X protein enhances transcriptional activity of hypoxia-inducible factor-1alpha through activation of mitogen-activated protein kinase pathway. J. Biol. Chem. 2003, 278, 39076–39084. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Hu, Z.; Feng, Y.; Hu, X.; Yuan, J.; Zhao, S.D.; Zhang, Y.; Yang, L.; Shan, W.; He, Q.; et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell 2015, 28, 529–540. [Google Scholar] [CrossRef] [Green Version]
- Brunner, A.L.; Beck, A.H.; Edris, B.; Sweeney, R.T.; Zhu, S.X.; Li, R.; Montgomery, K.; Varma, S.; Gilks, T.; Guo, X.; et al. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012, 13, R75. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Song, X.; Li, Y.; Chen, B.; Zhao, W.; Wang, L.; Zhang, H.; Liu, Y.; Han, D.; Zhang, N.; et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol. Cancer 2020, 19, 85. [Google Scholar] [CrossRef] [PubMed]
- Olivero, C.E.; Martinez-Terroba, E.; Zimmer, J.; Liao, C.; Tesfaye, E.; Hooshdaran, N.; Schofield, J.A.; Bendor, J.; Fang, D.; Simon, M.D.; et al. Activates the Long Noncoding RNA Pvt1b to Inhibit Myc and Suppress Tumorigenesis. Mol. Cell 2020, 77, 761–774. [Google Scholar] [CrossRef]
- Xu, M.; Xu, X.; Pan, B.; Chen, X.; Lin, K.; Zeng, K.; Liu, X.; Xu, T.; Sun, L.; Qin, J.; et al. LncRNA SATB2-AS1 inhibits tumor metastasis and affects the tumor immune cell microenvironment in colorectal cancer by regulating SATB2. Mol. Cancer 2019, 18, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Liu, X.; Zhou, J.; Hu, J.; Zhang, D.; Liu, J.; Qiao, Y.; Zhan, Q. Long noncoding RNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of extracellular signal-regulated kinase and YB-1 to enhance hepatocarcinogenesis. Hepatology 2017, 65, 1612–1627. [Google Scholar] [CrossRef] [Green Version]
- Hou, Z.; Xu, X.; Fu, X.; Tao, S.; Zhou, J.; Liu, S.; Tan, D. HBx-related long non-coding RNA MALAT1 promotes cell metastasis via up-regulating LTBP3 in hepatocellular carcinoma. Am. J. Cancer Res. 2017, 7, 845–856. [Google Scholar] [PubMed]
- Zhang, Y.; Li, J.; Wang, S.; Yang, F.; Zhou, Y.; Liu, Y.; Zhu, W.; Shi, X. HBx-associated long non-coding RNA activated by TGF-β promotes cell invasion and migration by inducing autophagy in primary liver cancer. Int. J. Oncol. 2020, 56, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.F.; Guo, Y.J.; Zhao, C.X.; Yuan, S.X.; Wang, Y.; Tang, G.N.; Zhou, W.P.; Sun, S.H. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology 2013, 57, 1882–1892. [Google Scholar] [CrossRef]
- Liu, N.; Liu, Q.; Yang, X.; Zhang, F.; Li, X.; Ma, Y.; Guan, F.; Zhao, X.; Li, Z.; Zhang, L.; et al. Hepatitis B Virus-Upregulated LNC-HUR1 Promotes Cell Proliferation and Tumorigenesis by Blocking p53 Activity. Hepatology 2018, 68, 2130–2144. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Liu, N.; Shangguan, Q.; Zhang, F.; Chai, W.; Tong, X.; Zhao, X.; Li, Z.; Qi, D.; Ye, X. LncRNA SAMD12-AS1 promotes cell proliferation and inhibits apoptosis by interacting with NPM1. Sci. Rep. 2019, 9, 11593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noh, J.H.; Kim, K.M.; McClusky, W.G.; Abdelmohsen, K.; Gorospe, M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip. Rev. RNA 2018, 9, e1471. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tian, H.; Yang, J.; Gong, Z. Long Noncoding RNAs Regulate Cell Growth, Proliferation, and Apoptosis. DNA Cell Biol. 2016, 35, 459–470. [Google Scholar] [CrossRef]
- Li, Q.F.; Spinelli, A.M.; Wang, R.; Anfinogenova, Y.; Singer, H.A.; Tang, D.D. Critical role of vimentin phosphorylation at Ser-56 by p21-activated kinase in vimentin cytoskeleton signaling. J. Biol. Chem. 2006, 281, 34716–34724. [Google Scholar] [CrossRef] [Green Version]
- Yasui, Y.; Goto, H.; Matsui, S.; Manser, E.; Lim, L.; Nagata, K.; Inagaki, M. Protein kinases required for segregation of vimentin filaments in mitotic process. Oncogene 2001, 20, 2868–2876. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Liu, W.; Yang, H.; Tan, L.; Ao, L.; Liu, J.; Cao, J.; Cui, Z. Inhibition of PPARalpha attenuates vimentin phosphorylation on Ser-83 and collapse of vimentin filaments during exposure of rat Sertoli cells in vitro to DBP. Reprod. Toxicol. 2014, 50, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Perez-Sala, D.; Oeste, C.L.; Martinez, A.E.; Carrasco, M.J.; Garzon, B.; Canada, F.J. Vimentin filament organization and stress sensing depend on its single cysteine residue and zinc binding. Nat. Commun. 2015, 6, 7287. [Google Scholar] [CrossRef] [Green Version]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- Muller, S.; Raulefs, S.; Bruns, P.; Afonso-Grunz, F.; Plotner, A.; Thermann, R.; Jager, C.; Schlitter, A.M.; Kong, B.; Regel, I.; et al. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol. Cancer 2015, 14, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Chen, L.; Gu, J.; Zhang, H.; Yuan, J.; Lian, Q.; Lv, G.; Wang, S.; Wu, Y.; Yang, Y.T.; et al. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat. Commun. 2017, 8, 14421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Z.; Zhou, J.K.; Peng, Y.; He, W.; Huang, C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol. Cancer 2020, 19, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, L.; Lin, P.; Chen, P.; Gao, R.Z.; Yang, H.; He, Y.; Chen, J.B.; Luo, Y.G.; Xu, Q.Q.; Liang, S.W.; et al. A novel risk signature that combines 10 long noncoding RNAs to predict neuroblastoma prognosis. J. Cell Physiol. 2020, 235, 3823–3834. [Google Scholar] [CrossRef]
- Cao, Q.; Dong, Z.; Liu, S.; An, G.; Yan, B.; Lei, L. Construction of a metastasis-associated ceRNA network reveals a prognostic signature in lung cancer. Cancer Cell Int. 2020, 20, 208. [Google Scholar] [CrossRef] [PubMed]
- Motavaf, M.; Safari, S.; Saffari, J.M.; Alavian, S.M. Hepatitis B virus-induced hepatocellular carcinoma: The role of the virus x protein. Acta Virol. 2013, 57, 389–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, H.; Yuan, D.; Bi, Y.; Zhang, N.; Li, Q.; Tu, T.; Wei, X.; Lian, Q.; Yu, T.; Kong, D.; et al. Hepatitis B virus X protein promotes vimentin expression via LIM and SH3 domain protein 1 to facilitate epithelial-mesenchymal transition and hepatocarcinogenesis. Cell Commun. Signal. 2021, 19, 33. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.J.; Song, W.; Zhang, S.D.; Shen, X.H.; Qiu, X.M.; Wu, H.Z.; Gong, P.H.; Lu, S.; Zhao, Z.J.; He, M.L.; et al. HBx-upregulated lncRNA UCA1 promotes cell growth and tumorigenesis by recruiting EZH2 and repressing p27Kip1/CDK2 signaling. Sci. Rep. 2016, 6, 23521. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.; Xu, Q.; Yan, Y.; Lu, Y.; Hu, Z.; Ou, B.; Zhang, H.; Mao, K.; Zhang, J.; Wang, J.; et al. HBx/ERalpha complex-mediated LINC01352 downregulation promotes HBV-related hepatocellular carcinoma via the miR-135b-APC axis. Oncogene 2020, 39, 3774–3789. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Ye, Z.; Hou, L.; Zhang, X.; Liu, Z.; Wu, R.; Huang, F.; Wang, G.; Geng, X.; Zhao, H. Hepatitis B virus x gene-downregulated growth-arrest specific 5 inhibits the cell viability and invasion of hepatocellular carcinoma cell lines by activating Y-box-binding protein 1/p21 signaling. J. Cell Commun. Signal. 2021, 18, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Satelli, A.; Li, S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol. Life Sci. 2011, 68, 3033–3046. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.Y.; Lin, H.H.; Tang, M.J.; Wang, Y.K. Vimentin contributes to epithelial-mesenchymal transition cancer cell mechanics by mediating cytoskeletal organization and focal adhesion maturation. Oncotarget 2015, 6, 15966–15983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilles, C.; Polette, M.; Piette, J.; Delvigne, A.C.; Thompson, E.W.; Foidart, J.M.; Birembaut, P. Vimentin expression in cervical carcinomas: Association with invasive and migratory potential. J. Pathol. 1996, 180, 175–180. [Google Scholar] [CrossRef]
- Singh, S.; Sadacharan, S.; Su, S.; Belldegrun, A.; Persad, S.; Singh, G. Overexpression of vimentin: Role in the invasive phenotype in an androgen-independent model of prostate cancer. Cancer Res. 2003, 63, 2306–2311. [Google Scholar] [PubMed]
- Burikhanov, R.; Sviripa, V.M.; Hebbar, N.; Zhang, W.; Layton, W.J.; Hamza, A.; Zhan, C.G.; Watt, D.S.; Liu, C.; Rangnekar, V.M. Arylquins target vimentin to trigger Par-4 secretion for tumor cell apoptosis. Nat. Chem. Biol. 2014, 10, 924–926. [Google Scholar] [CrossRef] [Green Version]
- Zamay, T.N.; Kolovskaya, O.S.; Glazyrin, Y.E.; Zamay, G.S.; Kuznetsova, S.A.; Spivak, E.A.; Wehbe, M.; Savitskaya, A.G.; Zubkova, O.A.; Kadkina, A.; et al. DNA-aptamer targeting vimentin for tumor therapy in vivo. Nucleic Acid Ther. 2014, 24, 160–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.P.; Raina, K.; Sharma, G.; Agarwal, R. Silibinin inhibits established prostate tumor growth, progression, invasion, and metastasis and suppresses tumor angiogenesis and epithelial-mesenchymal transition in transgenic adenocarcinoma of the mouse prostate model mice. Clin. Cancer Res. 2008, 14, 7773–7780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bargagna-Mohan, P.; Hamza, A.; Kim, Y.E.; Khuan, A.H.Y.; Mor-Vaknin, N.; Wendschlag, N.; Liu, J.; Evans, R.M.; Markovitz, D.M.; Zhan, C.G.; et al. The tumor inhibitor and antiangiogenic agent withaferin A targets the intermediate filament protein vimentin. Chem. Biol. 2007, 14, 623–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bollong, M.J.; Pietila, M.; Pearson, A.D.; Sarkar, T.R.; Ahmad, I.; Soundararajan, R.; Lyssiotis, C.A.; Mani, S.A.; Schultz, P.G.; Lairson, L.L. A vimentin binding small molecule leads to mitotic disruption in mesenchymal cancers. Proc. Natl. Acad. Sci. USA 2017, 114, E9903–E9912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaner, N.C.; Patterson, G.H.; Davidson, M.W. Advances in fluorescent protein technology. J. Cell Sci. 2007, 120, 4247–4260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, I.; Ostrowska-Podhorodecka, Z.; Lee, W.; Liu, R.; Carneiro, K.; Janmey, P.A.; McCulloch, C.A. Cooperative roles of PAK1 and filamin A in regulation of vimentin assembly and cell extension formation. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118739. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gan, L.; Shangguan, Q.; Zhang, F.; Tong, X.; Qi, D.; Zhao, Y.; Ye, X. HBV HBx-Downregulated lncRNA LINC01010 Attenuates Cell Proliferation by Interacting with Vimentin. Int. J. Mol. Sci. 2021, 22, 12497. https://doi.org/10.3390/ijms222212497
Gan L, Shangguan Q, Zhang F, Tong X, Qi D, Zhao Y, Ye X. HBV HBx-Downregulated lncRNA LINC01010 Attenuates Cell Proliferation by Interacting with Vimentin. International Journal of Molecular Sciences. 2021; 22(22):12497. https://doi.org/10.3390/ijms222212497
Chicago/Turabian StyleGan, Lipeng, Qilin Shangguan, Fang Zhang, Xiaomei Tong, Dandan Qi, Yan Zhao, and Xin Ye. 2021. "HBV HBx-Downregulated lncRNA LINC01010 Attenuates Cell Proliferation by Interacting with Vimentin" International Journal of Molecular Sciences 22, no. 22: 12497. https://doi.org/10.3390/ijms222212497




