Characterization and Genome Study of Novel Lytic Bacteriophages against Prevailing Saprophytic Bacterial Microflora of Minimally Processed Plant-Based Food Products
Abstract
1. Introduction
2. Results and Discussion
2.1. Microbiological Analysis of the RUC, MLSC, and MLSB Products
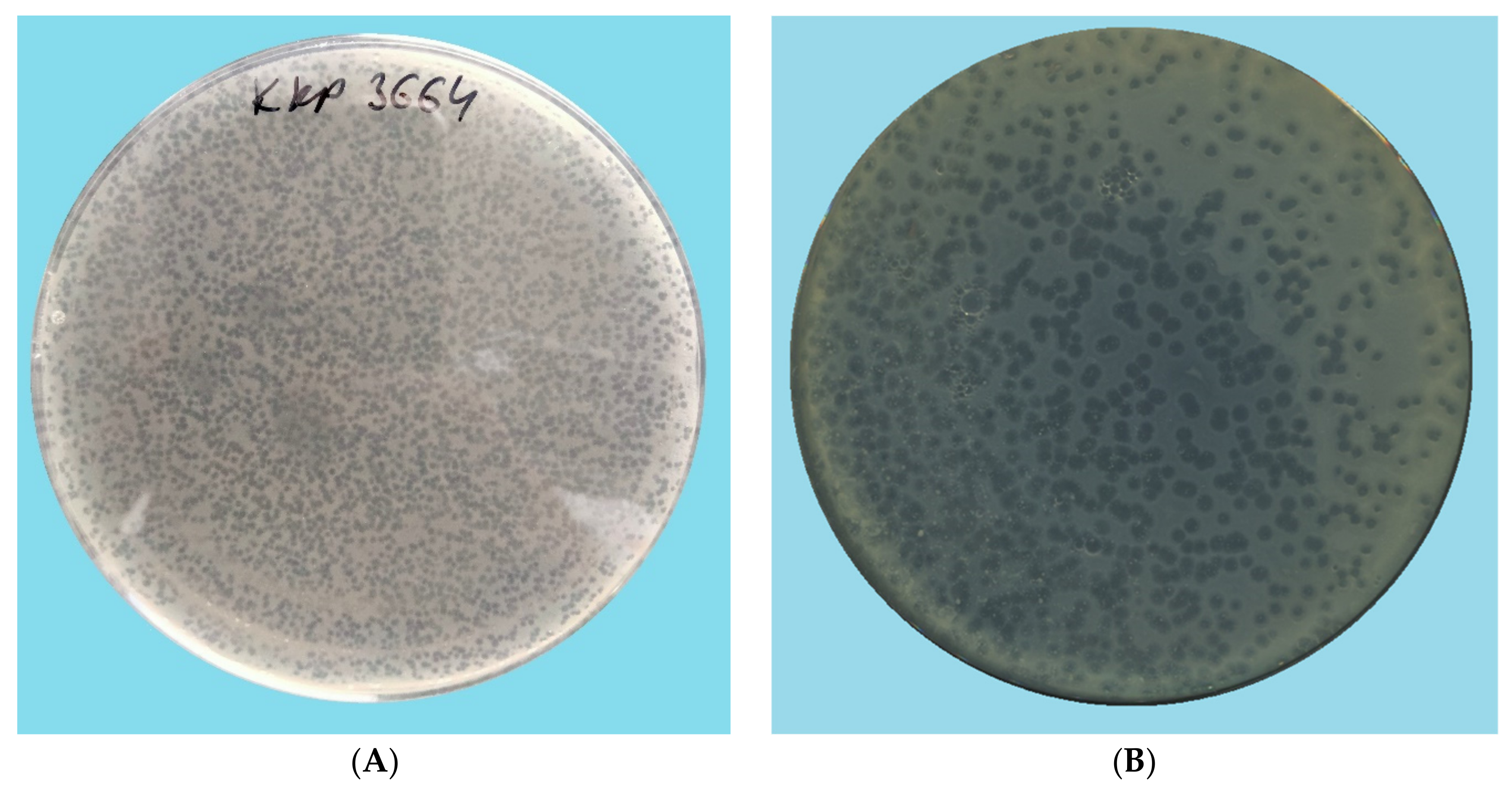
2.2. Evaluation of the Lytic Activity of Phages against Bacterial Hosts
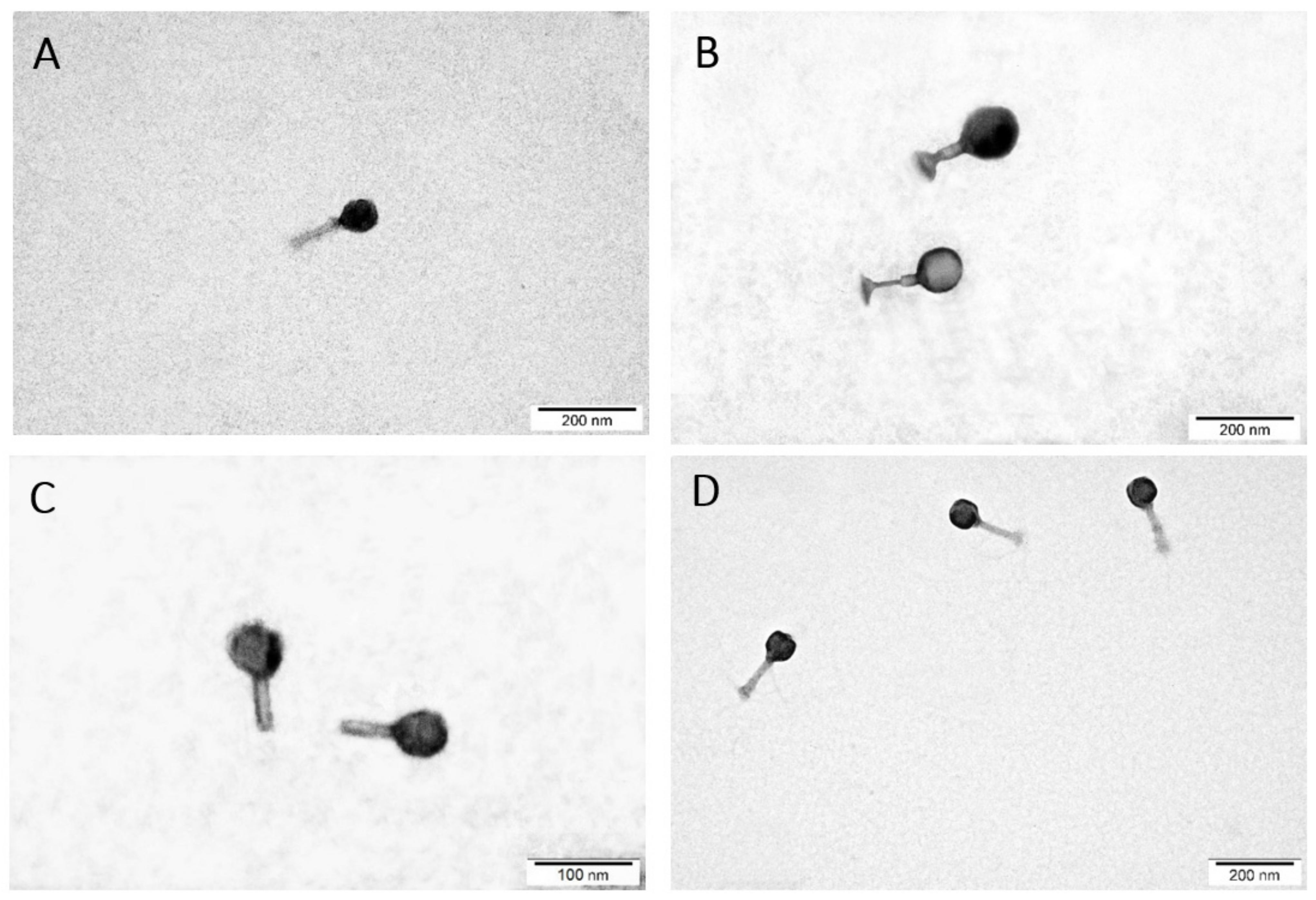
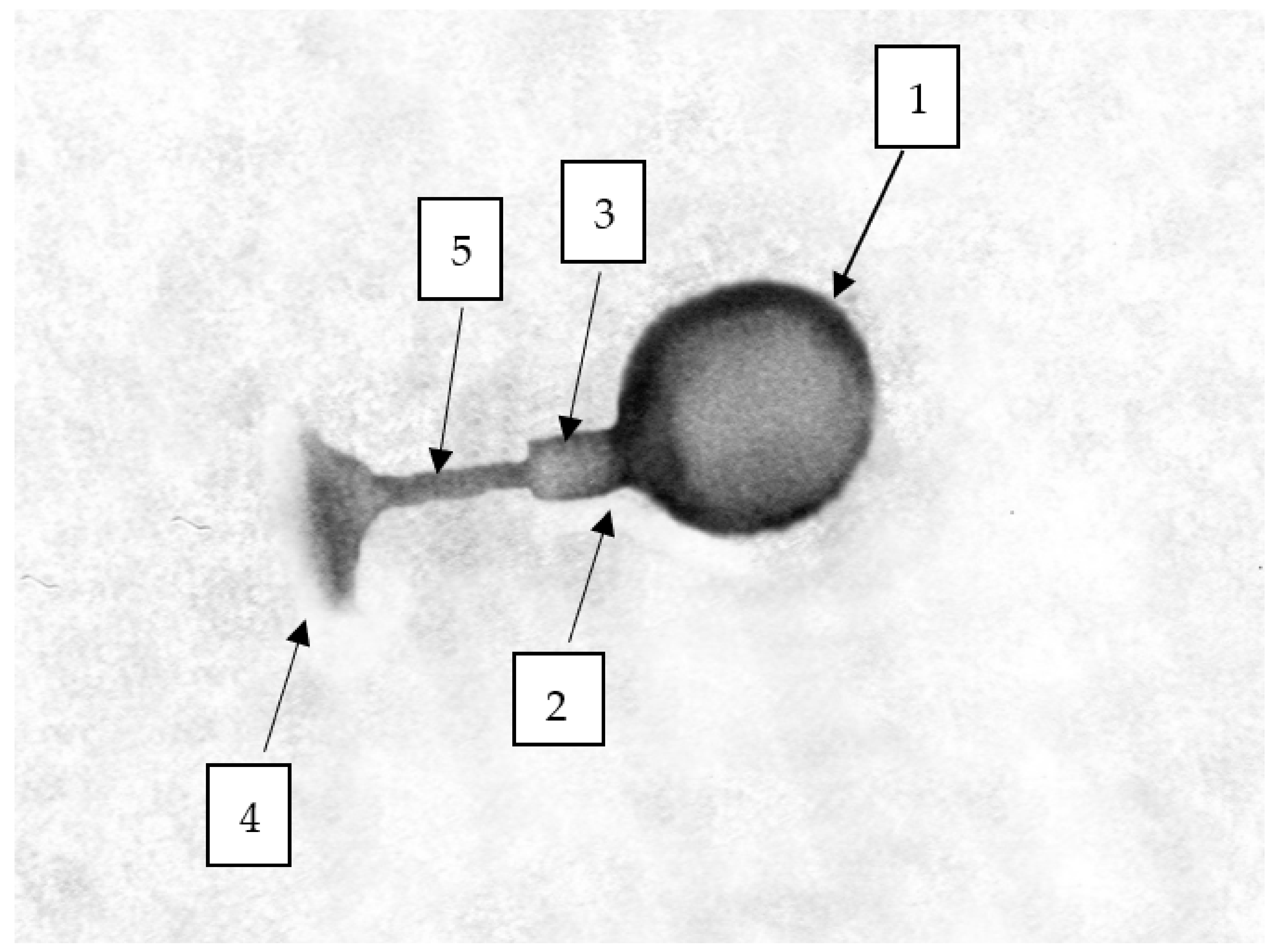
2.3. TEM Morphology of Selected Bacteriophages
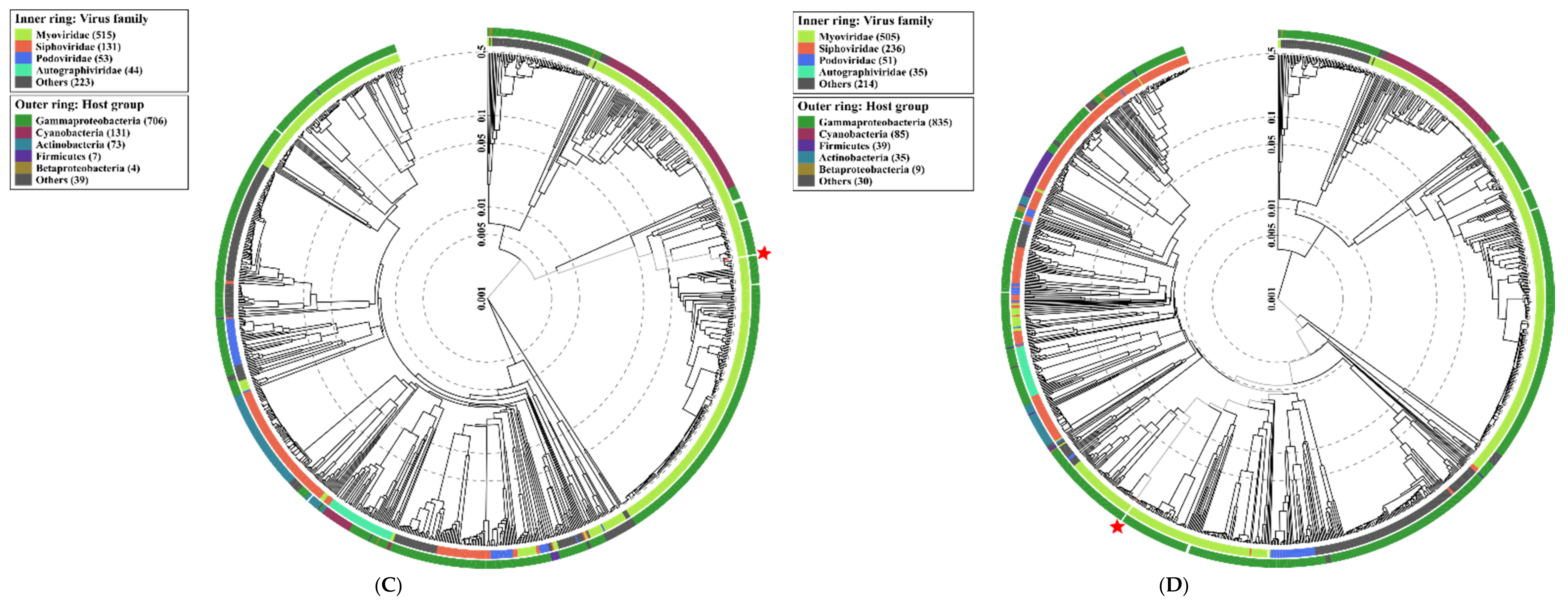
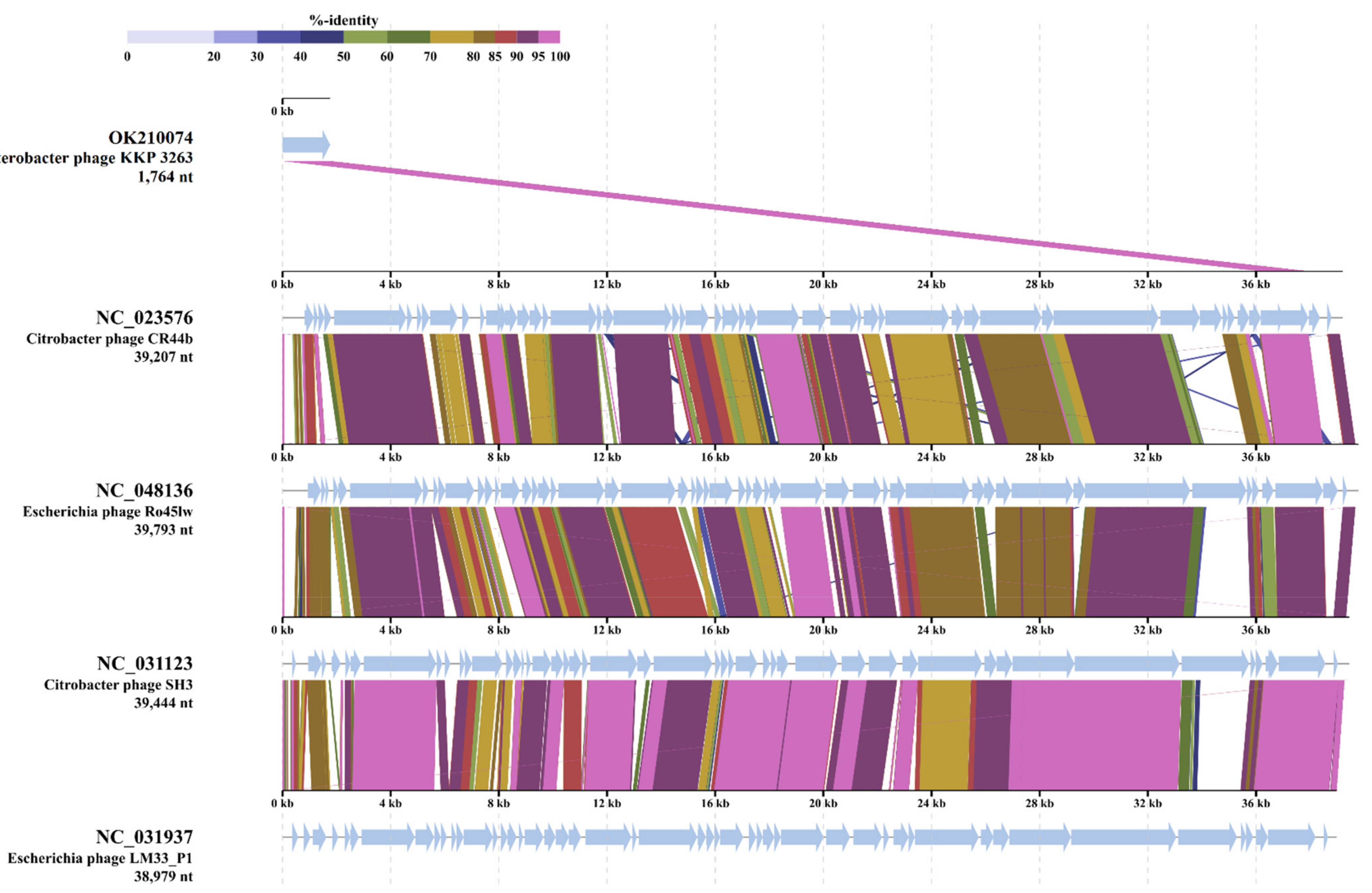
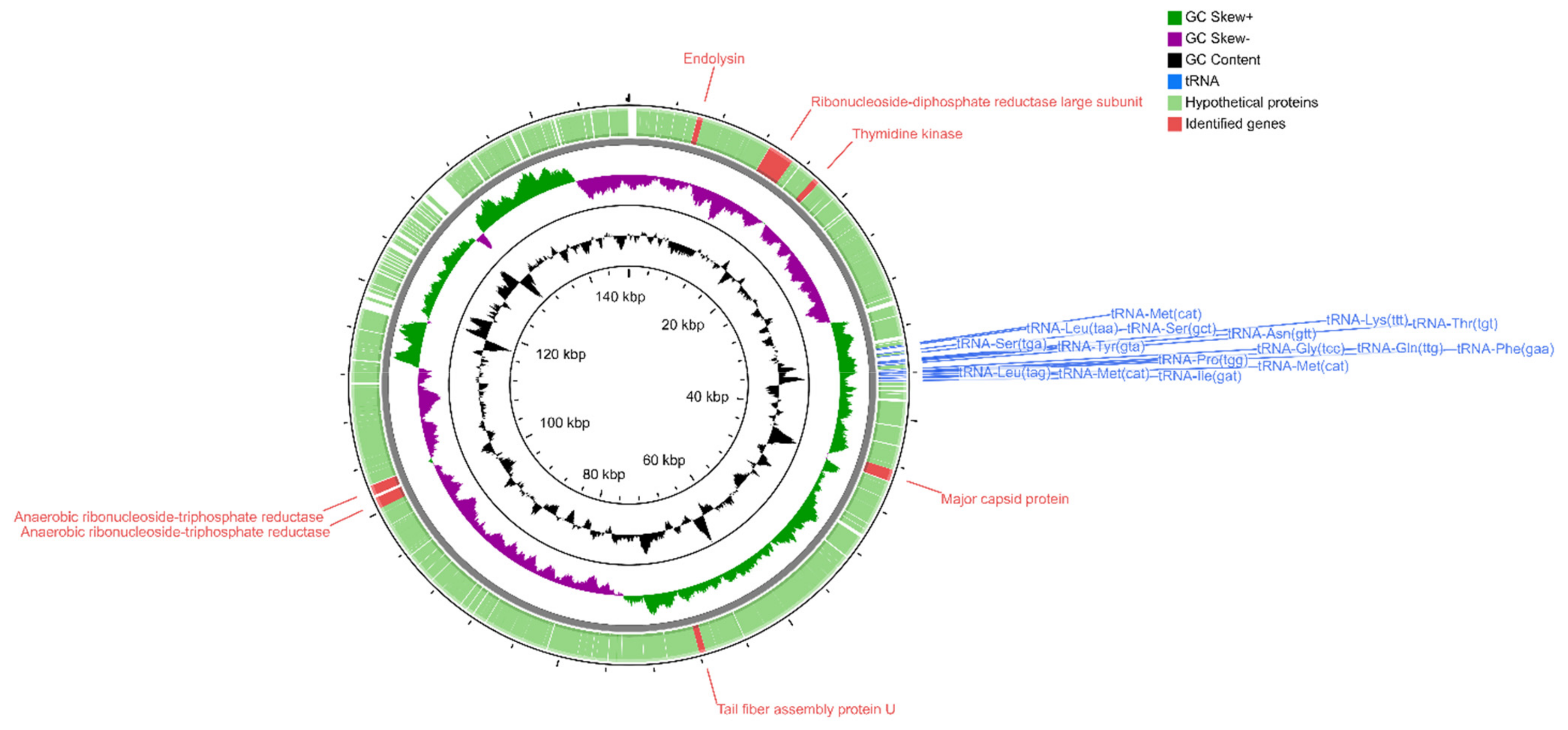
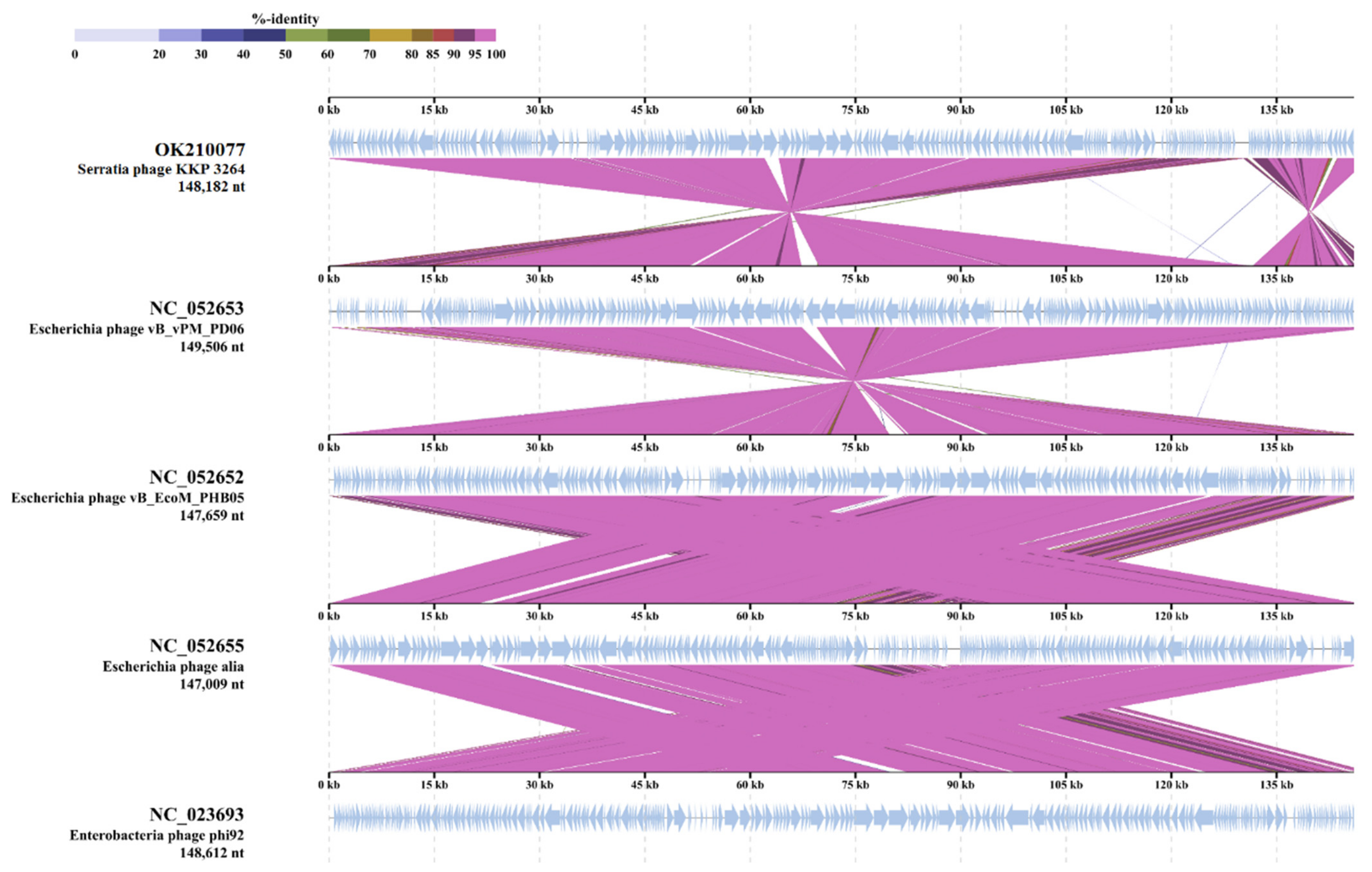
2.4. Analysis of Phage Genomes
2.5. Effect of Selected Environmental Factors on the Preservation of the Lytic Activity of Phages
2.5.1. Influence of Temperature on the Lytic Activity of Phages
2.5.2. Influence of pH on the Lytic Activity of Phages
3. Materials and Methods
3.1. Isolation and Taxonomic Identification of the Bacterial Strains from Food Products
3.2. Bacteriophage Isolation, Purification, and Propagation
3.3. Evaluation of the Lytic Activity of Phages against Bacterial Hosts
3.4. Determination of Morphological Features of Selected Phages
3.5. Extraction of Bacteriophage Genomic DNA
3.6. Genome Sequencing and Bioinformatics Analysis
3.7. Influence of Selected Factors on the Preservation of the Lytic Activity of Phages
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ragaert, P.; Devlieghere, F.; Debevere, J. Role of microbiological and physiological spoilage mechanisms during storage of minimally processed vegetables. Postharvest Biol. Technol. 2007, 44, 185–194. [Google Scholar]
- Bansal, V.; Siddiqui, M.W.; Rahman, M.S. Minimally processed foods: Overview. In Minimally Processed Foods; Food Engineering Series; Siddiqui, M.W., Rahman, M.S., Eds.; Springer International Publishing: Cham, Switzerland; New York, NY, USA, 2014; pp. 1–15. [Google Scholar]
- Meireles, A.; Giaouris, E.; Simões, M. Alternative disinfection methods to chlorine for use in the fresh-cut industry. Food Res. Int. 2016, 82, 71–85. [Google Scholar]
- Barba, F.J.; Koubaa, M.; Prado-Silva, L.; Orlien, V.; de SouzaSant’Ana, A. Mild processing applied to the inactivation of the main foodborne bacterial pathogens: A review. Trends Food Sci. Technol. 2017, 66, 20–35. [Google Scholar] [CrossRef]
- Lewis, R.; Bolocan, A.S.; Draper, L.A.; Ross, R.P.; Hill, C. The effect of a commercially available bacteriophage and bacteriocin on Listeria monocytogenes in Coleslaw. Viruses 2019, 11, 977. [Google Scholar] [CrossRef] [PubMed]
- Sandhya, M. Modified atmosphere packaging of fresh produce: Current status and future needs. LWT—Food Sci. Technol. 2010, 43, 381–392. [Google Scholar] [CrossRef]
- Mistriotis, A.; Briassoulis, D.; Giannoulis, A.; D’Aquino, S. Design of biodegradable bio-based equilibrium modified atmosphere packaging (EMAP) for fresh fruits and vegetables by using micro-perforated poly-lactic acid (PLA) films. Postharvest Biol. Technol. 2016, 111, 380–389. [Google Scholar] [CrossRef]
- Lee, S.; Kim, H.; Beuchat, L.R.; Kim, Y.; Ryu, J.H. Synergistic antimicrobial activity of oregano and thyme thymol essential oils against Leuconostoc citreum in a laboratory medium and tomato juice. Food Microbiol. 2020, 90, 103489. [Google Scholar]
- Lai, L.J.; Chiu, J.M.; Chiou, R.Y.Y. Fresh preservation of alfalfa sprouts and mushroom slices by soaking with thymol and resveratrol solutions. Food Sci. Nutr. 2017, 5, 776–783. [Google Scholar] [CrossRef]
- Kaliamurthi, S.; Selvaraj, G.; Hou, L.; Li, Z.; Wei, Y.; Gu, K.; Wei, D. Synergism of essential oils with lipid based nanocarriers: Emerging trends in preservation of grains and related food products. Grain Oil Sci. Technol. 2019, 2, 21–26. [Google Scholar] [CrossRef]
- Jeon, M.J.; Ha, J.W. Bactericidal and synergistic effects of X-ray irradiation and gallic acid against foodborne pathogens on lettuce. Food Microbiol. 2020, 92, 103584. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, C.; Sun, J.; Xin, M.; Yi, P.; He, X.; Sheng, J.; Zhou, Z.; Ling, D.; Zheng, F.; et al. Synergistic effects of ultraviolet light irradiation and high-oxygen modified atmosphere packaging on physiological quality, microbial growth and lignification metabolism of fresh-cut carrots. Postharvest Biol. Technol. 2021, 173, 111365. [Google Scholar] [CrossRef]
- Li, Y.; Brackett, R.E.; Shewfelt, R.L.; Beuchat, L.R. Changes in appearance and natural microflora on iceberg lettuce treated in warm, chlorinated water and then stored at refrigeration temperature. Food Microbiol. 2001, 18, 299–308. [Google Scholar] [CrossRef]
- Li, Y.; Brackett, R.E.; Chen, J.; Beuchat, L.R. Mild heat treatment of lettuce enhances growth of Listeria monocytogenes during subsequent storage at 5 °C or 15 °C. J. Appl. Microbiol. 2002, 92, 269–275. [Google Scholar] [CrossRef] [PubMed]
- McKellar, R.C.; Odumeru, J.; Zhou, T.; Harrison, A.; Mercer, D.G.; Toung, J.C.; Lu, X.; Boulter, J.; Piyasena, P.; Karr, S. Influence of a commercial warm chlorinated water treatment and packaging on the shelf-life of ready-to-use lettuce. Food Res. Int. 2004, 37, 343–354. [Google Scholar] [CrossRef]
- Martin-Diana, A.B.; Rico, D.; Frias, J.; Henehan, G.T.M.; Mulcahy, J.; Barat, J.M.; Barry-Ryan, C. Effect of calcium lactate and heat-shock on texture in fresh-cut lettuce during storage. J. Food Eng. 2006, 77, 1069–1077. [Google Scholar] [CrossRef]
- Abkas, M.Y.; Ölmez, H. Effectiveness of organic acid, ozonated water and chlorine dippings on microbial reduction and storage quality of fresh-cut iceberg lettuce. J. Sci. Food Agric. 2007, 87, 2609–2616. [Google Scholar]
- Huang, Y.; Chen, H. Effect of organic acids, hydrogen peroxide and mild heat on inactivation of Escherichia coli O157:H7 on baby spinach. Food Control 2011, 22, 1178–1183. [Google Scholar] [CrossRef]
- Pezzuto, A.; Belluco, S.; Losasso, C.; Patuzzi, I.; Bordin, P.; Piovesana, A.; Comin, D.; Mioni, R.; Ricci, A. Effectiveness of washing procedures in reducing Salmonella enterica and Listeria monocytogenes on a raw leafy green vegetables (Eruca vesicaria). Front. Microbiol. 2016, 7, 1663. [Google Scholar] [CrossRef] [PubMed]
- Wessels, K.; Rip, D.; Gouws, P. Salmonella in chicken meat: Consumption, outbreaks, characteristics, current control methods and the potential of bacteriophage use. Foods 2021, 10, 1742. [Google Scholar] [CrossRef]
- Deng, L.Z.; Mujumdar, A.S.; Zhang, Q.; Yang, X.H.; Wang, J.; Zheng, Z.A.; Gao, Z.J.; Xiao, H.W. Chemical and physical pretreatments of fruits and vegetables: Effects on drying characteristics and quality attributes—A comprehensive review. Crit. Rev. Food Sci. 2019, 59, 1408–1432. [Google Scholar] [CrossRef]
- Gientka, I.; Wójcicki, M.; Żuwalski, A.W.; Błażejak, S. Use of phage cocktail for improving the overall microbiological quality of sprouts—Two methods of application. Appl. Microbiol. 2021, 1, 21. [Google Scholar] [CrossRef]
- Wójcicki, M.; Błażejak, S.; Gientka, I.; Brzezicka, K. The concept of using bacteriophages to improve the microbiological quality of minimally processed foods. Acta Sci. Pol. Technol. Aliment. 2019, 18, 373–383. [Google Scholar] [PubMed]
- EFSA. The use and mode of action of bacteriophages in food production. Scientific opinion of the Panel on Biological Hazards. EFSA J. 2009, 1076, 1–26. [Google Scholar]
- Kosznik-Kwaśnicka, K.; Ciemińska, K.; Grabski, M.; Grabowski, Ł.; Górniak, M.; Jurczak-Kurek, A.; Węgrzyn, G.; Węgrzyn, A. Characteristics of a series of three bacteriophages infecting Salmonella enterica strains. Int. J. Mol. Sci. 2020, 21, 6152. [Google Scholar] [CrossRef] [PubMed]
- Żbikowska, K.; Michalczuk, M.; Dolka, B. The use of bacteriophages in the poultry industry. Animals 2020, 10, 872. [Google Scholar] [CrossRef]
- Upadhaya, S.D.; Ahn, J.M.; Cho, J.H.; Kim, J.Y.; Kang, D.K.; Kim, S.W.; Kim, H.B.; Kim, I.H. Bacteriophage cocktail supplementation improves growth performance, gut microbiome and production traits in broiler chickens. J. Anim. Sci. Biotechnol. 2021, 12, 49. [Google Scholar] [CrossRef]
- Kosznik-Kwaśnicka, K.; Grabowski, Ł.; Grabski, M.; Kaszubski, M.; Górniak, M.; Jurczak-Kurek, A.; Węgrzyn, G.; Węgrzyn, A. Bacteriophages vB_Sen-TO17 and vB_Sen-E22, newly isolated viruses from chicken feces, specific for several Salmonella enterica strains. Int. J. Mol. Sci. 2020, 21, 8821. [Google Scholar] [CrossRef]
- Oliveira, A.; Sousa, J.C.; Silva, A.C.; Melo, L.D.R.; Sillankorva, S. Chestnut honey and bacteriophage application to control Pseudomonas aeruginosa and Escherichia coli biofilms: Evaluation in an ex vivo wound model. Front. Microbiol. 2018, 9, 1725. [Google Scholar] [CrossRef] [PubMed]
- Hooton, S.P.T.; Atterbury, R.J.; Connerton, I.F. Application of a bacteriophage cocktail to reduce Salmonella Typhimurium U288 contamination on pig skin. Int. J. Food Microbiol. 2011, 151, 157–163. [Google Scholar] [CrossRef]
- Pao, S.; Randolph, S.; Westbrook, E.; Shen, H. Utilizing bacteriophages to control Salmonella in experimentally contaminated sprout seeds. J. Food Sci. 2004, 69, 127–130. [Google Scholar] [CrossRef]
- Fong, K.; LaBossiere, B.; Switt, A.I.M.; Delaquis, P.; Goodridge, L.; Levesque, R.C.; Danyluk, M.D.; Wang, S. Characterization of four novel bacteriophages isolated from British Columbia for control of non-typhoidal Salmonella in vitro and on sprouting alfalfa seeds. Front. Microbiol. 2017, 8, 2193. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.W.Y.; Delaquis, P.; Goodridge, L.; Lévesque, R.C.; Fong, K.; Wang, S. Inactivation of Salmonella enterica on post-harvest cantaloupe and lettuce by a lytic bacteriophage cocktail. Curr. Res. Food Sci. 2020, 2, 25–32. [Google Scholar] [CrossRef]
- Liu, A.; Liu, Y.; Peng, L.; Cai, X.; Shen, L.; Duan, M.; Ning, Y.; Liu, S.; Li, C.; Liu, Y.; et al. Characterization of the narrow-spectrum bacteriophage LSE7621 towards Salmonella enteritidis and its biocontrol potential on different foods. LWT—Food Sci. Technol. 2019, 118, 108791. [Google Scholar] [CrossRef]
- Biswas, B.; Adhya, S.; Washart, P.; Paul, B.; Trostel, A.N.; Powell, B.; Carlton, R.; Merril, C.R. Bacteriophage therapy rescues mice bacteriemic from a clinical isolate of vancomycin-resistant Enterococcus faecium. Infect. Immun. 2002, 70, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Guenther, S.; Huwyler, D.; Richard, S.; Loessner, M. Virulent bacteriophage for efficient biocontrol of Listeria monocytogenes in ready-to-eat foods. Appl. Environ. Microbiol. 2009, 75, 93–100. [Google Scholar] [CrossRef]
- Kazi, M.; Annapure, U.S. Bacteriophage biocontrol of foodborne pathogens. J. Food Sci. Technol. 2015, 53, 1355–1362. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Rodríguez-Rubio, L.; Martínez, B.; Rodríguez, A.; García, P. Bacteriophages as weapons against bacterial biofilms in the food industry. Front. Microb. 2016, 7, 825. [Google Scholar] [CrossRef] [PubMed]
- Spricigo, D.A.; Bardina, C.; Cortés, P.; Llagostera, M. Use of a bacteriophage cocktail to control Salmonella in food and the food industry. Int. J. Food Microbiol. 2013, 165, 169–174. [Google Scholar] [CrossRef]
- Połaska, M.; Sokołowska, B. Bacteriophages—A new hope or huge problem in the food industry. AIMS Microbiol. 2019, 5, 324–346. [Google Scholar] [CrossRef] [PubMed]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage application for food production and processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef]
- Svircev, A.; Roach, D.; Castle, A. Framing the future with bacteriophages in agriculture. Viruses 2018, 10, 218. [Google Scholar] [CrossRef] [PubMed]
- Kahn, L.H.; Bergeron, G.; Bourassa, M.W.; De Vegt, B.; Gill, J.; Gomes, F.; Malouin, F.; Opengart, K.; Ritter, G.D.; Singer, R.S.; et al. From farm management to bacteriophage therapy: Strategies to reduce antibiotic use in animal agriculture. Ann. N. Y. Acad. Sci. 2019, 1441, 31–39. [Google Scholar] [CrossRef] [PubMed]
- EFSA. Evaluation of the safety and efficacy of ListexTM P100 for reduction of pathogens on different ready-to-eat (RTE) food products. EFSA Panel on Biological Hazards. EFSA J. 2016, 14, 1–94. [Google Scholar]
- Arthur, T.M.; Kalchayanand, N.; Agga, G.E. Evaluation of bacteriophage application to cattle in lairage at beef processing plants to reduce Escherichia coli O157:H7 prevalence on hides and carcasses. Foodborne Pathog. Dis. 2016, 14, 17–22. [Google Scholar] [CrossRef]
- Oladunjoye, A.O.; Oyewole, S.A.; Singh, S.; Ijabadeniyi, O.A. Prediction of Listeria monocytogenes ATCC 7644 growth on fresh-cut produce treated with bacteriophage and sucrose monolaurate by using artificial neural network. Food Sci. Technol. 2017, 76, 9–17. [Google Scholar] [CrossRef]
- Garneau, E.J.; Moineau, S. Bacteriophages of lactic acid bacteria and their impact on milk fermentations. Microb. Cell Fact. 2001, 10, S20. [Google Scholar] [CrossRef]
- Pujato, S.A.; Quiberoni, A.; Mercanti, D.J. Bacteriophages on dairy foods. J. Appl. Microbiol. 2019, 126, 14–30. [Google Scholar] [CrossRef]
- Fernández, L.; Escobedo, S.; Gutiérrez, D.; Portilla, S.; Martínez, B.; García, P.; Rodríguez, A. Bacteriophages in the dairy environment: From enemies to allies. Antibiotics 2017, 6, 27. [Google Scholar] [CrossRef]
- Marcó, M.B.; Suárez, V.B.; Quiberoni, A.; Pujato, S.A. Inactivation of dairy bacteriophages by thermal and chemical treatments. Viruses 2019, 11, 480. [Google Scholar] [CrossRef]
- Yousuf, O.; Alam, I.; Bhat, M.I.; Titikshya, S. Assessment of microbial spoilage and techniques to avert the deterioration in fruits and vegetables. IJCS 2018, 6, 2230–2235. [Google Scholar]
- Rawat, S. Food spoilage: Microorganisms and their prevention. Asian J. Plant Sci. Res. 2015, 5, 47–56. [Google Scholar]
- Liu, H.K.; Li, Z.H.; Zhang, X.W.; Liu, Y.P.; Hu, J.P.; Yang, C.W.; Zhao, X.Y. The effects of ultrasound on the growth, nutritional quality and microbiological quality of sprouts. Trends Food Sci. Technol. 2021, 111, 292–300. [Google Scholar] [CrossRef]
- Salisbury, A.; Tsourkas, P.K. A method for improving the accuracy and efficiency of bacteriophage genome annotation. Int. J. Mol. Sci. 2019, 20, 3391. [Google Scholar] [CrossRef] [PubMed]
- Szymczak, P.; Rau, M.H.; Monteiro, J.M.; Pinho, M.G.; Filipe, S.R.; Vogensen, F.K.; Zeidan, A.A.; Janzen, T. A comparative genomics approach for identifying host-range determinants in Streptococcus thermophilus bacteriophages. Sci. Rep. 2019, 9, 7991. [Google Scholar] [CrossRef]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biot. 2017, 101, 3103. [Google Scholar] [CrossRef] [PubMed]
- Knecht, L.E.; Veljkovic, M.; Fieseler, L. Diversity and function of phage encoded depolymerases. Front. Microbiol. 2020, 10, 2949. [Google Scholar] [CrossRef]
- Cai, R.; Wang, G.; Le, S.; Wu, M.; Cheng, M.; Guo, Z.; Ji, Y.; Xi, H.; Zhao, C.; Wang, X.; et al. Three capsular polysaccharide synthesis-related glucosyltransferases, GT-1, GT-2 and WcaJ, are associated with virulence and phage sensitivity of Klebsiella pneumoniae. Front. Microbiol. 2019, 10, 1189. [Google Scholar] [CrossRef]
- Fernandes, S.; São-José, C. Enzymes and mechanisms employed by tailed bacteriophages to breach the bacterial cell barriers. Viruses 2018, 10, 396. [Google Scholar] [CrossRef]
- Drulis-Kawa, Z.; Majkowska-Skrobek, G.; Maciejewska, B. Bacteriophages and phage-derived proteins-application approaches. Curr. Med. Chem. 2015, 22, 1757–1773. [Google Scholar] [CrossRef]
- Cahill, J.; Young, R. Phage lysis: Multiple genes for multiple barriers. Adv. Virus Res. 2019, 103, 33–70. [Google Scholar]
- Abdelrahman, F.; Easwaran, M.; Daramola, O.I.; Ragab, S.; Lynch, S.; Oduselu, T.J.; Khan, F.M.; Ayobami, A.; Adnan, F.; Torrents, E.; et al. Phage-encoded endolysins. Antibiotics 2021, 10, 124. [Google Scholar] [CrossRef]
- Loessner, M.J. Bacteriophage endolysins—Current state of research and applications. Curr. Opin. Microbiol. 2005, 8, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Harper, D.R.; Parracho, H.M.R.T.; Walker, J.; Sharp, R.; Hughes, G.; Werthén, M.; Lehman, S.; Morales, S. Bacteriophages and biofilms. Antibiotics 2014, 3, 270–284. [Google Scholar] [CrossRef]
- Fischetti, V.A. Bacteriophage lytic enzymes: Novel anti-infectives. Trends Microbiol. 2005, 13, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Abedon, S.T. Bacteriophage and their enzymes in biofilm control. Curr. Pharm. Des. 2015, 21, 85–99. [Google Scholar] [CrossRef]
- Young, R. Bacteriophage lysis: Mechanism and regulation. Microbiol. Rev. 1992, 56, 430–481. [Google Scholar] [CrossRef] [PubMed]
- Park, S.C.; Shimamura, I.; Fukunaga, M.; Mori, K.I.; Nakai, T. Isolation of bacteriophages specific to a fish pathogen, Pseudomonas plecoglossicida, as a candidate for disease control. App. Environ. Microb. 2000, 66, 1416. [Google Scholar] [CrossRef]
- Zhao, J.; He, L.; Pan, L.; Liu, Y.; Yao, H.; Bao, G. Effect of a lytic bacteriophage on rabbits experimentally infected with pathogenic Escherichia coli. World Rabbit. Sci. 2017, 25, 273–279. [Google Scholar] [CrossRef][Green Version]
- Mahmoud, M.; Askora, A.; Barakat, A.B.; Rabie, O.E.-F.; Hassan, S.E. Isolation and characterization of polyvalent bacteriophages infecting multi drug resistant Salmonella serovars isolated from broilers in Egypt. Int. J. Food Microbiol. 2018, 266, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.-G.; Lim, J.A.; Song, Y.-R.; Heu, S.; Kim, G.H.; Koh, Y.J.; Oh, C.-S. Isolation and characterization of bacteriophages against Pseudomonas syringae pv. actinidiae causing bacterial canker disease in kiwifriut. J. Microbiol. Biotechnol. 2016, 26, 385–393. [Google Scholar]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018, 46, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Aprea, G.; D’Angelo, A.R.; Prencipe, V.A.; Migliorati, G. Bacteriophage morphological characterization by using Transmission Electron Microscopy. J. Life Sci. 2015, 9, 214–220. [Google Scholar]
- Zrelovs, N.; Dislers, A.; Kazaks, A. Motley crew: Overview of the currently available phage diversity. Front. Microbiol. 2020, 11, 579452. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, J.S.; Carstens, A.B.; Djurhuus, A.M.; Kot, W.; Neve, H.; Hansen, L.H. Pectobacterium Phage Jarilo Displays broad host range and represents a novel genus of bacteriophages within the family Autographiviridae. PHAGE 2020, 1, 237–244. [Google Scholar] [CrossRef]
- Timoshina, O.Y.; Shneider, M.M.; Evseev, P.V.; Shchurova, A.S.; Shelenkov, A.A.; Mikhaylova, Y.V.; Sokolova, O.S.; Kasimova, A.A.; Arbatsky, N.P.; Dmitrenok, A.S.; et al. Novel Acinetobacter baumannii bacteriophage aristophanes encoding structural polysaccharide deacetylase. Viruses 2021, 13, 1688. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.M.; Sullivan, M.B.; Knezevic, P.; van Zyl, L.J.; Sarkar, B.L.; Dutilh, B.E.; Alfenas-Zerbini, P.; Łobocka, M.; Tong, Y.; Brisner, J.R.; et al. Taxonomy of prokaryotic viruses: 2018–2019 update from the ICTV Bacterial and Archaeal Viruses Subcommittee. Arch. Virol. 2020, 165, 1253–1260. [Google Scholar] [CrossRef]
- Amarillas, L.; Lightbourn-Rojas, L.; Angulo-Gaxiola, A.K.; Heredia, J.B.; González-Robles, A.; León-Félix, J. The antibacterial effect of chitosan-based edible coating incorporated with a lytic bacteriophage against Escherichia coli O157:H7 on the surface of tomatoes. J. Food Saf. 2018, 38, e12571. [Google Scholar] [CrossRef]
- Chang, Y.; Bai, J.; Lee, J.-H.; Ryu, S. Mutation of a Staphylococcus aureus temperate bacteriophage to a virulent one and evaluation of its application. Food Microbiol. 2019, 82, 523–532. [Google Scholar] [CrossRef]
- Shahin, K.; Barazandeh, M.; Zhang, L.; Hedayatkhah, A.; He, T.; Bao, H.; Mansoorianfar, M.; Pang, M.; Wang, H.; Wei, R.; et al. Biodiversity of new lytic bacteriophages infecting Shigella spp. in a freshwater environment. Front. Microbiol. 2021, 12, 619323. [Google Scholar] [CrossRef]
- Leung, S.S.Y.; Carrigy, N.B.; Vehring, R.; Finlay, W.H.; Morales, S.; Carter, E.A.; Britton, W.J.; Kutter, E.; Chan, H.K. Jet nebulization of bacteriophages with different tail morphologies—Structural effects. Int. J. Pharm. 2019, 554, 322–326. [Google Scholar] [CrossRef]
- Gascuel, O. BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 1997, 14, 685–695. [Google Scholar] [CrossRef]
- Mihara, T.; Nishimura, Y.; Shimizu, Y.; Nishiyama, H.; Yoshikawa, G.; Uehara, H.; Hingamp, P.; Goto, S.; Ogata, H. Linking virus genomes with host taxonomy. Viruses 2016, 8, 66. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The viral proteomic tree server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Miroshnikov, K.A.; Evseev, P.V.; Lukianova, A.A.; Ignatov, A.N. Tailed lytic bacteriophages of soft rot Pectobacteriaceae. Microorganisms 2021, 9, 1819. [Google Scholar] [CrossRef] [PubMed]
- Merabishvili, M.; Vandenheuvel, D.; Kropinski, A.M.; Mast, J.; De Vos, D.; Verbeken, G.; Noben, J.P.; Lavigne, R.; Vaneechoutte, M.; Pirnay, J.P. Characterization of newly isolated lytic bacteriophages active against Acinetobacter baumannii. PLoS ONE 2014, 9, e104853. [Google Scholar] [CrossRef]
- Delesalle, V.A.; Tanke, N.T.; Vill, A.C.; Krukonis, G.P. Testing hypotheses for the presence of tRNA genes in mycobacteriophage genomes. Bacteriophage 2016, 6, e1219441. [Google Scholar] [CrossRef]
- Weiss, S.B.; Hsu, W.T.; Foft, J.W.; Schernerg, N.H. Transfer RNA coded by the T4 bacteriophage genome. Proc. Natl. Acad. Sci. USA 1968, 61, 114–121. [Google Scholar] [CrossRef]
- Morgado, S.; Vicente, A.C. Global in-silico scenario of tRNA genes and their organization in virus genomes. Viruses 2019, 11, 180. [Google Scholar] [CrossRef]
- Bonilla, B.E.; Costa, A.R.; Van Den Berg, D.F.; Van Rossum, T.; Hagedoorn, S.; Walinga, H.; Xiao, M.; Haas, P.J.; Brouns, S.J.J. Genomic characterization of four novel bacteriophages infecting the clinical pathogen Klebsiella pneumoniae. DNA Res. 2021, 28, dsab013. [Google Scholar] [CrossRef] [PubMed]
- Carbone, A. Codon bias is a major factor explaining phage evolution in translationally biased hosts. J. Mol. Evol. 2008, 66, 210–223. [Google Scholar] [CrossRef] [PubMed]
- Canchaya, C.; Fournous, G.; Brüssow, H. The impact of prophages on bacterial chromosomes. Mol. Microbiol. 2004, 53, 9–18. [Google Scholar] [CrossRef]
- Bailly-Bechet, M.; Vergassola, M.; Rocha, E. Causes for the intriguing presence of tRNAs in phages. Genome Res. 2007, 17, 1486–1495. [Google Scholar] [CrossRef] [PubMed]
- Roucourt, B.; Lavigne, R. The role of interactions between phage and bacterial proteins within the infected cell: A diverse and puzzling interactome. Environ. Microbiol. 2009, 11, 2789–2805. [Google Scholar] [CrossRef]
- Al-Shayeb, B.; Sachdeva, R.; Chen, L.X.; Ward, F.; Munk, P.; Devoto, A.; Castelle, C.J.; Olm, M.R.; Bouma-Gregson, K.; Amano, Y.; et al. Clades of huge phages from across Earth’s ecosystems. Nature 2020, 578, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Ku, H.J.; Lee, D.H.; Kim, Y.T.; Shin, H.; Ryu, S.; Lee, J.H. Characterization and genomic study of the novel bacteriophage HY01 infecting both Escherichia coli O157:H7 and Shigella flexneri: Potential as a biocontrol agent in food. PLoS ONE 2016, 11, e0168985. [Google Scholar]
- Duc, H.M.; Son, H.M.; Honjoh, K.I.; Miyamoto, T. Isolation and application of bacteriophages to reduce Salmonella contamination in raw chicken meat. LWT 2018, 91, 353–360. [Google Scholar] [CrossRef]
- Guo, Y.; Li, J.; Islam, M.S.; Yan, T.; Zhou, Y.; Liang, L.; Connerton, I.F.; Deng, K.; Li, J. Application of a novel phage vB_SalS-LPSTLL for the biological control of Salmonella in foods. Food Res. Int. 2021, 147, 110492. [Google Scholar] [CrossRef] [PubMed]
- Goodridge, L.D.; Bisha, B. Phage-based biocontrol strategies to reduce foodborne pathogens in foods. Bacteriophages 2013, 1, 130–137. [Google Scholar] [CrossRef]
- Tan, L.-H.; Chan, K.G.; Lee, L.H. Application of bacteriophages in biocontrol of major foodborne bacterial pathogens. J. Mol. Biol. Mol. Imaging 2014, 1, 9. [Google Scholar]
- Necel, A.; Bloch, S.; Nejman-Faleńczyk, B.; Grabski, M.; Topka, G.; Dydecka, A.; Kosznik-Kwaśnicka, K.; Grabowski, Ł.; Jurczak-Kurek, A.; Tomasz Wołkowicz, T.; et al. Characterization of a bacteriophage, vB_Eco4M-7, that effectively infects many Escherichia coli O157 strains. Sci. Rep. 2020, 10, 3743. [Google Scholar] [CrossRef]
- Brito, I.L. Examining horizontal gene transfer in microbial communities. Nat. Rev. Microbiol. 2021, 19, 442–453. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.L. Bacteriophage-mediated horizontal gene transfer: Transduction. In Bacteriophages: Biology, Technology, Therapy; Harper, D., Abedon, S., Burrowes, B., McConville, M., Eds.; Springer: Cham, Switzerland; New York, NY, USA, 2017; pp. 151–192. [Google Scholar]
- Castillo, D.; Kauffman, K.; Hussain, F.; Kalatzis, P.; Rørbo, N.; Polz, M.F.; Middelboe, M. Widespread distribution of prophage-encoded virulence factors in marine Vibrio communities. Sci. Rep. 2018, 8, 9973. [Google Scholar] [CrossRef]
- Moon, K.; Jeon, J.H.; Kang, I.; Park, K.S.; Lee, K.; Cha, C.J.; Lee, S.H.; Cho, J.C. Freshwater viral metagenome reveals novel and functional phage-borne antibiotic resistance genes. Microbiome 2020, 8, 75. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Adeyemi, D.E.; Park, M.K. Characterization of a new and efficient polyvalent phage infecting E. coli O157:H7, Salmonella spp., and Shigella sonnei. Microorganisms 2021, 9, 2105. [Google Scholar] [CrossRef] [PubMed]
- Gigante, A.M.; Hampton, C.M.; Dillard, R.S.; Gil, F.; Catalão, M.J.; Moniz-Pereira, J.; Wright, E.R.; Pimentel, M. The Ms6 mycolyl-arabinogalactan esterase LysB is essential for an efficient mycobacteriophage-induced lysis. Viruses 2017, 9, 343. [Google Scholar] [CrossRef]
- Bujak, K.; Decewicz, P.; Kamiński, J.; Radlińska, M. Identification, characterization, and genomic analysis of novel Serratia temperate phages from a gold mine. Int. J. Mol. Sci. 2020, 21, 6709. [Google Scholar] [CrossRef] [PubMed]
- Jończyk, E.; Kłak, M.; Międzybrodzki, R.; Górski, A. The influence of external factors on bacteriophages. Folia Microbiol. 2011, 56, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Olson, M.R.; Axler, R.P.; Hicks, R.E. Effects of freezing and storage temperature on MS2 viability. J. Virol. Methods 2004, 122, 147–152. [Google Scholar] [CrossRef]
- Tey, B.T.; Ooi, S.T.; Yong, K.C.; Tan Ng, M.Y.; Ling, T.C.; Tan, W.S. Production of fusion m13 phage bearing the disulphide constrained peptide sequence (C-WSFFSNI-C) that interacts with hepatitis B core antigen. Afr. J. Biotechnol. 2009, 8, 268–273. [Google Scholar]
- Donovan, D.M.; Dong, S.; Garrett, W.; Rousseau, G.M.; Moineau, S.; Pritchard, D.G. Peptidoglycan hydrolase fusions maintain their parental specificities. Appl. Environ. Microb. 2006, 72, 2988–2996. [Google Scholar] [CrossRef]
- Jończyk-Matysiak, E.; Łodej, N.; Kula, D.; Owczarek, B.; Orwat, F.; Międzybrodzki, R.; Neuberg, J.; Bagińska, N.; Weber-Dąbrowska, B.; Górski, A. Factors determining phage stability/activity: Challenges in practical phage application. Expert Rev. Anti-Infect. Ther. 2019, 17, 583–606. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.Y.; Li, W.W.; Christie, G.; Setlow, P.; Li, Y.Q. Characterization of heterogeneity and dynamics of lysis of single Bacillus subtilis cells upon prophage induction during spore germination, outgrowth, and vegetative growth using raman tweezers and live-cell phase-contrast microscopy. Anal. Chem. 2020, 93, 1443–1450. [Google Scholar] [CrossRef]
- Ding, F.; Allen, V.; Luo, W.; Zhang, S.; Duan, Y. Molecular mechanisms underlying heat or tetracycline treatments for citrus HLB control. Hortic. Res. 2018, 5, 30. [Google Scholar] [CrossRef]
- Shahin, K.; Bouzari, M. Bacteriophages application for biocontrolling Shigella flexneri in contminated foods. J. Food Sci. Technol. 2018, 55, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Thung, T.Y.; Premarathne, J.M.K.J.K.; Chang, W.S.; Loo, Y.Y.; Chin, Y.Z.; Kuan, C.H.; Tan, C.W.; Basri, D.F.; Radzi, C.W.J.W.M.; Radu, S. Use of lytic bacteriophage to control Salmonella Enteritidis in retail food. Food Sci. Technol. 2017, 78, 222–225. [Google Scholar] [CrossRef]
- Abd-Allah, I.M.; El-Housseiny, G.S.; Yahia, I.S.; Aboshanab, K.M.; Hassouna, N.A. Rekindling of a masterful precedent; Bacteriophage: Reappraisal and future pursuits. Front. Cell. Infect. Mi. 2021, 11, 477. [Google Scholar] [CrossRef] [PubMed]
- Fister, S.; Robben, C.; Witte, A.K.; Schoder, D.; Wagner, M.; Rossmanith, P. Influence of environmental factors on phage-bacteria interaction and on the efficacy and infectivity of phage P100. Front. Microbiol. 2016, 7, 1152. [Google Scholar] [CrossRef]
- Tokman, J.I.; Kent, D.J.; Wiedmann, M.; Denes, T. Temperature significantly affects the plaquing and adsorption efficiencies of Listeria phages. Front. Microbiol. 2016, 7, 631. [Google Scholar] [CrossRef]
- Silva, J.B.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef]
- Stone, E.; Campbell, K.; Grant, I.; McAuliffe, O. Understanding and exploiting phage-host interactions. Viruses 2019, 11, 567. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, F.; Helm, R.F.; Broadway, K.M.; Scharf, B.E. More than rotating flagella: Lipopolysaccharide as a secondary receptor for flagellotropic phage 7-7-1. J. Bacteriol. 2018, 200, e00363-18. [Google Scholar] [CrossRef] [PubMed]
- Denes, T.; Wiedmann, M. Environmental responses and phage susceptibility in foodborne pathogens: Implications for improving applications in food safety. Curr. Opin. Biotechnol. 2014, 26, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Jamal, M.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Das, C.R. Isolation and characterization of a bacteriophage and its utilization against multi-drug resistant Pseudomonas aeruginosa-2995. Life Sci. 2017, 190, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Shahin, K.; Bao, H.; Komijani, M.; Barazandeh, M.; Bouzari, M.; Hedayatkhah, A.; Zhang, I.; Zhao, H.; He, T.; Pang, M.; et al. Isolation, characterization and PCR-based molecular identification of a siphoviridae phage infecting Shigella dysenteriae. Microb. Pathog. 2019, 131, 175–180. [Google Scholar] [CrossRef]
- Li, Y.K.; Chen, H.; Shu, M.; Zhong, C.; Bi, Y.; Yang, H.H.; Wu, G.P. Isolation, characterization and application of an alkaline resistant virulent bacteriophage JN01 against Escherichia coli O157:H7 in milk and beef. LWT—Food Sci. Technol. 2021, 144, 111266. [Google Scholar] [CrossRef]
- Sváb, D.; Falgenhauer, L.; Rohde, M.; Szabó, J.; Chakraborty, T.; Tóth, I. Identification and characterization of T5-like bacteriophages representing two novel subgroups from food products. Front. Microbiol. 2018, 9, 222. [Google Scholar] [CrossRef] [PubMed]
- Thung, T.Y.; Lee, E.; Mahyudin, N.A.; Anuradha, K.; Mazlan, N.; Kuan, C.H.; Pui, C.F.; Ghazali, F.M.; Rashid, N.K.M.A.; Rollon, W.D.; et al. Evaluation of a lytic bacteriophage for bio-control of Salmonella typhimurium in different food matrices. Food Sci. Technol. 2019, 105, 211–214. [Google Scholar] [CrossRef]
- Międzybrodzki, R.; Kłak, M.; Jończyk-Matysiak, E.; Bubak, B.; Wójcik, A.; Kaszowska, M.; Weber-Dąbrowska, B.; Łobocka, M.; Górski, A. Means to facilitate the overcoming of gastric juice barier by a therapeutic staphylococcal bacteriophage A5/80. Front. Microbiol. 2017, 8, 467. [Google Scholar] [CrossRef]
- Wang, Q.; Sabour, P.M. Encapsulation and controlled release of bacteriophages. In Bacteriophages in the Control of Food and Waterborne Pathogens; Sabour, P.M., Griffiths, M.M.W., Eds.; ASM Press: Washington, DC, USA, 2010; pp. 237–255. [Google Scholar]
- Mirzaei, K.; Nilsson, A.S. Isolation of phage for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef]
- Ackermann, H.W. 5500 Phages examined in the electron microscope. Arch. Virol. 2007, 151, 227–243. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.W. Basic phage electron microscopy. In Bacteriophages. Methods and Protocols. Vol. 1: Isolation, Characterization and Interactions; Clokie, A.R.J., Kropinski, A.M., Eds.; Humana Press: Totowa, NJ, USA, 2009; pp. 113–126. [Google Scholar]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Ecale Zhou, C.L.; Malfatti, S.; Kimbrel, J.; Philipson, C.; McNair, K.; Hamilton, T.; Edwards, R.; Souza, B. multiPhATE: Bioinformatics pipeline for functional annotation of phage isolates. Bioinformatics 2019, 35, 4402–4404. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Hguyen, A.L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, 517–525. [Google Scholar] [CrossRef] [PubMed]










| Bacterial Strain Code | Bacterial Strain Number | Classification acc. to Bergey’s | Motility at a Temp. of 37 °C | Lactose Fermentation Capability | Bacteria Identification acc. to 16S rRNA Sequencing (GenBank No.) | Bacteria Identification acc. to MALDI-TOF MS | |||
|---|---|---|---|---|---|---|---|---|---|
| Gram Staining Result | Cell Shape | Oxygen Tolerance | Spore Formation | ||||||
| RUC-09 | KKP 3655 | Gram (−) | bacilli | relative anaerobes | non-spore-forming | motile | lactose-positive | Citrobacter freundii (MZ827001) | Citrobacter freundii |
| MLSC-11 | KKP 3082 | Gram (−) | bacilli | relative anaerobes | non-spore-forming | motile | lactose-positive | Enterobacter cloacae (MZ827006) | Enterobacter sp. |
| MLSC-21 | KKP 3083 | Gram (−) | bacilli | relative anaerobes | non-spore-forming | motile | lactose-positive | Enterobacter ludwigii (MZ827002) | Klebsiella oxytoca |
| MLSB-04 | KKP 3084 | Gram (−) | bacilli | relative anaerobes | non-spore-forming | motile | lactose-positive | Serratia fonticola (MZ827668) | Serratia fonticola |
| Bacterial Strain | Phage Strain | Phage Titer (MF) in the Lysate (PFU mL−1) | OD600 of Bacterial Culture at Which MOI 1.0 |
|---|---|---|---|
| Citrobacter freundii KKP 3655 | Citrobacter phage KKP 3664 | 6.2 × 109 | 0.302 |
| Enterobacter cloacae KKP 3082 | Enterobacter phage KKP 3262 | 1.4 × 1010 | 0.262 |
| Enterobacter ludwigii KKP 3083 | Enterobacter phage KKP 3263 | 7.2 × 108 | 0.234 |
| Serratia fonticola KKP 3084 | Serratia phage KKP 3264 | 4.4 × 108 | 0.208 |
| Bacterial Strain | Control Culture | MOI 1.0 | MOI 0.1 | |||
|---|---|---|---|---|---|---|
| ΔOD | μ [h−1] | ΔOD | μ [h−1] | ΔOD | μ [h−1] | |
| Citrobacter freundii KKP 3655 | 0.255 | 0.031 | 0.136 | 0.017 | 0.273 | 0.025 |
| Enterobacter cloacae KKP 3082 | 0.312 | 0.026 | 0.147 | 0.019 | 0.261 | 0.023 |
| Enterobacter ludwigii KKP 3083 | 0.486 | 0.035 | 0.160 | 0.014 | 0.206 | 0.022 |
| Serratia fonticola KKP 3084 | 0.464 | 0.039 | 0.102 | 0.008 | 0.173 | 0.019 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wójcicki, M.; Średnicka, P.; Błażejak, S.; Gientka, I.; Kowalczyk, M.; Emanowicz, P.; Świder, O.; Sokołowska, B.; Juszczuk-Kubiak, E. Characterization and Genome Study of Novel Lytic Bacteriophages against Prevailing Saprophytic Bacterial Microflora of Minimally Processed Plant-Based Food Products. Int. J. Mol. Sci. 2021, 22, 12460. https://doi.org/10.3390/ijms222212460
Wójcicki M, Średnicka P, Błażejak S, Gientka I, Kowalczyk M, Emanowicz P, Świder O, Sokołowska B, Juszczuk-Kubiak E. Characterization and Genome Study of Novel Lytic Bacteriophages against Prevailing Saprophytic Bacterial Microflora of Minimally Processed Plant-Based Food Products. International Journal of Molecular Sciences. 2021; 22(22):12460. https://doi.org/10.3390/ijms222212460
Chicago/Turabian StyleWójcicki, Michał, Paulina Średnicka, Stanisław Błażejak, Iwona Gientka, Monika Kowalczyk, Paulina Emanowicz, Olga Świder, Barbara Sokołowska, and Edyta Juszczuk-Kubiak. 2021. "Characterization and Genome Study of Novel Lytic Bacteriophages against Prevailing Saprophytic Bacterial Microflora of Minimally Processed Plant-Based Food Products" International Journal of Molecular Sciences 22, no. 22: 12460. https://doi.org/10.3390/ijms222212460
APA StyleWójcicki, M., Średnicka, P., Błażejak, S., Gientka, I., Kowalczyk, M., Emanowicz, P., Świder, O., Sokołowska, B., & Juszczuk-Kubiak, E. (2021). Characterization and Genome Study of Novel Lytic Bacteriophages against Prevailing Saprophytic Bacterial Microflora of Minimally Processed Plant-Based Food Products. International Journal of Molecular Sciences, 22(22), 12460. https://doi.org/10.3390/ijms222212460









