Functional Studies of Plant Latex as a Rich Source of Bioactive Compounds: Focus on Proteins and Alkaloids
Abstract
:1. Introduction
2. Diversity and Role of Latex in Plant Physiology
3. Main Components of Latex-Secondary Metabolites
4. Spectrum of Latex Proteins
| PR Proteins | Function | Latex-Bearing Plant Species | Reference |
|---|---|---|---|
| PR 2 | β-1,3-glucanases | Chelidonium majus Hevea brasiliensis | [78,83] |
| PR 3 | Class I, II, IV, V, VI, VII Chitinases | Chelidonium majus Hevea brasiliensis | [78,83] |
| PR 4 | Class I, II Chitinases | Chelidonium majus Hevea brasiliensis Carica papaya | [45,78,83] |
| PR 5 | Thaumatin-like proteins | Chelidonium majus Hevea brasiliensis | [78,83] |
| PR 6 | Proteinase inhibitor | Hevea brasiliensis Ficus carica Carica papaya | [44,45,83] |
| PR 7 | Endoproteinase | Chelidonium majus Hevea brasiliensis | [78,83] |
| PR 8 | Class III Chitinase | Hevea brasiliensis | [83] |
| PR 9 | Peroxidase | Chelidonium majus Hevea brasiliensis Papaver somniferum | [78,83,84] |
| PR 10 | Ribonuclease-like proteins | Chelidonium majus Papaver somniferum | [78,84] |
| PR 11 | Class I Chitinase | Chelidonium majus | [78] |
| PR 12 | Defensin | Chelidonium majus Hevea brasiliensis | [78,83] |
| PR 14 | Lipid-transfer protein | Chelidonium majus Hevea brasiliensis | [78,83] |
| PR 15 | Oxalate oxidase | Chelidonium majus | [78] |
| PR 16 | Oxidase-like | Chelidonium majus | [78] |
| PR 17 | Antifungal and antiviral | Chelidonium majus | [78] |
5. Biomedical Properties of Latex from Selected Plants with the Focus on Chelidonium majus L.
5.1. Antiviral Activity
5.2. Cytotoxicity
5.3. Antimicrobial Activity
5.4. Immunomodulatory Properties
6. CRISPR/Cas 9 System as Future Direction for Functional Analysis of Proteins
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jesus, A.; Bonhomme, V.; Evin, A.; Ivorra, S.; Soteras, R.; Salavert, A.; Antolín, F.; Bouby, L. A Morphometric Approach to Track Opium Poppy Domestication. Sci. Rep. 2021, 11, 9778. [Google Scholar] [CrossRef]
- Ren, G.; Zhang, X.; Li, Y.; Ridout, K.; Serrano-Serrano, M.L.; Yang, Y.; Liu, A.; Ravikanth, G.; Nawaz, M.A.; Mumtaz, A.S.; et al. Large-Scale Whole-Genome Resequencing Unravels the Domestication History of Cannabis sativa. Sci. Adv. 2021, 7, eabg2286. [Google Scholar] [CrossRef]
- Zielińska, S.; Jezierska-Domaradzka, A.; Wójciak-Kosior, M.; Sowa, I.; Junka, A.; Matkowski, A.M. Greater Celandine’s Ups and Downs—21 Centuries of Medicinal Uses of Chelidonium majus from the Viewpoint of Today’s Pharmacology. Front. Pharmacol. 2018, 9, 299. [Google Scholar] [CrossRef] [Green Version]
- Gilca, M.; Gaman, L.; Panait, E.; Stoian, I.; Atanasiu, V. Chelidonium maju—An Integrative Review: Traditional Knowledge versus Modern Findings. Complementary Med. Res. 2010, 17, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Maji, A.K.; Banerji, P. Chelidonium majus L. (Greater celandine—A Review on Its Phytochemical and Therapeutic Perspectives. Int. J. Herb. Med. 2015, 3, 10–27. [Google Scholar] [CrossRef]
- Nawrot, J.; Wilk-Jędrusik, M.; Nawrot, S.; Nawrot, K.; Wilk, B.; Dawid-Pać, R.; Urbańska, M.; Micek, I.; Nowak, G.; Gornowicz-Porowska, J. Milky Sap of Greater Celandine (Chelidonium majus L.) and Anti-Viral Properties. Int. J. Environ. Res. Public Health 2020, 17, 1540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salomé Abarca, L.F.; Klinkhamer, P.G.L.; Choi, Y.H. Plant Latex, from Ecological Interests to Bioactive Chemical Resources. Planta Med. 2019, 85, 856–868. [Google Scholar] [CrossRef] [Green Version]
- Nawrot, R. (Ed.) Latex, Laticifers and Their Molecular Components—From Functions to Possible Applications; Advances in Botanical Research Series; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Kekwick, R.G.O. Latex and Laticifers. In Encyclopedia of Life Sciences 2002; John Wiley & Sons: London, UK; New York, NY, USA, 2002. [Google Scholar]
- Ramos, M.V.; Demarco, D.; da Costa Souza, I.C.; de Freitas, C.D.T. Laticifers, Latex, and Their Role in Plant Defense. Trends Plant Sci. 2019, 24, 553–567. [Google Scholar] [CrossRef]
- Hagel, J.M.; Yeung, E.C.; Facchini, P.J. Got Milk? The Secret Life of Laticifers. Trends Plant Sci. 2008, 13, 631–639. [Google Scholar] [CrossRef]
- Prado, E.; Demarco, D. Laticifers and secretory ducts: Similarities and differences. In Ecosystem Services and Global Ecology; InTech: London, UK, 2018; ISBN 9781789237382. [Google Scholar]
- Marinho, C.R.; Teixeira, S.P. Cellulases and Pectinases Act Together on the Development of Articulated Laticifers in Ficus montana and Maclura tinctoria (Moraceae). Protoplasma 2019, 256, 1093–1107. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, S.; Taira, T.; Oda, K.; Yamato, K.T.; Inukai, Y.; Hori, Y. Comparative Study of Gene Expression and Major Proteins’ Function of Laticifers in Lignified and Unlignified Organs of Mulberry. Planta 2012, 235, 589–601. [Google Scholar] [CrossRef]
- Van Veenendaal, W.L.H.; Den Outer, R.W. Distribution and Development of the Non-Articulated Branched Laticifers of Morus nigra L. (Moraceae). Acta Bot. Neerl. 1990, 39, 285–296. [Google Scholar] [CrossRef]
- Webster, C.C. Natural Rubber: Biology, Cultivation and Technology. Agric. Syst. 1994, 45, 233–235. [Google Scholar] [CrossRef]
- Dussourd, D.E. Entrapment of Aphids and Whiteflies in Lettuce Latex. Ann. Entomol. Soc. Am. 1995, 88, 163–172. [Google Scholar] [CrossRef]
- Agrawal, A.A.; Konno, K. Latex: A Model for Understanding Mechanisms, Ecology, and Evolution of Plant Defense against Herbivory. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 311–331. [Google Scholar] [CrossRef] [Green Version]
- Nawrot, R.; Wołuń-Cholewa, M.; Goździcka-Józefiak, A. Nucleases Isolated from Chelidonium majus L. Milky Sap Can Induce Apoptosis in Human Cervical Carcinoma HeLa Cells but Not in Chinese Hamster Ovary CHO Cells. Folia Histochem. Cytobiol. 2008, 46, 79–83. [Google Scholar] [CrossRef] [Green Version]
- Konno, K. Plant Latex and Other Exudates as Plant Defense Systems: Roles of Various Defense Chemicals and Proteins Contained Therein. Phytochemistry 2011, 72, 1510–1530. [Google Scholar] [CrossRef] [PubMed]
- Wink, M. Plant Secondary Metabolism: Diversity, Function and Its Evolution. Nat. Prod. Commun. 2008, 3, 1205–1216. [Google Scholar] [CrossRef] [Green Version]
- Souza, D.P.; Freitas, C.D.T.; Pereira, D.A.; Nogueira, F.C.; Silva, F.D.A.; Salas, C.E.; Ramos, M.V. Laticifer Proteins Play a Defensive Role against Hemibiotrophic and Necrotrophic Phytopathogens. Planta 2011, 234, 183–193. [Google Scholar] [CrossRef]
- Ramos, M.V.; Grangeiro, T.B.; Freire, E.A.; Sales, M.P.; Souza, D.P.; Araújo, E.S.; Freitas, C.D.T. The Defensive Role of Latex in Plants: Detrimental Effects on Insects. Arthropod-Plant Interact. 2010, 4, 57–67. [Google Scholar] [CrossRef]
- Men, X.; Wang, F.; Chen, G.-Q.; Zhang, H.-B.; Xian, M. Biosynthesis of Natural Rubber: Current State and Perspectives. Int. J. Mol. Sci. 2018, 20, 50. [Google Scholar] [CrossRef] [Green Version]
- Tang, C.; Qi, J.; Li, H.; Zhang, C.; Wang, Y. A Convenient and Efficient Protocol for Isolating High-Quality RNA from Latex of Hevea brasiliensis (para Rubber Tree). J. Biochem. Biophys. Methods 2007, 70, 749–754. [Google Scholar] [CrossRef]
- Salvucci, M.E.; Barta, C.; Byers, J.A.; Canarini, A. Photosynthesis and Assimilate Partitioning between Carbohydrates and Isoprenoid Products in Vegetatively Active and Dormant Guayule: Physiological and Environmental Constraints on Rubber Accumulation in a Semiarid Shrub. Physiol. Plant. 2010, 140, 368–379. [Google Scholar] [CrossRef]
- Sessa, R.A.; Bennett, M.H.; Lewis, M.J.; Mansfield, J.W.; Beale, M.H. Metabolite Profiling of Sesquiterpene Lactones from Lactuca Species. Major Latex Components Are Novel Oxalate and Sulfate Conjugates of Lactucin and Its Derivatives. J. Biol. Chem. 2000, 275, 26877–26884. [Google Scholar] [CrossRef]
- Valitova, J.N.; Sulkarnayeva, A.G.; Minibayeva, F.V. Plant Sterols: Diversity, Biosynthesis, and Physiological Functions. Biochemistry 2016, 81, 819–834. [Google Scholar] [CrossRef] [PubMed]
- Che Sulaiman, I.S.; Basri, M.; Fard Masoumi, H.R.; Chee, W.J.; Ashari, S.E.; Ismail, M. Effects of Temperature, Time, and Solvent Ratio on the Extraction of Phenolic Compounds and the Anti-Radical Activity of Clinacanthus Nutans Lindau Leaves by Response Surface Methodology. Chem. Cent. J. 2017, 11, 54. [Google Scholar] [CrossRef]
- Snook, M.E.; Data, E.S.; Kays, S.J. Characterization and Quantitation of Hexadecyl, Octadecyl, and Eicosyl Esters of P-Coumaric Acid in the Vine and Root Latex of Sweetpotato [Ipomoea batatas (L.) Lam.]. J. Agric. Food Chem. 1994, 42, 2589–2595. [Google Scholar] [CrossRef]
- Ismun, A.; Ariffin, M.M.; Razak, S.B.A.; Wei, O.C.; Ahmad, F.T.; Mubarak, A. determination of polyphenol contents in Hevea brasiliensis and rubber-processing effluent. Malays. J. Anal. Sci. 2018, 22, 185–196. [Google Scholar] [CrossRef]
- Huber, M.; Triebwasser-Freese, D.; Reichelt, M.; Heiling, S.; Paetz, C.; Chandran, J.N.; Bartram, S.; Schneider, B.; Gershenzon, J.; Erb, M. Identification, Quantification, Spatiotemporal Distribution and Genetic Variation of Major Latex Secondary Metabolites in the Common Dandelion (Taraxacum officinale Agg.). Phytochemistry 2015, 115, 89–98. [Google Scholar] [CrossRef]
- Nguyen, T.-D.; Dang, T.-T.T. Cytochrome P450 Enzymes as Key Drivers of Alkaloid Chemical Diversification in Plants. Front. Plant Sci. 2021, 12, 682181. [Google Scholar] [CrossRef]
- Itenov, K.; Mølgaard, P.; Nyman, U. Diurnal Fluctuations of the Alkaloid Concentration in Latex of Poppy Papaver Somniferum Is due to Day–night Fluctuations of the Latex Water Content. Phytochemistry 1999, 52, 1229–1234. [Google Scholar] [CrossRef]
- Tomè, F.; Colombo, M.L. Distribution of Alkaloids in Chelidonium majus and Factors Affecting Their Accumulation. Phytochemistry 1995, 40, 37–39. [Google Scholar] [CrossRef]
- Mikołajczak, P.Ł.; Kędzia, B.; Ożarowski, M.; Kujawski, R.; Bogacz, A.; Bartkowiak-Wieczorek, J.; Białas, W.; Gryszczyńska, A.; Buchwald, W.; Szulc, M.; et al. Evaluation of Anti-Inflammatory and Analgesic Activities of Extracts from Herb of Chelidonium majus L. Cent. Eur. J. Immunol. 2015, 40, 400–410. [Google Scholar] [CrossRef] [Green Version]
- Kopp, B.; Bauer, W.P.; Bernkop-Schnürch, A. Analysis of Some Malaysian Dart Poisons. J. Ethnopharmacol. 1992, 36, 57–62. [Google Scholar] [CrossRef]
- Agrawal, A.A.; Petschenka, G.; Bingham, R.A.; Weber, M.G.; Rasmann, S. Toxic Cardenolides: Chemical Ecology and Coevolution of Specialized Plant-Herbivore Interactions. New Phytol. 2012, 194, 28–45. [Google Scholar] [CrossRef]
- Malcolm, S.B. Milkweeds, Monarch Butterflies and the Ecological Significance of Cardenolides. Chemoecology 1994, 5–6, 101–117. [Google Scholar] [CrossRef]
- Cho, W.K.; Jo, Y.; Chu, H.; Park, S.-H.; Kim, K.-H. Integration of Latex Protein Sequence Data Provides Comprehensive Functional Overview of Latex Proteins. Mol. Biol. Rep. 2014, 41, 1469–1481. [Google Scholar] [CrossRef]
- Balakireva, A.V.; Zamyatnin, A.A. Indispensable Role of Proteases in Plant Innate Immunity. Int. J. Mol. Sci. 2018, 19, 629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konno, K.; Hirayama, C.; Nakamura, M.; Tateishi, K.; Tamura, Y.; Hattori, M.; Kohno, K. Papain Protects Papaya Trees from Herbivorous Insects: Role of Cysteine Proteases in Latex. Plant J. 2004, 37, 370–378. [Google Scholar] [CrossRef]
- de Freitas, C.D.T.; de Freitas, C.D.T.; de Souza, D.P.; Araújo, E.S.; Cavalheiro, M.G.; Oliveira, L.S.; Ramos, M.V. Anti-Oxidative and Proteolytic Activities and Protein Profile of Laticifer Cells of Cryptostegia Grandiflora, Plumeria Rubra and Euphorbia Tirucalli. Braz. J. Plant Physiol. 2010, 22, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-Y.; Park, S.-C.; Hwang, I.; Cheong, H.; Nah, J.-W.; Hahm, K.-S.; Park, Y. Protease Inhibitors from Plants with Antimicrobial Activity. Int. J. Mol. Sci. 2009, 10, 2860–2872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.S.; Kim, Y.O.; Ryu, H.J.; Kwak, Y.S.; Lee, J.Y.; Kang, H. Isolation of Stress-Related Genes of Rubber Particles and Latex in Fig Tree (Ficus Carica) and Their Expressions by Abiotic Stress or Plant Hormone Treatments. Plant Cell Physiol. 2003, 44, 412–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azarkan, M.; Wintjens, R.; Looze, Y.; Baeyens-Volant, D. Detection of Three Wound-Induced Proteins in Papaya Latex. Phytochemistry 2004, 65, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Gidrol, X.; Chrestin, H.; Tan, H.L.; Kush, A. Hevein, a Lectin-like Protein from Hevea Brasiliensis (rubber Tree) Is Involved in the Coagulation of Latex. J. Biol. Chem. 1994, 269, 9278–9283. [Google Scholar] [CrossRef]
- Barre, A.; Van Damme, E.J.M.; Simplicien, M.; Benoist, H.; Rougé, P. Are Dietary Lectins Relevant Allergens in Plant Food Allergy? Foods 2020, 9, 1724. [Google Scholar] [CrossRef]
- Vandenborre, G.; Smagghe, G.; Van Damme, E.J.M. Plant Lectins as Defense Proteins against Phytophagous Insects. Phytochemistry 2011, 72, 1538–1550. [Google Scholar] [CrossRef]
- Lehrman, A. Does Pea Lectin Expressed Transgenically in Oilseed Rape (Brassica napus) Influence Honey Bee (Apis mellifera) Larvae? Environ. Biosafety Res. 2007, 6, 271–278. [Google Scholar] [CrossRef] [Green Version]
- John, K.S.; Bhat, S.G.; Prasada Rao, U.J.S. Biochemical Characterization of Sap (latex) of a Few Indian Mango Varieties. Phytochemistry 2003, 62, 13–19. [Google Scholar] [CrossRef]
- Wititsuwannakul, D.; Chareonthiphakorn, N.; Pace, M.; Wititsuwannakul, R. Polyphenol Oxidases from Latex of Hevea Brasiliensis: Purification and Characterization. Phytochemistry 2002, 61, 115–121. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, X. Recent Advances in Polyphenol Oxidase-Mediated Plant Stress Responses. Phytochemistry 2021, 181, 112588. [Google Scholar] [CrossRef]
- Rajput, V.D.; Harish; Singh, R.K.; Verma, K.K.; Sharma, L.; Quiroz-Figueroa, F.R.; Meena, M.; Gour, V.S.; Minkina, T.; Sushkova, S.; et al. Recent Developments in Enzymatic Antioxidant Defence Mechanism in Plants with Special Reference to Abiotic Stress. Biology 2021, 10, 267. [Google Scholar] [CrossRef]
- Kidwai, M.; Ahmad, I.Z.; Chakrabarty, D. Class III Peroxidase: An Indispensable Enzyme for Biotic/abiotic Stress Tolerance and a Potent Candidate for Crop Improvement. Plant Cell Rep. 2020, 39, 1381–1393. [Google Scholar] [CrossRef]
- Bishop, J.G.; Dean, A.M.; Mitchell-Olds, T. Rapid Evolution in Plant Chitinases: Molecular Targets of Selection in Plant-Pathogen Coevolution. Proc. Natl. Acad. Sci. USA 2000, 97, 5322–5327. [Google Scholar] [CrossRef] [Green Version]
- Kitajima, S.; Kamei, K.; Taketani, S.; Yamaguchi, M.; Kawai, F.; Komatsu, A.; Inukai, Y. Two Chitinase-like Proteins Abundantly Accumulated in Latex of Mulberry Show Insecticidal Activity. BMC Biochem. 2010, 11, 6. [Google Scholar] [CrossRef] [Green Version]
- Gursharan, S.; Shailendra Kumar, A. Antifungal and Insecticidal Potential of Chitinases: A Credible Choice for the Eco-Friendly Farming. Biocatal. Agric. Biotechnol. 2019, 20, 101289. [Google Scholar]
- Taira, T.; Ohdomari, A.; Nakama, N.; Shimoji, M.; Ishihara, M. Characterization and Antifungal Activity of Gazyumaru (Ficus Microcarpa) Latex Chitinases: Both the Chitin-Binding and the Antifungal Activities of Class I Chitinase Are Reinforced with Increasing Ionic Strength. Biosci. Biotechnol. Biochem. 2005, 69, 811–818. [Google Scholar] [CrossRef] [Green Version]
- Martin, M.N. The Latex of Hevea Brasiliensis Contains High Levels of Both Chitinases and Chitinases/Lysozymes. Plant Physiol. 1991, 95, 469–476. [Google Scholar] [CrossRef] [Green Version]
- Freitas, C.D.T.; Viana, C.A.; Vasconcelos, I.M.; Moreno, F.B.B.; Lima-Filho, J.V.; Oliveira, H.D.; Moreira, R.A.; Monteiro-Moreira, A.C.O.; Ramos, M.V. First Insights into the Diversity and Functional Properties of Chitinases of the Latex of Calotropis procera. Plant Physiol. Biochem. 2016, 108, 361–371. [Google Scholar] [CrossRef]
- Musidlak, O.; Nawrot, R.; Goździcka-Józefiak, A. Which Plant Proteins Are Involved in Antiviral Defense? Review on In Vivo and In Vitro Activities of Selected Plant Proteins against Viruses. Int. J. Mol. Sci. 2017, 18, 2300. [Google Scholar] [CrossRef] [Green Version]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P.; et al. Pathogenesis-Related Proteins and Peptides as Promising Tools for Engineering Plants with Multiple Stress Tolerance. Microbiol. Res. 2018, 212–213, 29–37. [Google Scholar] [CrossRef]
- Nessler, C.L. Sequence Analysis of Two New Members of the Major Latex Protein Gene Family Supports the Triploid-Hybrid Origin of the Opium Poppy. Gene 1994, 139, 207–209. [Google Scholar] [CrossRef]
- Nessler, C.L.; Allen, R.D.; Galewsky, S. Identification and Characterization of Latex-Specific Proteins in Opium Poppy. Plant Physiol. 1985, 79, 499–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nessler, C.L.; Vonder Haar, R.A. Cloning and Expression Analysis of DNA Sequences for the Major Latex Protein of Opium Poppy. Planta 1990, 180, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Ruperti, B.; Bonghi, C.; Ziliotto, F.; Pagni, S.; Rasori, A.; Varotto, S.; Tonutti, P.; Giovannoni, J.J.; Ramina, A. Characterization of a Major Latex Protein (MLP) Gene down-Regulated by Ethylene during Peach Fruitlet Abscission. Plant Sci. 2002, 163, 265–272. [Google Scholar] [CrossRef]
- Lytle, B.L.; Song, J.; de la Cruz, N.B.; Peterson, F.C.; Johnson, K.A.; Bingman, C.A.; Phillips, G.N., Jr.; Volkman, B.F. Structures of Two Arabidopsis Thaliana Major Latex Proteins Represent Novel Helix-Grip Folds. Proteins 2009, 76, 237–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chruszcz, M.; Ciardiello, M.A.; Osinski, T.; Majorek, K.A.; Giangrieco, I.; Font, J.; Breiteneder, H.; Thalassinos, K.; Minor, W. Structural and Bioinformatic Analysis of the Kiwifruit Allergen Act D 11, a Member of the Family of Ripening-Related Proteins. Mol. Immunol. 2013, 56, 794–803. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Kim, M.-K.; Pulla, R.K.; Kim, Y.-J.; Yang, D.-C. Isolation and Expression Analysis of a Novel Major Latex-like Protein (MLP151) Gene from Panax Ginseng. Mol. Biol. Rep. 2010, 37, 2215–2222. [Google Scholar] [CrossRef] [PubMed]
- Osmark, P.; Boyle, B.; Brisson, N. Sequential and Structural Homology between Intracellular Pathogenesis-Related Proteins and a Group of Latex Proteins. Plant Mol. Biol. 1998, 38, 1243–1246. [Google Scholar] [CrossRef]
- Park, C.-J.; Kim, K.-J.; Shin, R.; Park, J.M.; Shin, Y.-C.; Paek, K.-H. Pathogenesis-Related Protein 10 Isolated from Hot Pepper Functions as a Ribonuclease in an Antiviral Pathway. Plant J. 2004, 37, 186–198. [Google Scholar] [CrossRef]
- Yang, C.-L.; Liang, S.; Wang, H.-Y.; Han, L.-B.; Wang, F.-X.; Cheng, H.-Q.; Wu, X.-M.; Qu, Z.-L.; Wu, J.-H.; Xia, G.-X. Cotton Major Latex Protein 28 Functions as a Positive Regulator of the Ethylene Responsive Factor 6 in Defense against Verticillium dahliae. Mol. Plant 2015, 8, 399–411. [Google Scholar] [CrossRef] [Green Version]
- Nawrot, R. Defense-Related Proteins from Chelidonium majus L. as Important Components of Its Latex. Curr. Protein Pept. Sci. 2017, 18, 864–880. [Google Scholar] [CrossRef] [PubMed]
- Chadha, P.; Das, R.H. A Pathogenesis Related Protein, AhPR10 from Peanut: An Insight of Its Mode of Antifungal Activity. Planta 2006, 225, 213–222. [Google Scholar] [CrossRef]
- Michalska, K.; Fernandes, H.; Sikorski, M.; Jaskolski, M. Crystal Structure of Hyp-1, a St. John’s Wort Protein Implicated in the Biosynthesis of Hypericin. J. Struct. Biol. 2010, 169, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Bantignies, B.; Séguin, J.; Muzac, I.; Dédaldéchamp, F.; Gulick, P.; Ibrahim, R. Direct Evidence for Ribonucleolytic Activity of a PR-10-like Protein from White Lupin Roots. Plant Mol. Biol. 2000, 42, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Nawrot, R.; Barylski, J.; Lippmann, R.; Altschmied, L.; Mock, H.-P. Combination of Transcriptomic and Proteomic Approaches Helps to Unravel the Protein Composition of Chelidonium majus L. Milky Sap. Planta 2016, 244, 1055–1064. [Google Scholar] [CrossRef] [Green Version]
- Song, L.; Wang, J.; Jia, H.; Kamran, A.; Qin, Y.; Liu, Y.; Hao, K.; Han, F.; Zhang, C.; Li, B.; et al. Identification and Functional Characterization of NbMLP28, a Novel MLP-like Protein 28 Enhancing Potato Virus Y Resistance in Nicotiana Benthamiana. BMC Microbiol. 2020, 20, 55. [Google Scholar] [CrossRef] [PubMed]
- Malter, D.; Wolf, S. Melon Phloem-Sap Proteome: Developmental Control and Response to Viral Infection. Protoplasma 2011, 248, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Onoyovwe, A.; Hagel, J.M.; Chen, X.; Khan, M.F.; Schriemer, D.C.; Facchini, P.J. Morphine Biosynthesis in Opium Poppy Involves Two Cell Types: Sieve Elements and Laticifers. Plant Cell 2013, 25, 4110–4122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawrot, R.; Lippmann, R.; Matros, A.; Musidlak, O.; Nowicki, G.; Mock, H.-P. Proteomic Comparison of Chelidonium majus L. Latex in Different Phases of Plant Development. Plant Physiol. Biochem. 2017, 112, 312–325. [Google Scholar] [CrossRef]
- D’Amato, A.; Bachi, A.; Fasoli, E.; Boschetti, E.; Peltre, G.; Sénéchal, H.; Sutra, J.P.; Citterio, A.; Righetti, P.G. In-Depth Exploration of Hevea Brasiliensis Latex Proteome and “Hidden Allergens” via Combinatorial Peptide Ligand Libraries. J. Proteom. 2010, 73, 1368–1380. [Google Scholar] [CrossRef] [PubMed]
- Zulak, K.G.; Khan, M.F.; Alcantara, J.; Schriemer, D.C.; Facchini, P.J. Plant Defense Responses in Opium Poppy Cell Cultures Revealed by Liquid Chromatography-Tandem Mass Spectrometry Proteomics. Mol. Cell. Proteom. 2009, 8, 86–98. [Google Scholar] [CrossRef] [Green Version]
- Samatadze, T.E.; Zoshchuk, S.A.; Khomik, A.S.; Amosova, A.V.; Svistunova, N.Y.; Suslina, S.N.; Hazieva, F.M.; Yurkevich, O.Y.; Muravenko, O.V. Molecular Cytogenetic Characterization, Leaf Anatomy and Ultrastructure of the Medicinal Plant Potentilla alba L. Genet. Resour. Crop. Evol. 2018, 65, 1637–1647. [Google Scholar] [CrossRef]
- Gardin, N.E.; Braga, A.J. Greater Celandine (Chelidonium majus L.) for COVID-19: A Twenty-Case Series. Phytother. Res. 2021, 35, 3792–3798. [Google Scholar] [CrossRef]
- Foster, S.; Bradshaw, R.H.; McLanahan, S.; Redwood, D.; Upton, R. Book Reviews: The Complete German Commission E Monographs: Therapeutic Guide to Herbal Medicines. J. Altern. Complementary Med. 1998, 4, 479–486. [Google Scholar] [CrossRef]
- Lee, E.-J.; Hagel, J.M.; Facchini, P.J. Role of the Phloem in the Biochemistry and Ecophysiology of Benzylisoquinoline Alkaloid Metabolism. Front. Plant Sci. 2013, 4, 182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mithöfer, A.; Boland, W. Plant Defense against Herbivores: Chemical Aspects. Annu. Rev. Plant Biol. 2012, 63, 431–450. [Google Scholar] [CrossRef] [Green Version]
- Upadhyay, R.K. Plant Latex: A Natural Source of Pharmaceuticals and Pesticides. Int. J. Green Pharm. (IJGP) 2011, 5, 169–180. [Google Scholar] [CrossRef]
- Gurkok, T.; Turktas, M.; Parmaksiz, I.; Unver, T. Transcriptome Profiling of Alkaloid Biosynthesis in Elicitor Induced Opium Poppy. Plant Mol. Biol. Rep. 2015, 33, 673–688. [Google Scholar] [CrossRef]
- Labanca, F.; Ovesnà, J.; Milella, L. Papaver Somniferum L. Taxonomy, Uses and New Insight in Poppy Alkaloid Pathways. Phytochem. Rev. 2018, 17, 853–871. [Google Scholar] [CrossRef]
- De Petrocellis, L.; Ligresti, A.; Moriello, A.S.; Allarà, M.; Bisogno, T.; Petrosino, S.; Stott, C.G.; Di Marzo, V. Effects of Cannabinoids and Cannabinoid-Enriched Cannabis Extracts on TRP Channels and Endocannabinoid Metabolic Enzymes. Br. J. Pharmacol. 2011, 163, 1479–1494. [Google Scholar] [CrossRef] [Green Version]
- Pacher, P.; Bátkai, S.; Kunos, G. The Endocannabinoid System as an Emerging Target of Pharmacotherapy. Pharmacol. Rev. 2006, 58, 389–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burstein, S. Cannabidiol (CBD) and Its Analogs: A Review of Their Effects on Inflammation. Bioorg. Med. Chem. 2015, 23, 1377–1385. [Google Scholar] [CrossRef]
- Etxenagusia, M.A.; Anda, M.; González-Mahave, I.; Fernández, E.; Fernández de Corrès, L. Contact Dermatitis from Chelidonium majus (greater Celandine). Contact Dermat. 2000, 43, 47. [Google Scholar]
- Monavari, S.H.; Shahrabadi, M.S.; Keyvani, H.; Bokharaei-Salim, F. Evaluation of In Vitro Antiviral Activity of Chelidonium majus L. against Herpes Simplex Virus Type-1. Afr. J. Microbiol. Res. 2012, 6, 4360–4364. [Google Scholar]
- Horvath, J.; Kery, A.; Kulcsar, G.; Dan, P.; Nasz, J. Antiviral Effect of Chelidonium Extracts. In Spitzy ILH, Karrev K (eds) 13–14 Proc Int Congr Chemother; Egermann: Vienna, Austria, 1983; Volume 9, pp. 124/106–124/110. [Google Scholar]
- Gerencer, M.; Turecek, P.L.; Kistner, O.; Mitterer, A.; Savidis-Dacho, H.; Barrett, N.P. In Vitro and in Vivo Anti-Retroviral Activity of the Substance Purified from the Aqueous Extract of Chelidonium majus L. Antivir. Res. 2006, 72, 153–156. [Google Scholar] [CrossRef]
- Mandadi, K.K.; Scholthof, K.-B.G. Plant Immune Responses against Viruses: How Does a Virus Cause Disease? Plant Cell 2013, 25, 1489–1505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wojtaszek, P. Oxidative Burst: An Early Plant Response to Pathogen Infection. Biochem. J. 1997, 322 Pt 3, 681–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawrot, R.; Kalinowski, A.; Gozdzicka-Jozefiak, A. Proteomic Analysis of Chelidonium majus Milky Sap Using Two-Dimensional Gel Electrophoresis and Tandem Mass Spectrometry. Phytochemistry 2007, 68, 1612–1622. [Google Scholar] [CrossRef]
- Nawrot, R.; Tomaszewski, Ł.; Czerwoniec, A.; Goździcka-Józefiak, A. Identification of a Coding Sequence and Structure Modeling of a Glycine-Rich RNA-Binding Protein (CmGRP1) from Chelidonium majus L. Plant Mol. Biol. Report. 2013, 31, 470–476. [Google Scholar] [CrossRef] [Green Version]
- Huh, S.U.; Paek, K.-H. Plant RNA Binding Proteins for Control of RNA Virus Infection. Front. Physiol. 2013, 4, 397. [Google Scholar] [CrossRef] [Green Version]
- Zvereva, A.S.; Pooggin, M.M. Silencing and Innate Immunity in Plant Defense against Viral and Non-Viral Pathogens. Viruses 2012, 4, 2578–2597. [Google Scholar] [CrossRef] [Green Version]
- Kéry, A.; Horváth, J.; Nász, I.; Verzár-Petri, G.; Kulcsár, G.; Dán, P. Antiviral Alkaloid in Chelidonium majus L. Acta Pharm. Hung. 1987, 57, 19–25. [Google Scholar]
- Lozjuk, R.M.; Lisnyak, O.I.; Lozjuk, L.V. Theoretical Grounds and Experimental Confirmation of the Antiviral Effect of the Preparation Ukrain. Drugs Exp. Clin. Res. 1996, 22, 213–217. [Google Scholar] [PubMed]
- Camero, M.; Marinaro, M.; Lovero, A.; Elia, G.; Losurdo, M.; Buonavoglia, C.; Tempesta, M. In Vitroantiviral Activity ofFicus Caricalatex against Caprine Herpesvirus-1. Nat. Prod. Res. 2014, 28, 2031–2035. [Google Scholar] [CrossRef] [PubMed]
- Aref, H.L.; Gaaliche, B.; Fekih, A.; Mars, M.; Aouni, M.; Chaumon, J.P.; Said, K. In Vitrocytotoxic and Antiviral Activities ofFicus Caricalatex Extracts. Nat. Prod. Res. 2011, 25, 310–319. [Google Scholar] [CrossRef]
- Beloin, N.; Gbeassor, M.; Akpagana, K.; Hudson, J.; de Soussa, K.; Koumaglo, K.; Thor Arnason, J. Ethnomedicinal Uses of Momordica Charantia (Cucurbitaceae) in Togo and Relation to Its Phytochemistry and Biological Activity. J. Ethnopharmacol. 2005, 96, 49–55. [Google Scholar] [CrossRef]
- Pengsuparp, T.; Serit, M.; Hughes, S.H.; Soejarto, D.D.; Pezzuto, J.M. Specific Inhibition of Human Immunodeficiency Virus Type 1 Reverse Transcriptase Mediated by Soulattrolide, a Coumarin Isolated from the Latex of Calophyllum Teysmannii. J. Nat. Prod. 1996, 59, 839–842. [Google Scholar] [CrossRef]
- Ubillas, R.; Jolad, S.D.; Bruening, R.C.; Kernan, M.R.; King, S.R.; Sesin, D.F.; Barrett, M.; Stoddart, C.A.; Flaster, T.; Kuo, J.; et al. SP-303, an Antiviral Oligomeric Proanthocyanidin from the Latex of Croton Lechleri (Sangre de Drago). Phytomedicine 1994, 1, 77–106. [Google Scholar] [CrossRef]
- Parhira, S.; Yang, Z.-F.; Zhu, G.-Y.; Chen, Q.-L.; Zhou, B.-X.; Wang, Y.-T.; Liu, L.; Bai, L.-P.; Jiang, Z.-H. In Vitro Anti-Influenza Virus Activities of a New Lignan Glycoside from the Latex of Calotropis Gigantea. PLoS ONE 2014, 9, e104544. [Google Scholar] [CrossRef]
- Kim, H.K.; Farnsworth, N.R.; Blomster, R.N.; Fong, H.H. Biological and Phytochemical Evaluation of Plants. V. Isolation of Two Cytotoxic Alkaloids from Chelidonium majus. J. Pharm. Sci. 1969, 58, 372–374. [Google Scholar] [CrossRef]
- Vavrečková, C.; Gawlik, I.; Müller, K. Benzophenanthridine Alkaloids of Chelidonium majus; II. Potent Inhibitory Action Against the Growth of Human Keratinocytes. Planta Med. 1996, 62, 491–494. [Google Scholar] [CrossRef]
- Rogelj, B.; Popovic, T.; Ritonja, A.; Strukelj, B.; Brzin, J. Chelidocystatin, a Novel Phytocystatin from Chelidonium majus. Phytochemistry 1998, 49, 1645–1649. [Google Scholar] [CrossRef]
- Philchenkov, A.; Kaminskyy, V.; Zavelevich, M.; Stoika, R. Apoptogenic Activity of Two Benzophenanthridine Alkaloids from Chelidonium majus L. Does Not Correlate with Their DNA Damaging Effects. Toxicol. In Vitro 2008, 22, 287–295. [Google Scholar] [CrossRef]
- Paul, A.; Bishayee, K.; Ghosh, S.; Mukherjee, A.; Sikdar, S.; Chakraborty, D.; Boujedaini, N.; Khuda-Bukhsh, A.R. Chelidonine Isolated from Ethanolic Extract of Chelidonium majus Promotes Apoptosis in HeLa Cells through p38-p53 and PI3K/AKT Signalling Pathways. Zhong Xi Yi Jie He Xue Bao 2012, 10, 1025–1038. [Google Scholar] [CrossRef]
- Kulp, M.; Bragina, O. Capillary Electrophoretic Study of the Synergistic Biological Effects of Alkaloids from Chelidonium majus L. in Normal and Cancer Cells. Anal. Bioanal. Chem. 2013, 405, 3391–3397. [Google Scholar] [CrossRef]
- Riede, F.v.D.I. Chelidonium majus in der Therapie Prämaligner Hautveränderungen. Available online: https://www.researchgate.net/publication/260197370_Chelidonium_majus_in_der_Therapie_pramaligner_Hautveranderungen (accessed on 15 November 2021).
- Abdel-Aty, A.M.; Hamed, M.B.; Salama, W.H.; Ali, M.M.; Fahmy, A.S.; Mohamed, S.A. Ficus Carica, Ficus Sycomorus and Euphorbia Tirucalli Latex Extracts: Phytochemical Screening, Antioxidant and Cytotoxic Properties. Biocatal. Agric. Biotechnol. 2019, 20, 101199. [Google Scholar] [CrossRef]
- Tulasi, C.; Lakshmi Narasu, M.; Saida, L. Cytotoxic Effect of Ficus Religiosa and Ficus Benghalensis Latex Extracts on MCF-7 Cell Line. Int. J. Sci. Res. Biol. Sci. 2019, 5, 96–100. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://www.researchgate.net/profile/Flavio-Beltrame/publication/350549554_TERPENES_OF_EUPHORBIA_UMBELLATA_LATEX_ARE_INVOLVED_IN_CYTOTOXIC_EFFECT_AGAINST_MELANOMA_CELLS/links/60b52790a6fdcc476bda6c8d/TERPENES-OF-EUPHORBIA-UMBELLATA-LATEX-ARE-INVOLVED-IN-CYTOTOXIC-EFFECT-AGAINST-MELANOMA-CELLS.pdf (accessed on 30 September 2021).
- Cheng, J.; Han, W.; Wang, Z.; Shao, Y.; Wang, Y.; Zhang, Y.; Li, Z.; Xu, X.; Zhang, Y. Hepatocellular Carcinoma Growth Is Inhibited by Euphorbia Helioscopia L. Extract in Nude Mice Xenografts. Biomed Res. Int. 2015, 2015, 601015. [Google Scholar] [CrossRef]
- Sadeghi-Aliabadi, H.; Sajjadi, S.E.; Khodamoradi, M. Cytotoxicity of Euphorbia Macroclada on MDA-MB-468 Breast Cancer Cell Line. Iran. J. Pharm. Sci. 2009, 5, 103–108. [Google Scholar]
- Choedon, T.; Mathan, G.; Arya, S.; Kumar, V.L.; Kumar, V. Anticancer and Cytotoxic Properties of the Latex of Calotropis Procera in a Transgenic Mouse Model of Hepatocellular Carcinoma. World J. Gastroenterol. 2006, 12, 2517–2522. [Google Scholar] [CrossRef]
- Colombo, M.L.; Bosisio, E. Pharmacological Activities of Chelidonium Majus L. (Papaveraceae). Pharmacol. Res. 1996, 33, 127–134. [Google Scholar] [CrossRef]
- Ciric, A.; Vinterhalter, B.; Savikin-Fodulovic, K.; Sokovic, M.; Vinterhalter, D. Chemical Analysis and Antimicrobial Activity of Methanol Extracts of Celandine (Chelidonium majus L.) Plants Growing in Nature and Cultured in Vitro. Arch. Biol. Sci. 2008, 60, 7–8. [Google Scholar] [CrossRef]
- Kokoska, L.; Polesny, Z.; Rada, V.; Nepovim, A.; Vanek, T. Screening of Some Siberian Medicinal Plants for Antimicrobial Activity. J. Ethnopharmacol. 2002, 82, 51–53. [Google Scholar] [CrossRef]
- Recio, M.C.; Rios, J.L.; Villar, A. Antimicrobial Activity of Selected Plants Employed in the Spanish Mediterranean Area. Part II. Phytother. Res. 1989, 3, 77–80. [Google Scholar] [CrossRef]
- Miao, F.; Yang, X.-J.; Zhou, L.; Hu, H.-J.; Zheng, F.; Ding, X.-D.; Sun, D.-M.; Zhou, C.-D.; Sun, W. Structural Modification of Sanguinarine and Chelerythrine and Their Antibacterial Activity. Nat. Prod. Res. 2011, 25, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Pavão, M.L.; Pinto, R.E. Sensivity of Bacillus Subtilis to Water Soluble Alkaloid Extracts of Chelidonium majus L. (Papaveracea) Roots from Azores. ARQUIPÉLAGO. Ciências Biológicas e Marinhas (Life Mar. Sci.) 1995, 13, 93–97. [Google Scholar]
- Asamenew, G.; Bisrat, D.; Mazumder, A.; Asres, K. In Vitro Antimicrobial and Antioxidant Activities of Anthrone and Chromone from the Latex of Aloe Harlana Reynolds. Phytother. Res. 2011, 25, 1756–1760. [Google Scholar] [CrossRef]
- Emiru, Y.K.; Siraj, E.A.; Teklehaimanot, T.T.; Amare, G.G. Antibacterial Potential of Aloe weloensis (Aloeacea) Leaf Latex against Gram-Positive and Gram-Negative Bacteria Strains. Int. J. Microbiol. 2019, 2019, 5328238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siritapetawee, J.; Thammasirirak, S.; Samosornsuk, W. Antimicrobial Activity of a 48-kDa Protease (AMP48) from Artocarpus heterophyllus Latex. Eur. Rev. Med. Pharmacol. Sci. 2012, 16, 132–137. [Google Scholar] [PubMed]
- Raghavendra, R.; Mahadevan, G.D. In Vitro Antimicrobial Activity of Various Plant Latex against Resistant Human Pathogens. Int. J. Pharm. Pharm. Sci. 2011, 3, 70–72. [Google Scholar]
- Nenaah, E.G.; Ahmed, M.E. Antimicrobial Activity of Extracts and Latex of Calotropis procera (Ait.) and Synergistic Effect with Reference Antimicrobials. J. Med. Plant Res. 2011, 5, 706–716. [Google Scholar] [CrossRef] [Green Version]
- Ishnava, K.B.; Chauhan, J.B.; Garg, A.A.; Thakkar, A.M. Antibacterial and Phytochemical Studies on Calotropis gigantia (L.) R. Br. Latex against Selected Cariogenic Bacteria. Saudi J. Biol. Sci. 2012, 19, 87–91. [Google Scholar] [CrossRef] [Green Version]
- Kori, P.; Alawa, P. Antimicrobial Activity and Phytochemical Analysis of Calotropis Gigantea Root, Latex Extracts. IOSR J. Pharm. (IOSRPHR) 2014, 4, 7–11. [Google Scholar] [CrossRef]
- Ml, P.; Mk, M.; Mary, R.S. Efficacy of Euphorbia Heterophylla Latex against Pathogenic Bacteria and Fungi. Asian J. Pharm. Clin. Res. 2020, 13, 141–145. [Google Scholar] [CrossRef]
- Lazreg-Aref, H.; Mars, M.; Fekih, A.; Aouni, M.; Said, K. Chemical Composition and Antibacterial Activity of a Hexane Extract of Tunisian Caprifig Latex from the Unripe Fruit of Ficus Carica. Pharm. Biol. 2012, 50, 407–412. [Google Scholar] [CrossRef] [Green Version]
- Prastiyanto, M.E.; Tama, P.D.; Ananda, N.; Wilson, W.; Mukaromah, A.H. Antibacterial Potential of Jatropha Sp. Latex against Multidrug-Resistant Bacteria. Int. J. Microbiol. 2020, 2020, 8509650. [Google Scholar] [CrossRef]
- Arun, K.; Anu, B.; Ruchika, G. A Comparative Study of Antibacterial Activity of Leaves and Latex of Jatropha Curcas L. Int. J. Pharmacogn. Phytochem. Res. 2012, 4, 190–194. [Google Scholar]
- Arekemase, M.O.; Kayode, R.M.O.; Ajiboye, A.E. Antimicrobial Activity and Phytochemical Analysis of Jatropha Curcas Plant against Some Selected Microorganisms. Int. J. Biol. 2011, 3, 52. [Google Scholar] [CrossRef]
- Hoekou, P.Y.; Tchacondo, T.; Gbogbo, K.A.; Tchelougou, D.; Pissang, P.; Karou, S.D.; Améyapohm, Y.A.; Batawila, K.; Annigoni, P.; Faso, B. Antibacterial Activities of Three Latex Plants of Asclepiadaceae Family Used in Traditional Medicine in South Togo. Int. J. Curr. Microbiol. App. Sci. 2015, 4, 882–891. [Google Scholar]
- Liu, C.-B.; Shan, B.; Bai, H.-M.; Tang, J.; Yan, L.-Z.; Ma, Y.-B. Hydrophilic/hydrophobic Characters of Antimicrobial Peptides Derived from Animals and Their Effects on Multidrug Resistant Clinical Isolates. Dongwuxue Yanjiu 2015, 36, 41–47. [Google Scholar]
- Fox, J.L. Antimicrobial Peptides Stage a Comeback: Better Understanding of the Mechanisms of Action, Modification and Synthesis of Antimicrobial Peptides Is Reigniting Commercial Development. Nat. Biotechnol. 2013, 31, 379–382. [Google Scholar] [CrossRef]
- Bahar, A.A.; Ren, D. Antimicrobial Peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, O.N.; de la Fuente-Núñez, C.; Haney, E.F.; Fensterseifer, I.C.M.; Ribeiro, S.M.; Porto, W.F.; Brown, P.; Faria-Junior, C.; Rezende, T.M.B.; Moreno, S.E.; et al. An Anti-Infective Synthetic Peptide with Dual Antimicrobial and Immunomodulatory Activities. Sci. Rep. 2016, 6, 35465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Parijs, J.; Broekaert, W.F.; Goldstein, I.J.; Peumans, W.J. Hevein: An Antifungal Protein from Rubber-Tree (Hevea Brasiliensis) Latex. Planta 1991, 183, 258–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.-Y.; Yang, H.-O.; Pyo, S.-N.; Jung, I.-S.; Yi, S.-Y.; Yun, Y.-S. Immunomodulatory Activity of Protein-Bound Polysaccharide Extracted from Chelidonium majus. Arch. Pharm. Res. 2002, 25, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.-S.; An, H.-J.; Jeong, H.-J.; Won, J.-H.; Hong, S.-H.; Kim, H.-M. Water Extract Isolated from Chelidonium majus Enhances Nitric Oxide and Tumour Necrosis Factor-α Production via Nuclear Factor-κB Activation in Mouse Peritoneal Macrophages. J. Pharm. Pharmacol. 2010, 56, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Khmel’nitskaia, N.M.; Vorob’ev, K.V.; Kliachko, L.L.; Ankhimova, E.S.; Kosenko, V.A.; Tyrnova, E.V.; Mal’tseva, G.S.; Medvedev, E.A. A comparative study of conservative treatment schemes in chronic tonsillitis in children. Vestn. Otorinolaringol. 1998, 4, 39–42. [Google Scholar]
- Nascimento, D.C.d.O.; de Oliveira Nascimento, D.C.; Ralph, M.T.; Batista, J.E.C.; Silva, D.M.F.; Gomes-Filho, M.A.; Alencar, N.M.; Leal, N.C.; Ramos, M.V.; Lima-Filho, J.V. Latex Protein Extracts from Calotropis Procera with Immunomodulatory Properties Protect against Experimental Infections with Listeria Monocytogenes. Phytomedicine 2016, 23, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; Kim, D.; Jiang, Z.; Ueno, M.; Okimura, T.; Yamaguchi, K.; Oda, T. Immunostimulatory Activities of the Sulfated Polysaccharide Ascophyllan from Ascophyllum Nodosum in in Vivo and in Vitro Systems. Biosci. Biotechnol. Biochem. 2012, 76, 1573–1576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, V.K.; Rani, K.V.; Sehgal, N.; Prakash, O. Immunostimulatory Response Induced by Supplementation of Ficus Benghalensis Root Powder, in the Artificial Feed the Indian Freshwater Murrel, Channa Punctatus. Fish Shellfish Immunol. 2012, 33, 590–596. [Google Scholar] [CrossRef]
- Akram, M.; Hamid, A.; Khalil, A.; Ghaffar, A.; Tayyaba, N.; Saeed, A.; Ali, M.; Naveed, A. Review on Medicinal Uses, Pharmacological, Phytochemistry and Immunomodulatory Activity of Plants. Int. J. Immunopathol. Pharmacol. 2014, 27, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Kukhetpitakwong, R.; Hahnvajanawong, C.; Homchampa, P.; Leelavatcharamas, V.; Satra, J.; Khunkitti, W. Immunological Adjuvant Activities of Saponin Extracts from the Pods of Acacia Concinna. Int. Immunopharmacol. 2006, 6, 1729–1735. [Google Scholar] [CrossRef] [PubMed]
- Hueza, I.M.; Gorniak, S.L. The Immunomodulatory Effects of Ipomoea Carnea in Rats Vary Depending on Life Stage. Hum. Exp. Toxicol. 2011, 30, 1690–1700. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, K.V.; Padmavathi, K. Assessment of Immunomodulatory Activity of Euphorbia hirta L. Indian J. Pharm. Sci. 2010, 72, 621–625. [Google Scholar]
- Shah, A.S.; Wakade, A.S.; Juvekar, A.R. Immunomodulatory Activity of Methanolic Extract of Murraya koenigii (L) Spreng. Leaves. Indian J. Exp. Biol. 2008, 46, 505–509. [Google Scholar]
- Benninger, J.; Thomas Schneider, H.; Schuppan, D.; Kirchner, T.; Hahn, E.G. Acute Hepatitis Induced by Greater Celandine (Chelidonium majus). Gastroenterology 1999, 117, 1234–1237. [Google Scholar] [CrossRef]
- Moro, P.A.; Cassetti, F.; Giugliano, G.; Falce, M.T.; Mazzanti, G.; Menniti-Ippolito, F.; Raschetti, R.; Santuccio, C. Hepatitis from Greater Celandine (Chelidonium majus L.): Review of Literature and Report of a New Case. J. Ethnopharmacol. 2009, 124, 328–332. [Google Scholar] [CrossRef]
- Jakovljevic, Z.D.; Stankovic, S.M.; Topuzovic, D.M. Seasonal Variability of Chelidonium majus L. Secondary Metabolites Content and Antioxidant Activity. EXCLI J. 2013, 12, 260–268. [Google Scholar] [PubMed]
- Rao, M.J.; Wang, L. CRISPR/Cas9 Technology for Improving Agronomic Traits and Future Prospective in Agriculture. Planta 2021, 254, 68. [Google Scholar] [CrossRef] [PubMed]
- Alok, A.; Jain, P.; Kumar, J.; Yajnik, K.; Bhalothia, P. Genome Engineering in Medicinally Important Plants Using CRISPR/Cas9 Tool. In Genome Engineering via CRISPR-Cas9 System; Academic Press: Cambridge, MA, USA, 2020; pp. 155–161. [Google Scholar]
- Dey, A. CRISPR/Cas Genome Editing to Optimize Pharmacologically Active Plant Natural Products. Pharmacol. Res. 2021, 164, 105359. [Google Scholar] [CrossRef]
- Shen, C.; Que, Z.; Xia, Y.; Tang, N.; Li, D.; He, R.; Cao, M. Knock out of the Annexin Gene OsAnn3 via CRISPR/Cas9-Mediated Genome Editing Decreased Cold Tolerance in Rice. J. Plant Biol. 2017, 60, 539–547. [Google Scholar] [CrossRef]
- Li, J.-F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and Homologous Recombination-Mediated Genome Editing in Arabidopsis and Nicotiana Benthamiana Using Guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Wei, P.; Yang, J. Use of CRISPR/Cas Genome Editing Technology for Targeted Mutagenesis in Rice. Methods Mol. Biol. 2017, 1498, 33–40. [Google Scholar]
- Liang, Z.; Zhang, K.; Chen, K.; Gao, C. Targeted Mutagenesis in Zea Mays Using TALENs and the CRISPR/Cas System. J. Genet. Genom. 2014, 41, 63–68. [Google Scholar] [CrossRef]
- Michno, J.-M.; Wang, X.; Liu, J.; Curtin, S.J.; Kono, T.J.; Stupar, R.M. CRISPR/Cas Mutagenesis of Soybean and Medicago Truncatula Using a New Web-Tool and a Modified Cas9 Enzyme. GM Crops Food 2015, 6, 243–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nekrasov, V.; Staskawicz, B.; Weigel, D.; Jones, J.D.G.; Kamoun, S. Targeted Mutagenesis in the Model Plant Nicotiana Benthamiana Using Cas9 RNA-Guided Endonuclease. Nat. Biotechnol. 2013, 31, 691–693. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, S.; Wang, W.; Xiong, X.; Meng, F.; Cui, X. Efficient Targeted Mutagenesis in Potato by the CRISPR/Cas9 System. Plant Cell Rep. 2015, 34, 1473–1476. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Xin, S.; Dai, X.; Yang, X.; Huang, H.; Hua, Y. Efficient Genome Editing of Rubber Tree (Hevea brasiliensis) Protoplasts Using CRISPR/Cas9 Ribonucleoproteins. Ind. Crops Prod. 2020, 146, 112146. [Google Scholar] [CrossRef]
- Van Laere, A.; Van Den Ende, W. Inulin Metabolism in Dicots: Chicory as a Model System. Plant Cell Environ. 2002, 25, 803–813. [Google Scholar] [CrossRef]
- Iaffaldano, B.; Zhang, Y.; Cornish, K. CRISPR/Cas9 Genome Editing of Rubber Producing Dandelion Taraxacum Kok-Saghyz Using Agrobacterium Rhizogenes without Selection. Ind. Crops Prod. 2016, 89, 356–362. [Google Scholar] [CrossRef] [Green Version]
- Bergonci, T.; Ribeiro, B.; Ceciliato, P.H.O.; Guerrero-Abad, J.C.; Silva-Filho, M.C.; Moura, D.S. Arabidopsis Thaliana RALF1 Opposes Brassinosteroid Effects on Root Cell Elongation and Lateral Root Formation. J. Exp. Bot. 2014, 65, 2219–2230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wieghaus, A.; Prüfer, D.; Schulze Gronover, C. Loss of Function Mutation of the Rapid Alkalinization Factor (RALF1)-like Peptide in the Dandelion Taraxacum Koksaghyz Entails a High-Biomass Taproot Phenotype. PLoS ONE 2019, 14, e0217454. [Google Scholar] [CrossRef] [Green Version]
- Alagoz, Y.; Gurkok, T.; Zhang, B.; Unver, T. Manipulating the Biosynthesis of Bioactive Compound Alkaloids for Next-Generation Metabolic Engineering in Opium Poppy Using CRISPR-Cas 9 Genome Editing Technology. Sci. Rep. 2016, 6, 30910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Xu, G.; Cheng, C.; Lei, L.; Sun, J.; Xu, Y.; Deng, C.; Dai, Z.; Yang, Z.; Chen, X.; et al. Establishment of an Agrobacterium-Mediated Genetic Transformation and CRISPR/Cas9-Mediated Targeted Mutagenesis in Hemp (Cannabis sativa L.). Plant Biotechnol. J. 2021, 19, 1979–1987. [Google Scholar] [CrossRef] [PubMed]
- Ekor, M. The Growing Use of Herbal Medicines: Issues Relating to Adverse Reactions and Challenges in Monitoring Safety. Front. Pharmacol. 2014, 4, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]

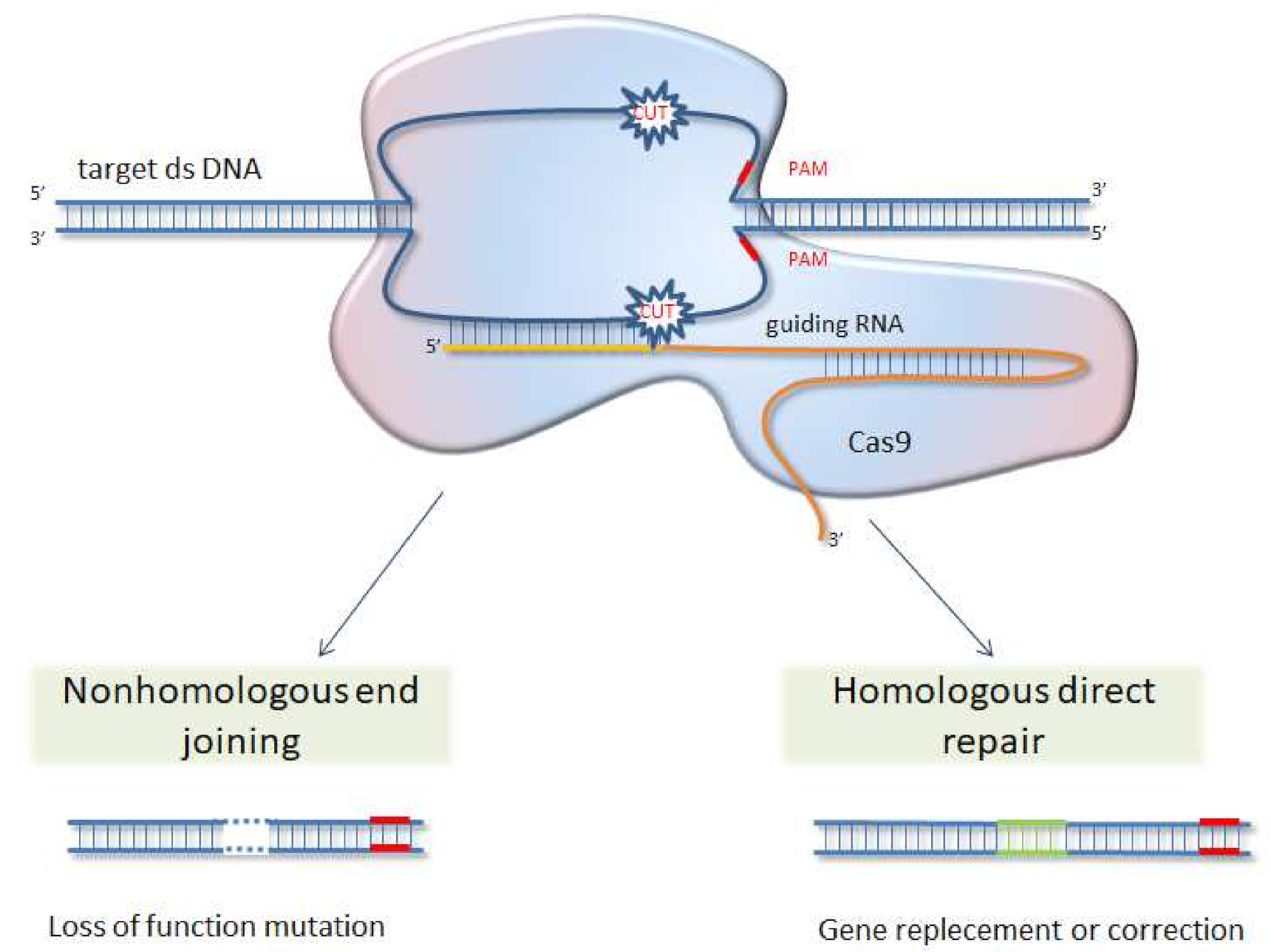
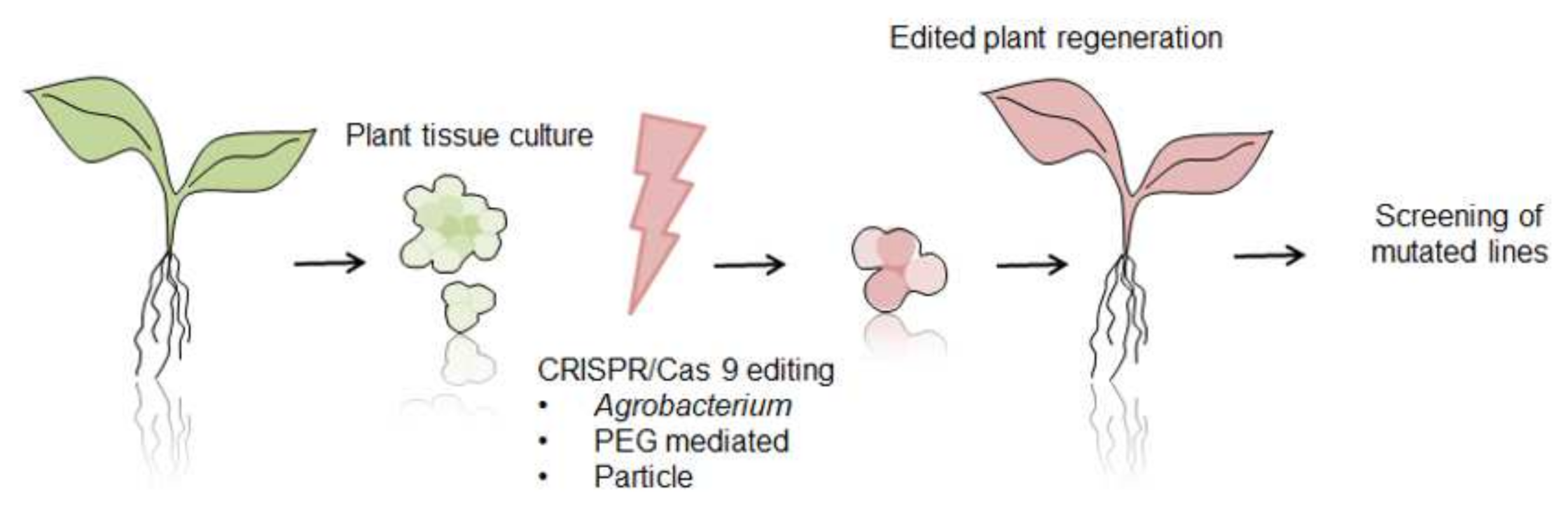
| Line of Defense | Type of Action | Predominant Proteins and Compounds |
|---|---|---|
| 1st | Mechanical damage | PPO, LOX (latex stickiness, different chemicals) |
| 2nd | Oxidative burst | POX, LOX et al. |
| 3rd | Antiviral activity | MLP, GRP (RNase/DNase activity, nucleic acid binding) |
| Latex-Bearing Plant Species | Examined Bacteria | Examined Fungi | Bioactive Compounds | Reference |
|---|---|---|---|---|
| Aloe harlana Reynolds | Bacillus pumilus (82) Bacillus subtilis (ATCC 6633) Escherichia coli (CD/99/1, K88, K99, LT37, ROW 7/12, 3:37C, 306, 872) Salmonella typhi (Ty2) Shigella boydii (D13629) Shigella dysentery 1 Shigella dysentery 8 Shigella flexneri (Type 6) Shigella sonnei 1 Staphylococcus aureus (ML267) Vibrio cholerae (85, 293, 1313, 1315) | Aspergillus niger (ATCC 6275) Candida albicans (ATCC 10231) Penicillium funiculosum (NCTC 287) Penicillium notatum (ATCC 11625) | Anthrone (aloin) Chromone (7-O-methylaloeresin A) | [133] |
| Aloe weloensis Sebsebe | Enterococcus faecalis Escherichia coli Pseudomonas aeruginosa Staphylococcus aureus | - | Alkaloids Anthraquinone Flavonoids Glycosides Tannins Terpenoids | [134] |
| Artocarpus heterophyllus Lam. | Bacillus subtilis Klebsiella Pneumoniae Pseudomonas aeruginosa (ATCC 27853) Streptococcus haemolyticus Salmonella typhi | Aspergillus niger Candida albicans | 48-kDa protease (AMP48) | [135,136] |
| Calotropis procera (Aiton) W.T.Aiton | Bacillus cereus Bacillus subtilis Escherichia coli Klebsiella Pneumoniae Pseudomonas aeruginosa Salmonella typhi Staphylococcus aureus Staphylococcus epidermidis Streptococcus haemolyticus Streptococcus pneumoniae | Aspergillus flavus Aspergillus niger Candida albicans Candida tropicalis Penicillium chrysogenum Saccharomyces cereviciae | - | [137] |
| Calotropis gigantea L. | Bacillus cereus Escherichia coli Lactobacillus acidophilus Micrococcus luteus Staphylococcus aureus Streptococcus mutans | Candida krusei | Alkaloids Phenolic Steroids Terpenes Cardiac glycoside | [138,139] |
| Carica papaya L. | Bacillus subtilis Klebsiella Pneumoniae Streptococcus haemolyticus Salmonella typhi | Aspergillus niger Candida albicans | - | [136] |
| Chelidonium majus L. | Aeromonas hydrophila Agrobacterium tumefacians Bacillus cereus Bacillus subtilis Candida albicans Escherichia coli Micrococcus luteus Mycobacterium phlei Salmonella enteritidis Sarcina lutea Staphylococcus aureus | Candida albicans | [127,128,129,130] | |
| Euphorbia heterophylla L. | Bacillus subtilis Proteus vulgaris Pseudomonas aeruginosa Staphylococcus aureus | Aspergillus niger Fusarium oxysporum Penicillium sp. | Alkaloids Flavonoids Phenols Saponins Steroids Tannins | [140] |
| Ficus carica L. | Enterobacter cloacae Enterococcus faecalis (ATCC 29212) Escherichia coli Escherichia coli ATCC 25922 Pseudomonas aeruginosa (ATCC 2783, ATCC 27950) Staphylococcus aureus Staphylococcus aureus (ATCC 25923) Staphylococcus epidermidis Staphylococcus saprophyticus | - | alpha-Amyrenyl acetate Aristolone Bornanone-3 Lanosta-8 Olean-12-en-3-ol, acetate Urs-12-en-24-oic acid | [141] |
| Jatropha gossypifolia L. | Pseudomonas aeruginosa (CRPA) Staphylococcus aureus (MRSA) | - | Flavonoids | [142] |
| Jatropha multifida L. | Pseudomonas aeruginosa (CRPA) Staphylococcus aureus (MRSA) | - | Flavonoids | [142] |
| Jatropa carcass L. | Bacillus subtilis Escherichia coli Klebsiella Pneumoniae Neisseria gonorrhoea Pseudomonas aeruginosa Salmonella typhi Staphylococcus aureus Streptococcus haemolyticus | Aspergillus niger Candida albicans | Alkaloid Glycoside Saponin Steroid Tannin | [136,143,144] |
| Leptadenia hastata (Pers.) Decne. | Klebsiella Pneumoniae Staphylococcus aureus Staphylococcus aureus (ATCC 29213) Salmonella typhi | - | Alkaloids Flavonoids Glycosides Phenolic Proanthocyanidins Saponins Tannins Triterpene | [145] |
| Pergularia daemia (Forssk.) Chiov. | Escherichia coli (ATCC 25922) Klebsiella pneumoniae Pseudomonas aeruginosa (ATCC 27853) Staphylococcus aureus Staphylococcus aureus (ATCC 29213) Salmonella typhi | - | Alkaloids Flavonoids Phenols Saponins Steroids Tannins Terpenoids | [145] |
| Secamone afzelii (Schult.) K.Schum. | Escherichia coli ATCC 25922 Klebsiella pneumoniae Pseudomonas aeruginosa ATCC 27853 Secamone afzelii Staphylococcus aureus Staphylococcus aureus ATCC 29213 Salmonella typhi | - | Alkaloids Cardiac glycosides Saponins Tannins | [145] |
| Thevetia peruviana L. | Bacillus subtilis Klebsiella Pneumoniae Streptococcus haemolyticus Salmonella typhi | Aspergillus niger Candida albicans | - | [136] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gracz-Bernaciak, J.; Mazur, O.; Nawrot, R. Functional Studies of Plant Latex as a Rich Source of Bioactive Compounds: Focus on Proteins and Alkaloids. Int. J. Mol. Sci. 2021, 22, 12427. https://doi.org/10.3390/ijms222212427
Gracz-Bernaciak J, Mazur O, Nawrot R. Functional Studies of Plant Latex as a Rich Source of Bioactive Compounds: Focus on Proteins and Alkaloids. International Journal of Molecular Sciences. 2021; 22(22):12427. https://doi.org/10.3390/ijms222212427
Chicago/Turabian StyleGracz-Bernaciak, Joanna, Oliwia Mazur, and Robert Nawrot. 2021. "Functional Studies of Plant Latex as a Rich Source of Bioactive Compounds: Focus on Proteins and Alkaloids" International Journal of Molecular Sciences 22, no. 22: 12427. https://doi.org/10.3390/ijms222212427






