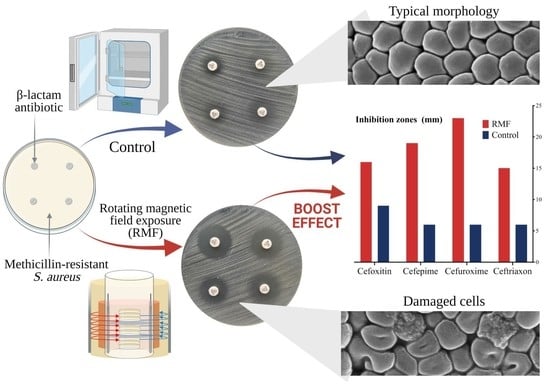
Rotating Magnetic Field Increases β-Lactam Antibiotic Susceptibility of Methicillin-Resistant Staphylococcus aureus Strains
Abstract
:1. Introduction
2. Results and Discussion
2.1. Analysis of Changes in Antibiotic Susceptibility of S. aureus Strains under the Influence of RMF
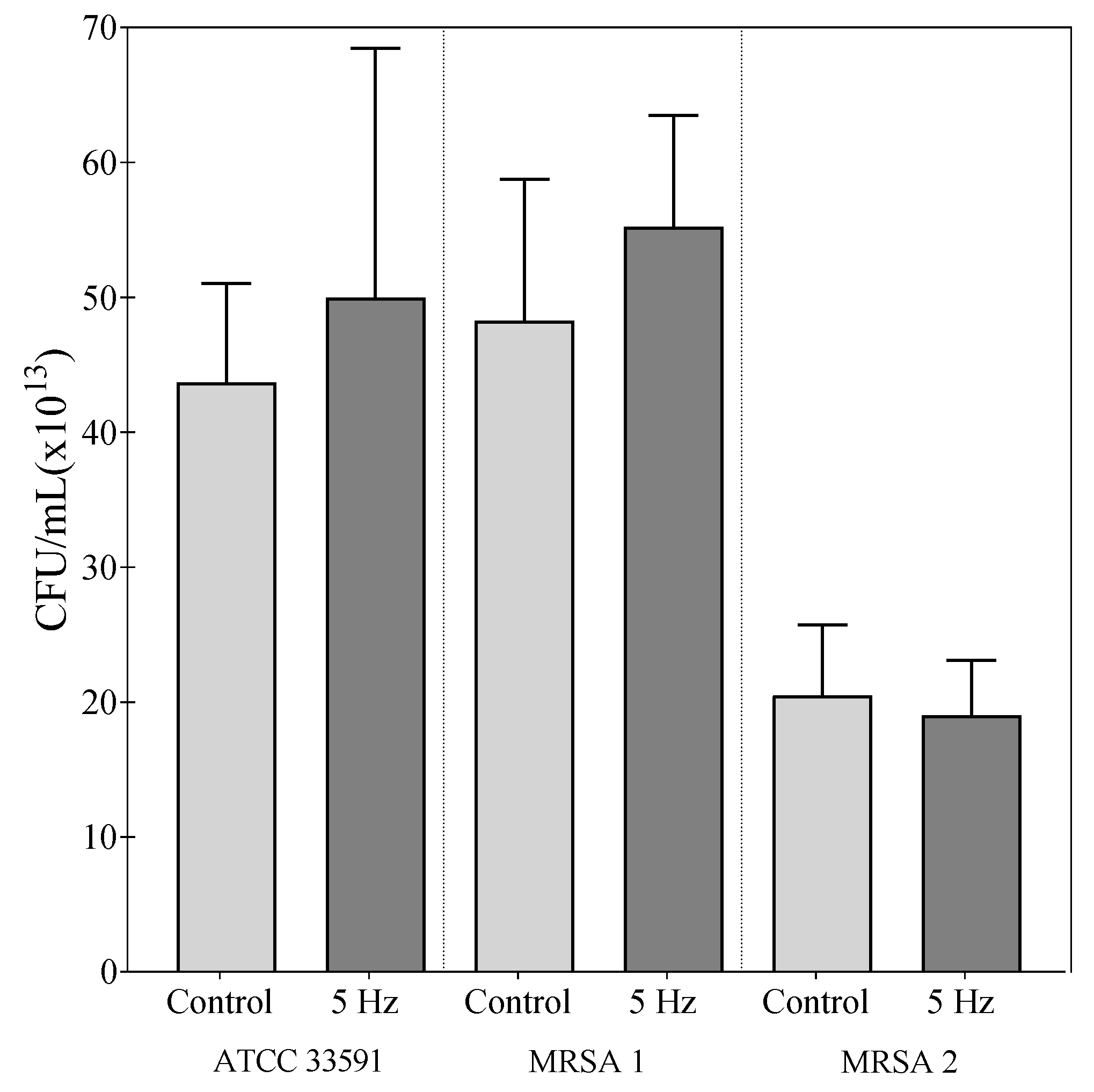
2.2. Effect of RMF on Changes in Number of Culturable Bacteria
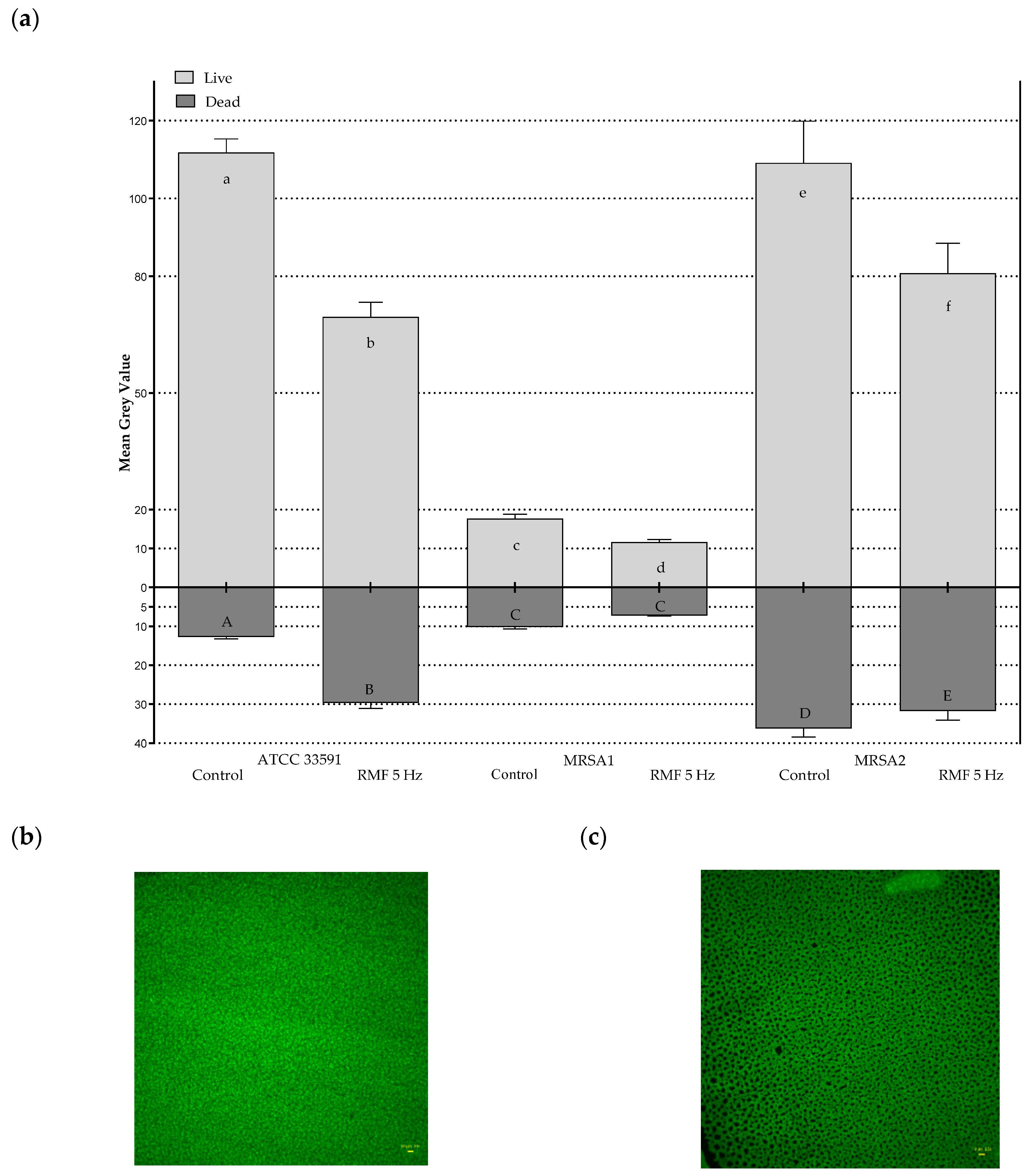
2.3. Effect of RMF on Changes in Relative Number of Live and Dead Bacterial Cells
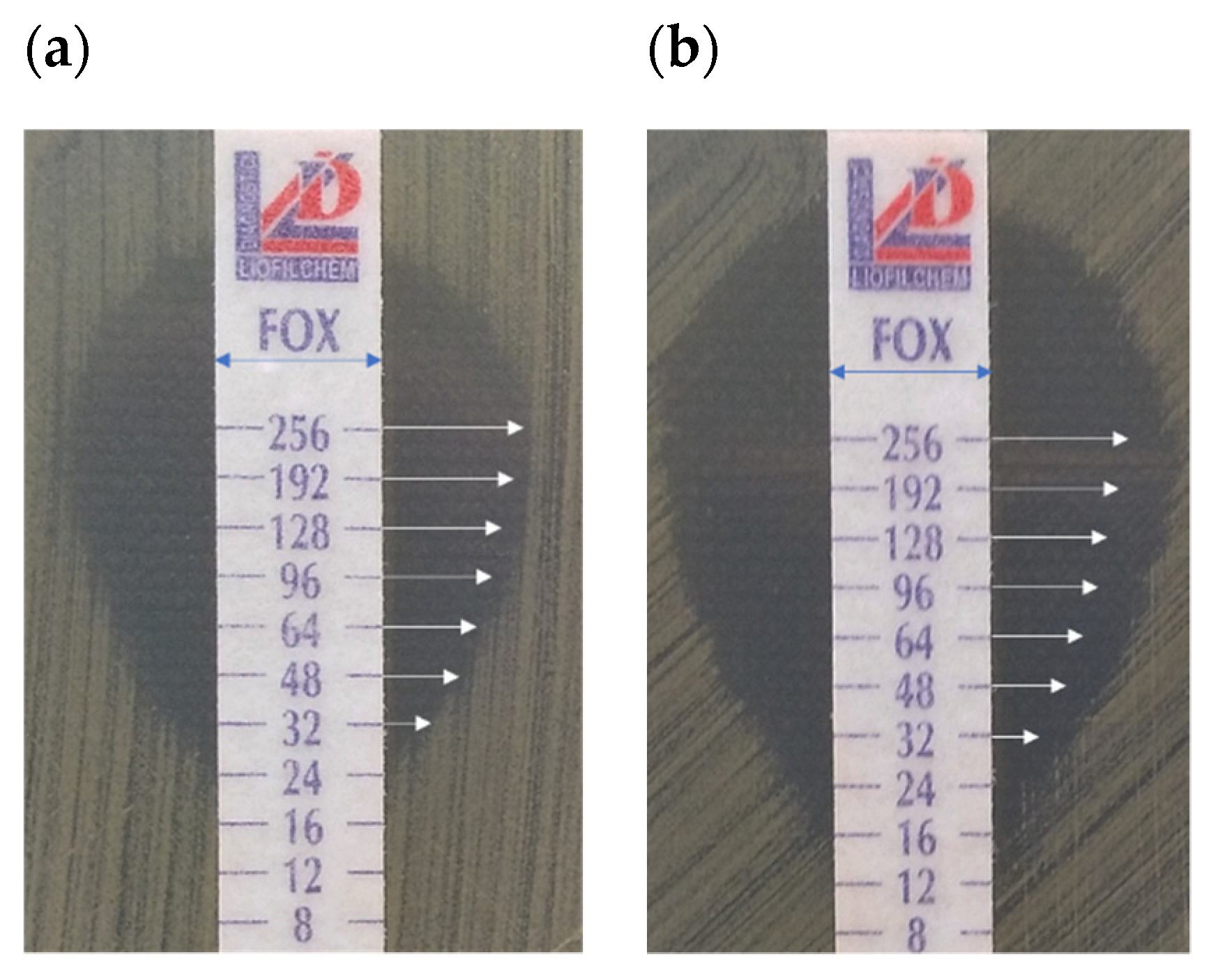
2.4. Effect of RMF on Diffusion of Antibiotics
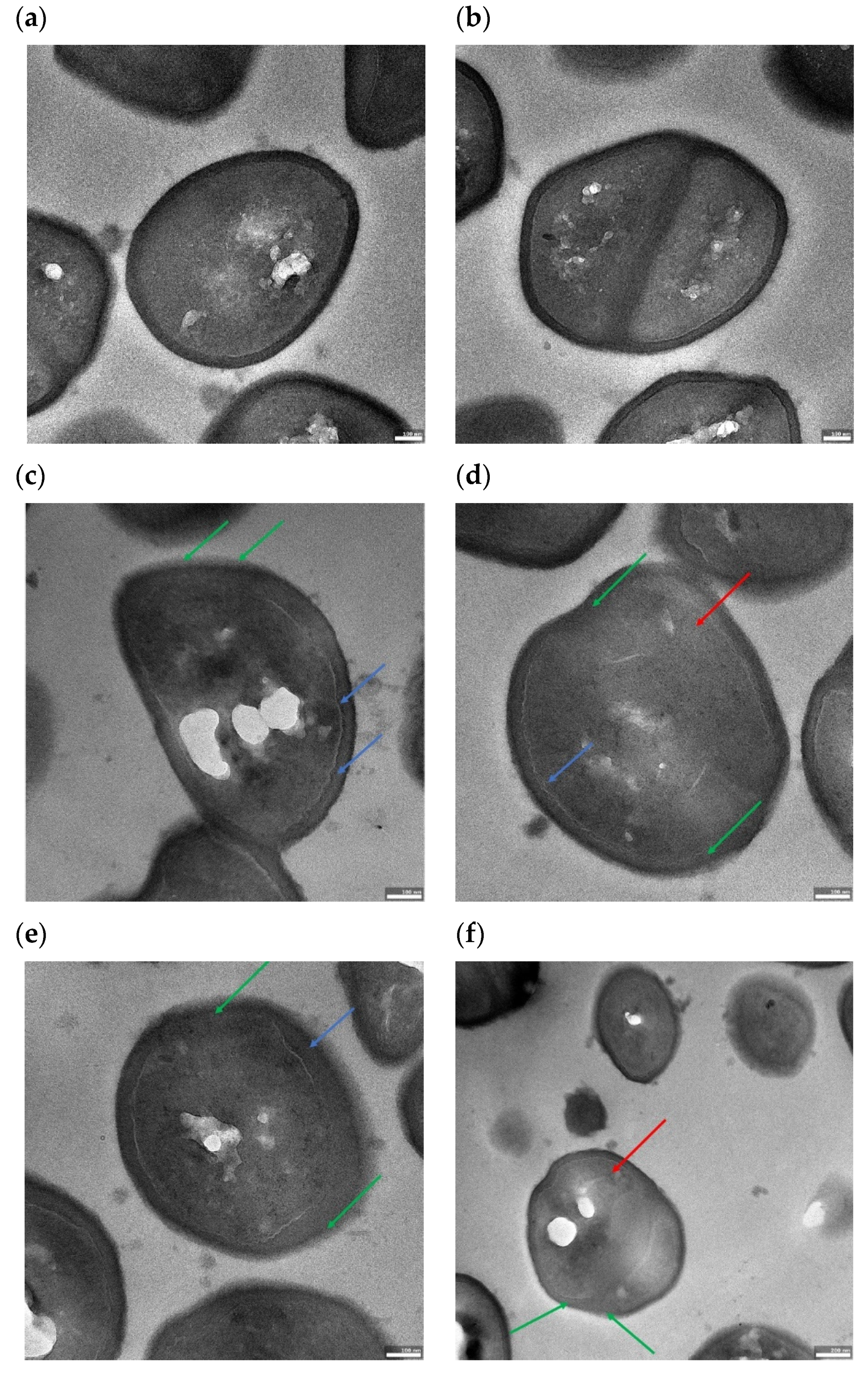
2.5. Effect of RMF on Cell Morphology
3. Conclusions
4. Materials and Methods
4.1. Microorganisms
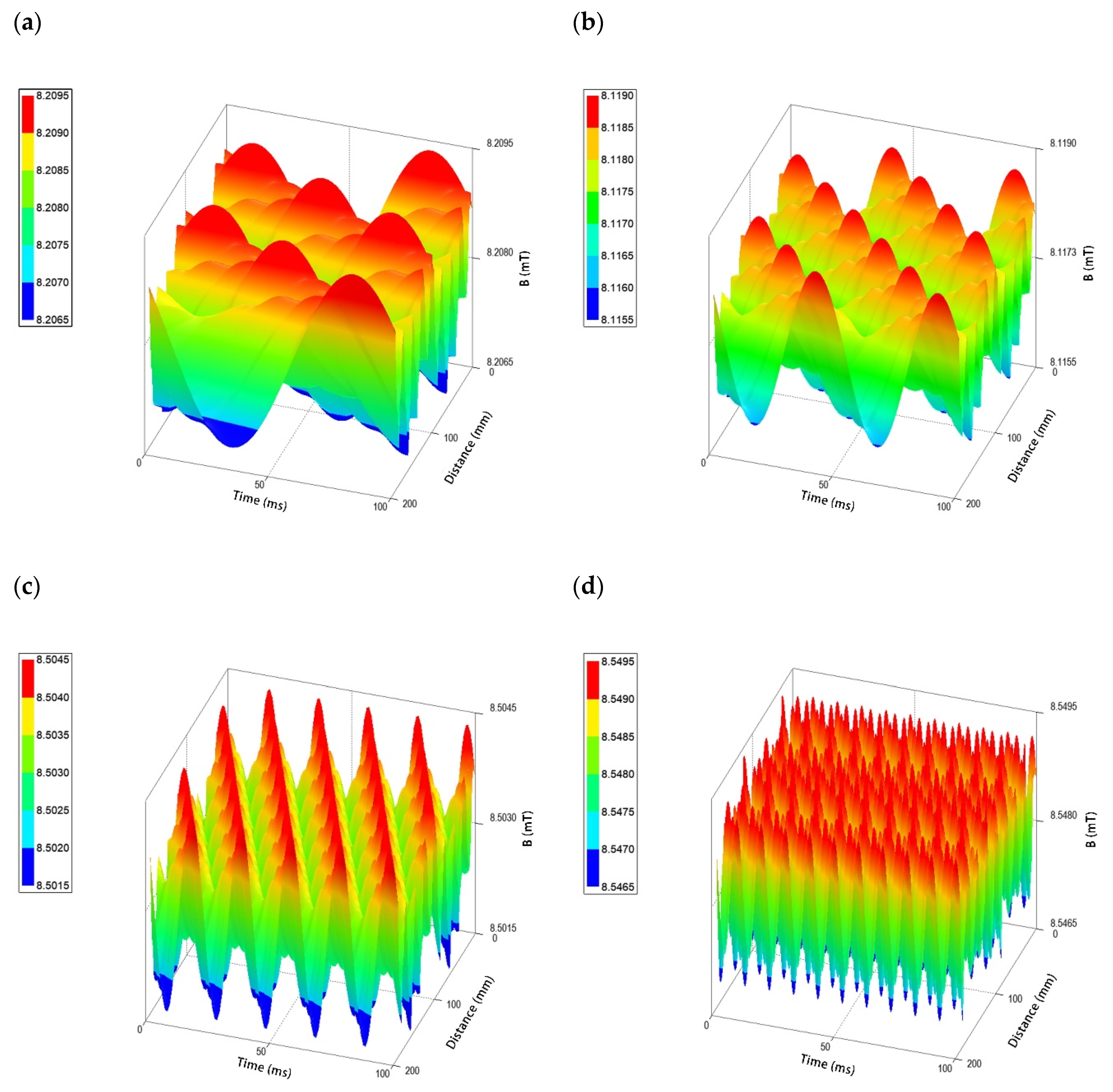
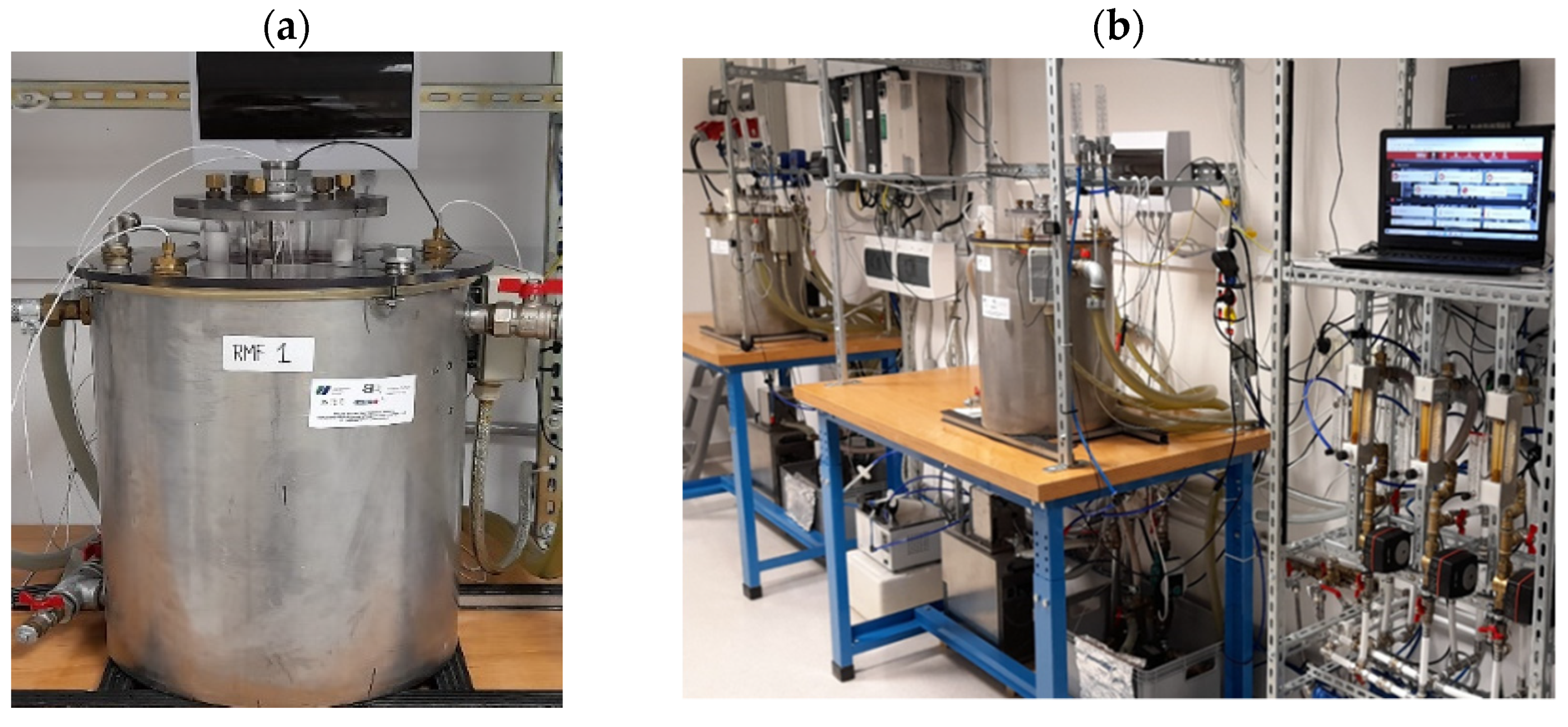
4.2. Rotating Magnetic Field Generator
4.3. Detection of mecA Gene
4.4. Analysis of the Impact of RMF on Changes in Antibiotic Susceptibility
4.4.1. Disc Diffusion Method
4.4.2. Gradient MIC Strips (E-Test)
4.5. Analysis of the Impact of RMF on Changes in the Number of Culturable Bacteria
4.6. Analysis of the Impact of RMF on Changes in the Relative Number of Live and Dead Bacterial Cells Using Fluorescence Microscopy and Picture Processing
4.7. Analysis of the Impact of RMF on the Diffusion of Antibiotics in the Agar Medium
4.8. Analysis of the Impact of RMF on Staphylococcal Cell Morphology
4.8.1. Scanning Electron Microscopy
4.8.2. Transmission Electron Microscopy
4.9. Analysis of the RMF Post-Exposure Effect
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, A.S.; De Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Prim. 2018, 4, 18033. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/mrsa/healthcare/inpatient.html (accessed on 31 February 2021).
- Styers, D.; Sheehan, D.J.; Hogan, P.; Sahm, D.F. Laboratory-based surveillance of current antimicrobial resistance patterns and trends among Staphylococcus aureus: 2005 status in the United States. Ann. Clin. Microbiol. Antimicrob. 2006, 5, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solis-Velazquez, O.A.; Gutiérrez-Lomelí, M.; Guerreo-Medina, P.J.; Rosas-García, M.D.L.; Iñiguez-Moreno, M.; Avila-Novoa, M.G. Nosocomial pathogen biofilms on biomaterials: Different growth medium conditions and components of biofilms produced in vitro. J. Microbiol. Immunol. Infect. 2020. [Google Scholar] [CrossRef]
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 2019, 17, 203–218. [Google Scholar] [CrossRef]
- Becker, K.; van Alen, S.; Idelevich, E.A.; Schleimer, N.; Seggewiß, J.; Mellmann, A.; Kaspar, U.; Peters, G. Plasmid-Encoded Transferable mecB-mediated methicillin resistance in Staphylococcus aureus. Emerg. Infect. Dis. 2018, 24, 242–248. [Google Scholar] [CrossRef] [Green Version]
- Cikman, A.; Aydin, M.; Gulhan, B.; Karakecili, F.; Kurtoglu, M.G.; Yuksekkaya, S.; Parlak, M.; Gultepe, B.S.; Cicek, A.C.; Bilman, F.B.; et al. Absence of the mecC gene in methicillin-resistant Staphylococcus aureus isolated from various clinical samples: The first multi-centered study in Turkey. J. Infect. Public Health 2019, 12, 528–533. [Google Scholar] [CrossRef] [PubMed]
- Fuda, C.; Suvorov, M.; Vakulenko, S.B.; Mobashery, S. The basis for resistance to beta-lactam antibiotics by penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. J. Biol. Chem. 2004, 279, 40802–40806. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.-H.; Hsieh, Y.-H.; Powers, Z.M.; Kao, C.-Y. Defeating antibiotic-resistant bacteria: Exploring alternative therapies for a post-antibiotic era. Int. J. Mol. Sci. 2020, 21, 1061. [Google Scholar] [CrossRef] [Green Version]
- Junka, A.F.; Rakoczy, R.; Szymczyk, P.; Bartoszewicz, M.; Sedghizadeh, P.P.; Fijałkowski, K. Application of rotating magnetic fields increase the activity of antimicrobials against wound biofilm pathogens. Sci. Rep. 2018, 8, 167. [Google Scholar] [CrossRef] [Green Version]
- Novickij, V.; Stanevičienė, R.; Vepštaitė-Monstavičė, I.; Gruškienė, R.; Krivorotova, T.; Sereikaitė, J.; Novickij, J.; Servienė, E. Overcoming antimicrobial resistance in bacteria using bioactive magnetic nanoparticles and pulsed electromagnetic fields. Front. Microbiol. 2017, 8, 2678. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Deng, A.; Cao, W.; Li, Q.; Wang, L.; Zhou, J.; Hu, B.; Xing, X. Synthesis of chitosan/poly (ethylene glycol)-modified magnetic nanoparticles for antibiotic delivery and their enhanced anti-biofilm activity in the presence of magnetic field. J. Mater. Sci. 2018, 53, 6433–6449. [Google Scholar] [CrossRef]
- Wang, X.; Wu, J.; Li, P.; Wang, L.; Zhou, J.; Zhang, G.; Li, X.; Hu, B.; Xing, X. Microenvironment-responsive magnetic nanocomposites based on silver nanoparticles/gentamicin for enhanced biofilm disruption by magnetic field. ACS Appl. Mater. Interfaces 2018, 10, 34905–34915. [Google Scholar] [CrossRef]
- Redlarski, G.; Lewczuk, B.; Żak, A.; Koncicki, A.; Krawczuk, M.; Piechocki, J.; Jakubiuk, K.; Tojza, P.; Jaworski, J.; Ambroziak, D.; et al. The influence of electromagnetic pollution on living organisms: Historical trends and forecasting changes. Biomed Res. Int. 2015, 2015, 234098. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, U.; Schulz, J.; Pilwat, G. Transcellular ion flow in Escherichia coli B and electrical sizing of bacterias. Biophys. J. 1973, 13, 1005–1013. [Google Scholar] [CrossRef] [Green Version]
- Luo, Q.; Wang, H.; Zhang, X.; Qian, Y. Effect of direct electric current on the cell surface properties of phenol-degrading bacteria. Appl. Environ. Microbiol. 2005, 71, 423–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golzio, M.; Rols, M.P.; Teissié, J. In vitro and in vivo electric field-mediated permeabilization, gene transfer, and expression. Methods 2004, 33, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Obermeier, A.; Matl, F.D.; Friess, W.; Stemberger, A. Growth inhibition of Staphylococcus aureus induced by low-frequency electric and electromagnetic fields. Bioelectromagnetics 2009, 30, 270–279. [Google Scholar] [CrossRef]
- Nawrotek, P.; Fijałkowski, K.; Struk, M.; Kordas, M.; Rakoczy, R. Effects of 50 Hz rotating magnetic field on the viability of Escherichia coli and Staphylococcus aureus . Electromagn. Biol. Med. 2014, 33, 29–34. [Google Scholar] [CrossRef]
- Fijałkowski, K.; Nawrotek, P.; Struk, M.; Kordas, M.; Rakoczy, R. Effects of rotating magnetic field exposure on the functional parameters of different species of bacteria. Electromagn. Biol. Med. 2015, 34, 48–55. [Google Scholar] [CrossRef]
- Konopacki, M.; Rakoczy, R. The analysis of rotating magnetic field as a trigger of Gram-positive and Gram-negative bacteria growth. Biochem. Eng. J. 2019, 141, 259–267. [Google Scholar] [CrossRef]
- Fijalkowski, K.; Nawrotek, P.; Struk, M.; Kordas, M.; Rakoczy, R. The effects of rotating magnetic Field on growth rate, cell metabolic activity and biofilm formation by Staphylococcus aureus and Escherichia coli. J. Magn. 2013, 18, 289–296. [Google Scholar] [CrossRef] [Green Version]
- Perez-Roa, R.E.; Tompkins, D.T.; Paulose, M.; Grimes, C.A.; Anderson, M.A.; Noguera, D.R. Effects of localised, low-voltage pulsed electric fields on the development and inhibition of Pseudomonas aeruginosa biofilms. Biofouling 2006, 22, 383–390. [Google Scholar] [CrossRef]
- Huwiler, S.G.; Beyer, C.; Fröhlich, J.; Hennecke, H.; Egli, T.; Schürmann, D.; Rehrauer, H.; Fischer, H.-M. Genome-wide transcription analysis of Escherichia coli in response to extremely low-frequency magnetic fields. Bioelectromagnetics 2012, 33, 488–496. [Google Scholar] [CrossRef]
- Yoshie, S.; Ikehata, M.; Hirota, N.; Takemura, T.; Minowa, T.; Hanagata, N.; Hayakawa, T. Evaluation of mutagenicity and co-mutagenicity of strong static magnetic fields up to 13 Tesla in Escherichia coli deficient in superoxide dismutase. J. Magn. Reson. Imaging 2012, 35, 731–736. [Google Scholar] [CrossRef] [PubMed]
- Filipič, J.; Kraigher, B.; Tepuš, B.; Kokol, V.; Mandic-Mulec, I. Effects of low-density static magnetic fields on the growth and activities of wastewater bacteria Escherichia coli and Pseudomonas putida. Bioresour. Technol. 2012, 120, 225–232. [Google Scholar] [CrossRef]
- Sakakibara, Y.; Kuroda, M. Electric prompting and control of denitrification. Biotechnol. Bioeng. 1993, 42, 535–537. [Google Scholar] [CrossRef] [PubMed]
- Nakasono, S.; Matsumoto, N.; Saiki, H. Electrochemical cultivation of Thiobacillus ferrooxidans by potential control. Bioelectrochem. Bioenerg. 1997, 43, 61–66. [Google Scholar] [CrossRef]
- Chen, X.; Chen, G.; Yue, P.L. Investigation on the electrolysis voltage of electrocoagulation. Chem. Eng. Sci. 2002, 57, 2449–2455. [Google Scholar] [CrossRef]
- Diao, H.; Li, X.; Gu, J.; Shi, H.; Xie, Z. Electron microscopic investigation of the bactericidal action of electrochemical disinfection in comparison with chlorination, ozonation and Fenton reaction. Process Biochem. 2004, 39, 1421–1426. [Google Scholar] [CrossRef]
- Zituni, D.; Schütt-Gerowitt, H.; Kopp, M.; Krönke, M.; Addicks, K.; Hoffmann, C.; Hellmich, M.; Faber, F.; Niedermeier, W. The growth of Staphylococcus aureus and Escherichia coli in low-direct current electric fields. Int. J. Oral Sci. 2014, 6, 7–14. [Google Scholar] [CrossRef] [Green Version]
- Fojt, L.; Strasák, L.; Vetterl, V.; Smarda, J. Comparison of the low-frequency magnetic field effects on bacteria Escherichia coli, Leclercia adecarboxylata and Staphylococcus aureus. Bioelectrochemistry 2004, 63, 337–341. [Google Scholar] [CrossRef]
- Piatti, E.; Cristina Albertini, M.; Baffone, W.; Fraternale, D.; Citterio, B.; Piera Piacentini, M.; Dachà, M.; Vetrano, F.; Accorsi, A. Antibacterial effect of a magnetic field on Serratia marcescens and related virulence to Hordeum vulgare and Rubus fruticosus callus cells. Comp. Biochem. Physiol. Part B 2002, 132, 359–365. [Google Scholar] [CrossRef]
- Strašák, L.; Vetterl, V.; Fojt, L. Effects of 50 Hz Magnetic fields on the viability of different bacterial strains. Electromagn. Biol. Med. 2005, 24, 293–300. [Google Scholar] [CrossRef]
- Strašák, L.; Vetterl, V.; Šmarda, J. Effects of low-frequency magnetic fields on bacteria Escherichia coli. Bioelectrochemistry 2002, 55, 161–164. [Google Scholar] [CrossRef]
- Gao, W.; Liu, Y.; Zhou, J.; Pan, H. Effects of a strong static magnetic field on bacterium Shewanella oneidensis: An assessment by using whole genome microarray. Bioelectromagnetics 2005, 26, 558–563. [Google Scholar] [CrossRef]
- László, J.; Kutasi, J. Static magnetic field exposure fails to affect the viability of different bacteria strains. Bioelectromagnetics 2010, 31, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Grosman, Z.; Kolár, M.; Tesaríková, E. Effects of static magnetic field on some pathogenic microorganisms. Acta Univ. Palacki. Olomuc. Fac. Med. 1992, 134, 7–9. [Google Scholar] [PubMed]
- Poortinga, A.T.; Bos, R.; Busscher, H.J. Lack of effect of an externally applied electric field on bacterial adhesion to glass. Coll. Surf. B 2001, 20, 189–194. [Google Scholar] [CrossRef]
- Babushkina, I.V.; Borodulin, V.B.; Shmetkova, N.A.; Morrison, V.V.; Usanov, A.D.; Skripal’, A.V.; Usanov, D.A. The Influence of alternating magnetic field on Escherichia coli bacterial cells. Pharm. Chem. J. 2005, 39, 398–400. [Google Scholar] [CrossRef]
- Justo, O.R.; Pérez, V.H.; Alvarez, D.C.; Alegre, R.M. Growth of Escherichia coli under extremely low-frequency electromagnetic fields. ABAB 2006, 134, 155–164. [Google Scholar] [CrossRef]
- Cellini, L.; Grande, R.; Di Campli, E.; Di Bartolomeo, S.; Di Giulio, M.; Robuffo, I.; Trubiani, O.; Mariggiò, M.A. Bacterial response to the exposure of 50 Hz electromagnetic fields. Bioelectromagnetics 2008, 29, 302–311. [Google Scholar] [CrossRef]
- Dunca, S.; Creanga, D.-E.; Ailiesei, O.; Nimitan, E. Microorganisms growth with magnetic fluids. J. Magn. Magn. Mater. 2005, 289, 445–447. [Google Scholar] [CrossRef]
- Segatore, B.; Setacci, D.; Bennato, F.; Cardigno, R.; Amicosante, G.; Iorio, R. Evaluations of the effects of extremely low-frequency electromagnetic fields on growth and antibiotic susceptibility of Escherichia coli and Pseudomonas aeruginosa. Int. J. Microbiol. 2012, 2012, 587293. [Google Scholar] [CrossRef] [Green Version]
- Golberg, A.; Broelsch, G.F.; Vecchio, D.; Khan, S.; Hamblin, M.R.; Austen, W.G.; Sheridan, R.L.; Yarmush, M.L. Eradication of multidrug-resistant A. baumannii in burn wounds by antiseptic pulsed electric field. Technology 2014, 2, 153–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tagourti, J.; El May, A.; Aloui, A.; Chatti, A.; Ben Aissa, R.; Landoulsi, A. Static magnetic field increases the sensitivity of Salmonella to gentamicin. Ann. Microbiol. 2010, 60, 519–522. [Google Scholar] [CrossRef]
- Ahmed, I.; Istivan, T.; Cosic, I.; Pirogova, E. Evaluation of the effects of Extremely Low Frequency (ELF) Pulsed Electromagnetic Fields (PEMF) on survival of the bacterium Staphylococcus aureus. EPJ Nonlinear Biomed Phys. 2013, 1, 5. [Google Scholar] [CrossRef] [Green Version]
- Del Re, B.; Bersani, F.; Agostini, C.; Mesirca, P.; Giorgi, G. Various effects on transposition activity and survival of Escherichia coli cells due to different ELF-MF signals . Radiat. Environ. Biophys. 2004, 43, 265–270. [Google Scholar] [CrossRef]
- Del Re, B.; Garoia, F.; Mesirca, P.; Agostini, C.; Bersani, F.; Giorgi, G. Extremely low frequency magnetic fields affect transposition activity in Escherichia coli. Radiat. Environ. Biophys. 2003, 42, 113–118. [Google Scholar] [CrossRef] [PubMed]
- EUCAST Disk Diffusion Method for Antimicrobial Susceptibility Testing-Version 9.0. January 2021. Available online: https://www.eucast.org/ast_of_bacteria/disk_diffusion_methodology/ (accessed on 31 February 2021).
- Bjedov, I.; Tenaillon, O.; Gérard, B.; Souza, V.; Denamur, E.; Radman, M.; Taddei, F.; Matic, I. Stress-induced mutagenesis in bacteria. Science 2003, 300, 1404–1409. [Google Scholar] [CrossRef]
- Eichenbaum, Z.; Livneh, Z. UV light induces IS10 transposition in Escherichia coli. Genetics 1998, 149, 1173–1181. [Google Scholar] [CrossRef]
- Lamrani, S.; Ranquet, C.; Gama, M.J.; Nakai, H.; Shapiro, J.A.; Toussaint, A.; Maenhaut-Michel, G. Starvation-induced Mucts62-mediated coding sequence fusion: A role for ClpXP, Lon, RpoS and Crp. Mol. Microbiol. 1999, 32, 327–343. [Google Scholar] [CrossRef] [Green Version]
- Avery, S.V. Cell individuality: The bistability of competence development. Trends Microbiol. 2005, 13, 459–462. [Google Scholar] [CrossRef]
- Stepanian, R.S.; Barsegian, A.A.; Alaverdian, Z.R.; Oganesian, G.G.; Markosian, L.S.; Aĭrapetian, S.N. Vliianie magnitnykh poleĭ na rost i delenie lon mutanta Escherichia coli K-12. Radiats. Biol. Radioecol. 2000, 40, 319–322. [Google Scholar]
- Kohno, M.; Yamazaki, M.; Kimura, I.; Wada, M. Effect of static magnetic fields on bacteria: Streptococcus mutans, Staphylococcus aureus, and Escherichia coli. Pathophysiology 2000, 7, 143–148. [Google Scholar] [CrossRef]
- Jin, F.; Liu, T.; Li, F.; He, J. Effects of static magnetic fields on aerobes: Escherichia coli, Staphylococcus aureus and Bacillus subtilis. J. Biomed. Eng. 2009, 26, 757–760. [Google Scholar]
- Mittenzwey, R.; Süßmuth, R.; Mei, W. Effects of extremely low-frequency electromagnetic fields on bacteria—the question of a co-stressing factor. Bioelectrochem. Bioenerg. 1996, 40, 21–27. [Google Scholar] [CrossRef]
- Bajpai, I.; Saha, N.; Basu, B. Moderate intensity static magnetic field has bactericidal effect on E. coli and S. epidermidis on sintered hydroxyapatite. J. Biomed. Mater. Res. B Appl. Biomater. 2012, 100, 1206–1217. [Google Scholar] [CrossRef]
- Mohamed, S.B.; Adlan, T.A.; Khalafalla, N.A.; Abdalla, N.I.; Ali, Z.S.; Munir Ka, A.; Hassan, M.M.; Elnour, M.-A.B. Proteomics and docking study targeting penicillin-binding protein and penicillin-binding protein2a of methicillin-resistant Staphylococcus aureus Strain SO-1977 Isolated from Sudan. Evol. Bioinform. 2019, 15, 1176934319864945. [Google Scholar] [CrossRef] [Green Version]
- Harrison, E.M.; Ba, X.; Coll, F.; Blane, B.; Restif, O.; Carvell, H.; Köser, C.U.; Jamrozy, D.; Reuter, S.; Lovering, A.; et al. Genomic identification of cryptic susceptibility to penicillins and β-lactamase inhibitors in methicillin-resistant Staphylococcus aureus. Nat. Microbiol. 2019, 4, 1680–1691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torgomyan, H.; Ohanyan, V.; Blbulyan, S.; Kalantaryan, V.; Trchounian, A. Electromagnetic irradiation of Enterococcus hirae at low-intensity 51.8- and 53.0-GHz frequencies: Changes in bacterial cell membrane properties and enhanced antibiotics effects. FEMS Microbiol. Lett. 2012, 329, 131–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rakoczy, R.; Przybył, A.; Kordas, M.; Konopacki, M.; Drozd, R.; Fijałkowski, K. The study of influence of a rotating magnetic field on mixing efficiency. Chem. Eng. Process 2017, 112, 1–8. [Google Scholar] [CrossRef]
- Kim, C.; Mwangi, M.; Chung, M.; Milheiriço, C.; Milheirço, C.; de Lencastre, H.; Tomasz, A. The mechanism of heterogeneous beta-lactam resistance in MRSA: Key role of the stringent stress response. PLoS ONE 2013, 8, e82814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kermanshahi, R.K.; Sailani, M.R. Effect of static electric field treatment on multiple antibiotic-resistant pathogenic strains of Escherichia coli and Staphylococcus aureus. J. Microbiol. Immunol. Infect. 2005, 38, 394–398. [Google Scholar]
- Robertson, J.; McGoverin, C.; Vanholsbeeck, F.; Swift, S. Optimisation of the Protocol for the LIVE/DEAD® BacLightTM Bacterial Viability Kit for rapid determination of bacterial load. Front. Microbiol. 2019, 10, 801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emerson, J.B.; Adams, R.I.; Román, C.M.B.; Brooks, B.; Coil, D.A.; Dahlhausen, K.; Ganz, H.H.; Hartmann, E.M.; Hsu, T.; Justice, N.B.; et al. Schrödinger’s microbes: Tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 2017, 5, 86. [Google Scholar] [CrossRef] [PubMed]
- Fijałkowski, K.; Żywicka, A.; Drozd, R.; Junka, A.F.; Peitler, D.; Kordas, M.; Konopacki, M.; Szymczyk, P.; El Fray, M.; Rakoczy, R. Increased yield and selected properties of bacterial cellulose exposed to different modes of a rotating magnetic field. Eng. Life Sci. 2016, 16, 483–493. [Google Scholar] [CrossRef]
- Sutton, J.A.F.; Carnell, O.T.; Lafage, L.; Gray, J.; Biboy, J.; Gibson, J.F.; Pollitt, E.J.G.; Tazoll, S.C.; Turnbull, W.; Hajdamowicz, N.H.; et al. Staphylococcus aureus cell wall structure and dynamics during host-pathogen interaction. PLoS Pathog. 2021, 17, e1009468. [Google Scholar] [CrossRef] [PubMed]
- Wong, F.; Stokes, J.M.; Cervantes, B.; Penkov, S.; Friedrichs, J.; Renner, L.D.; Collins, J.J. Cytoplasmic condensation induced by membrane damage is associated with antibiotic lethality. Nat. Commun. 2021, 12, 2321. [Google Scholar] [CrossRef]
- Mahasenan, K.V.; Molina, R.; Bouley, R.; Batuecas, M.T.; Fisher, J.F.; Hermoso, J.A.; Chang, M.; Mobashery, S. Conformational dynamics in penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus, allosteric communication network and enablement of catalysis. J. Am. Chem. Soc. 2017, 139, 2102–2110. [Google Scholar] [CrossRef] [Green Version]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. January 2021. Available online: http://www.eucast.org (accessed on 31 February 2021).
- Drozd, R.; Szymańska, M.; Żywicka, A.; Kowalska, U.; Rakoczy, R.; Kordas, M.; Konopacki, M.; Junka, A.F.; Fijałkowski, K. Exposure to non-continuous rotating magnetic field induces metabolic strain-specific response of Komagataeibacter xylinus. Biochem. Eng. J. 2021, 166, 107855. [Google Scholar] [CrossRef]
- Oliveira, D.C.; Lencastre, H.D. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2002, 46, 2155–2161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fijałkowski, K.; Peitler, D.; Karakulska, J. Staphylococci isolated from ready-to-eat meat - identification, antibiotic resistance and toxin gene profile. Int. J. Food Microbiol. 2016, 238, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, J.; Jeong, S.-I.; Jahng, K.Y.; Yu, K.-Y. Antimicrobial effects and resistant regulation of magnolol and honokiol on methicillin-resistant Staphylococcus aureus. Biomed Res. Int. 2015, 2015, 283630. [Google Scholar] [CrossRef] [Green Version]
- Makarova, O.; Johnston, P.; Walther, B.; Rolff, J.; Roesler, U. Complete genome sequence of the disinfectant susceptibility testing reference strain Staphylococcus aureus subsp. aureus ATCC 6538. Genome Announc. 2017, 5, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, T.; Rasband, W. ImageJ User Guide. Available online: https://imagej.nih.gov/ij/docs/guide/user-guide.pdf (accessed on 31 February 2021).
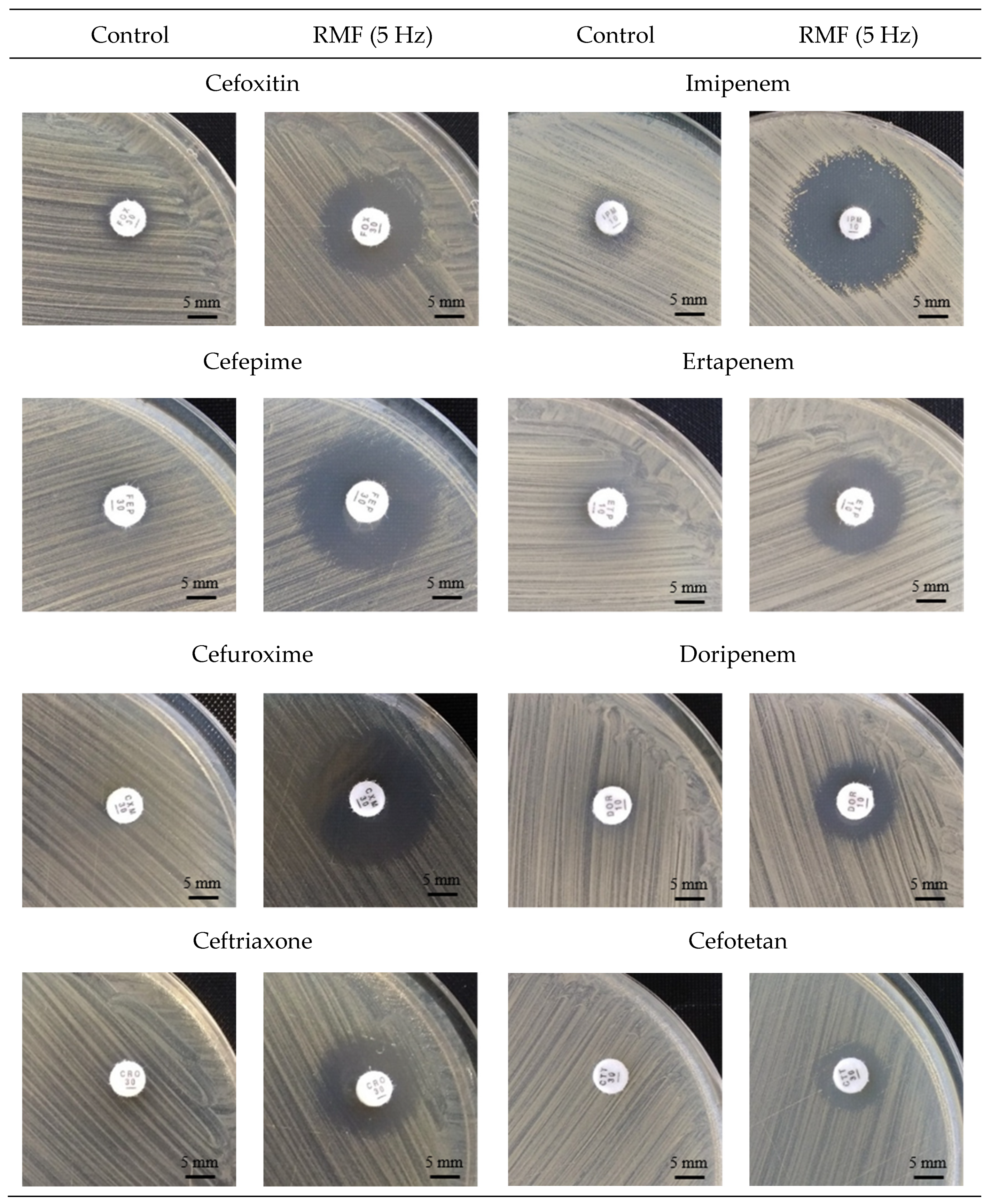




| Strain | RMF Frequency | ||||
|---|---|---|---|---|---|
| C | 5 Hz | 10 Hz | 25 Hz | 50 Hz | |
| Cefoxitin | |||||
| ATTC 33591 | 12 | 18 | 15 | 16 | 15 |
| MRSA 1 | 6 | 16 | 14 | 16 | 14 |
| MRSA 2 | 7 | 21 | 15 | 17 | 15 |
| Cefepime | |||||
| ATTC 33591 | 10 | 16 | 15 | 16 | 14 |
| MRSA 1 | 6 | 19 | 14 | 18 | 13 |
| MRSA 2 | 6 | 18 | 6 (14p.) | 6 (16p.) | 6 |
| Cefuroxime | |||||
| ATTC 33591 | 6 | 22 | 20 | 20 | 18 |
| MRSA 1 | 6 | 23 | 19 | 21 | 18 |
| MRSA 2 | 6 | 15 | 6 (14p.) | 14 | 6 |
| Ceftriaxone | |||||
| ATTC 33591 | 6 | 18 | 15 | 17 | 16 |
| MRSA 1 | 6 | 15 | 14 | 15 | 13 |
| MRSA 2 | 6 | 15 | 6 (13p.) | 15 | 6 |
| Strain | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATCC 33591 | MRSA 1 | MRSA 2 | ||||||||||||||||||||||
| FOX | FEP | CXM | CRO | FOX | FEP | CXM | CRO | FOX | FEP | CXM | CRO | |||||||||||||
| t | C | RMF | C | RMF | C | RMF | C | RMF | C | RMF | C | RMF | C | RMF | C | RMF | C | RMF | C | RMF | C | RMF | C | RMF |
| 1 | 13 | 13 | 10 | 10 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 7 | 7 | 6 | 6 | 6 | 6 | 6 | 6 |
| 2 | 12 | 13 | 9 | 11 | 6 | 6 | 6 | 10 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 10 | 6 | 6 | 6 | 6 | 6 | 6 |
| 3 | 12 | 13 | 9 | 12 | 6 | 16 | 6 | 12 | 6 | 11 | 6 | 6 | 6 | 6 | 6 | 6 | 7 | 12 | 6 | 6 | 6 | 6 | 6 | 6 |
| 4 | 12 | 14 | 10 | 12 | 6 | 15 | 6 | 12 | 6 | 14 | 6 | 14 | 6 | 19 | 6 | 14 | 7 | 13 | 6 | 13 | 6 | 14 | 6 | 6 |
| 5 | 12 | 15 | 9 | 13 | 6 | 16 | 6 | 13 | 6 | 15 | 6 | 13 | 6 | 20 | 6 | 15 | 7 | 14 | 6 | 15 | 6 | 15 | 6 | 13 |
| 6 | 12 | 15 | 9 | 13 | 6 | 17 | 6 | 13 | 6 | 15 | 6 | 15 | 6 | 19 | 6 | 15 | 7 | 14 | 6 | 17 | 6 | 15 | 6 | 13 |
| 7 | 13 | 16 | 10 | 14 | 6 | 17 | 6 | 13 | 6 | 16 | 6 | 15 | 6 | 21 | 6 | 15 | 7 | 13 | 6 | 17 | 6 | 15 | 6 | 13 |
| 8 | 12 | 16 | 10 | 15 | 6 | 18 | 6 | 13 | 6 | 16 | 6 | 15 | 6 | 20 | 6 | 15 | 7 | 14 | 6 | 16 | 6 | 15 | 6 | 12 |
| 9 | 12 | 16 | 10 | 14 | 6 | 18 | 6 | 15 | 6 | 16 | 6 | 16 | 6 | 20 | 6 | 15 | 7 | 16 | 6 | 18 | 6 | 15 | 6 | 13 |
| 10 | 12 | 17 | 10 | 15 | 6 | 19 | 6 | 16 | 6 | 16 | 6 | 17 | 6 | 21 | 6 | 15 | 7 | 18 | 6 | 18 | 6 | 15 | 6 | 13 |
| 11 | 12 | 18 | 10 | 16 | 6 | 20 | 6 | 17 | 6 | 16 | 6 | 18 | 6 | 22 | 6 | 15 | 7 | 20 | 6 | 18 | 6 | 15 | 6 | 14 |
| 12 | 12 | 18 | 10 | 16 | 6 | 22 | 6 | 18 | 6 | 16 | 6 | 19 | 6 | 23 | 6 | 15 | 7 | 21 | 6 | 18 | 6 | 15 | 6 | 15 |
| 18 * | 12 | 18 | 10 | 16 | 6 | 22 | 6 | 18 | 6 | 16 | 6 | 19 | 6 | 23 | 6 | 15 | 7 | 21 | 6 | 18 | 6 | 15 | 6 | 15 |
| Strain | C | RMF | C | RMF | C | RMF | C | RMF |
|---|---|---|---|---|---|---|---|---|
| Cefazolin | Cefotetan | Cefradine | Cefalexin | |||||
| ATCC 33591 | 11 | 18 | 9 | 12 | 16 | 16 | 10 | 15 |
| MRSA 1 | 6 | 18 | 6 | 11 | 6 | 12 | 6 | 9 |
| MRSA 2 | 6 | 12 | 6 | 14 | 6 | 6 | 6 | 6 |
| Ceftaroline | Meropenem | Imipenem | Ertapenem | |||||
| ATCC 33591 | 21 | 23 | 9 | 15 | 31 | 35 | 6 | 16 |
| MRSA 1 | 19 | 22 | 6 | 18 | 6 | 22 | 6 | 15 |
| MRSA 2 | 20 | 22 | 6 | 15 | 10 | 20 | 6 | 17 |
| Ceftazidime | Doripenem | Amoxicilin | ||||||
| ATCC 33591 | 6 | 6 | 6 | 13 | 6 | 6 | ||
| MRSA 1 | 6 | 6 | 6 | 12 | 6 | 6 | ||
| MRSA 2 | 6 | 6 | 6 | 12 | 6 | 6 | ||
| Strain | ||||||
|---|---|---|---|---|---|---|
| ATCC 33591 | MRSA 1 | MRSA 2 | ||||
| C | RMF | C | RMF | C | RMF | |
| Cefoxitin | 24 | 16 | 256 | 24 | 256 | 6 |
| Amoxicilin | 8 | 6 | 24 | 6 | 24 | 12 |
| Imipenem | 1.5 | 1 | 32 | 0.19 | 32 | 1.5 |
| Meropenem | 1 | 0.75 | 32 | 1.5 | 32 | 4 |
| Ceftriaxone | 32 | 24 | 256 | 16 | 256 | 24 |
| Cefuroxime | 12 | 8 | 256 | 8 | 256 | 12 |
| Ceftazidime | 32 | 32 | 256 | 32 | 256 | 96 |
| Cefepime | 32 | 16 | 256 | 16 | 256 | 64 |
| Strain | C | RMF | Strain | C | RMF |
|---|---|---|---|---|---|
| ATCC 33591 | 12 | 18 | MRSA 12 | 12 | 16 |
| MRSA 1 | 6 | 16 | MRSA 13 | 6 | 20 |
| MRSA 2 | 7 | 21 | MRSA 14 | 9 | 15 |
| MRSA 3 | 6 | 17 | MRSA 15 | 12 | 18 |
| MRSA 4 | 6 | 18 | MRSA 16 | 6 | 14 |
| MRSA 5 | 6 | 16 | MRSA 17 | 7 | 18 |
| MRSA 6 | 6 | 16 | MRSA 18 | 11 | 16 |
| MRSA 7 | 6 | 21 | MRSA 19 | 6 | 14 |
| MRSA 8 | 12 | 16 | MRSA 20 | 6 | 16 |
| MRSA 9 | 6 | 13 | MRSA 21 | 15 | 19 |
| MRSA 10 | 6 | 19 | MRSA 22 | 6 | 14 |
| MRSA 11 | 12 | 19 | MRSA 23 | 6 | 12 |
| Strain | Cefoxitin | Cefepime | Cefuroxime | Ceftriaxone | ||||
|---|---|---|---|---|---|---|---|---|
| C | RMF | C | RMF | C | RMF | C | RMF | |
| ATCC 6538 | 3 | 3 | 3 | 2 | 1.5 | 1 | 3 | 3 |
| MSSA 1 | 3 | 3 | 4 | 3 | 1 | 0.75 | 2 | 2 |
| MSSA 2 | 3 | 3 | 4 | 3 | 2 | 1 | 6 | 3 |
| MSSA 3 | 3 | 3 | 4 | 3 | 1.5 | 1 | 6 | 4 |
| MSSA 4 | 3 | 3 | 3 | 2 | 1 | 0.75 | 3 | 2 |
| MSSA 5 | 3 | 3 | 8 | 4 | 2 | 1.5 | 8 | 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woroszyło, M.; Ciecholewska-Juśko, D.; Junka, A.; Drozd, R.; Wardach, M.; Migdał, P.; Szymczyk-Ziółkowska, P.; Styburski, D.; Fijałkowski, K. Rotating Magnetic Field Increases β-Lactam Antibiotic Susceptibility of Methicillin-Resistant Staphylococcus aureus Strains. Int. J. Mol. Sci. 2021, 22, 12397. https://doi.org/10.3390/ijms222212397
Woroszyło M, Ciecholewska-Juśko D, Junka A, Drozd R, Wardach M, Migdał P, Szymczyk-Ziółkowska P, Styburski D, Fijałkowski K. Rotating Magnetic Field Increases β-Lactam Antibiotic Susceptibility of Methicillin-Resistant Staphylococcus aureus Strains. International Journal of Molecular Sciences. 2021; 22(22):12397. https://doi.org/10.3390/ijms222212397
Chicago/Turabian StyleWoroszyło, Marta, Daria Ciecholewska-Juśko, Adam Junka, Radosław Drozd, Marcin Wardach, Paweł Migdał, Patrycja Szymczyk-Ziółkowska, Daniel Styburski, and Karol Fijałkowski. 2021. "Rotating Magnetic Field Increases β-Lactam Antibiotic Susceptibility of Methicillin-Resistant Staphylococcus aureus Strains" International Journal of Molecular Sciences 22, no. 22: 12397. https://doi.org/10.3390/ijms222212397
APA StyleWoroszyło, M., Ciecholewska-Juśko, D., Junka, A., Drozd, R., Wardach, M., Migdał, P., Szymczyk-Ziółkowska, P., Styburski, D., & Fijałkowski, K. (2021). Rotating Magnetic Field Increases β-Lactam Antibiotic Susceptibility of Methicillin-Resistant Staphylococcus aureus Strains. International Journal of Molecular Sciences, 22(22), 12397. https://doi.org/10.3390/ijms222212397