Distinct and Overlapping Functions of Miscanthus sinensis MYB Transcription Factors SCM1 and MYB103 in Lignin Biosynthesis
Abstract
1. Introduction
2. Results and Discussion
2.1. Miscanthus MYB103 and SCM1 Are Expressed in Tissues Undergoing Lignification
2.2. Ectopic Expression of MsSCM1 and MsMYB103 Lead to Lignin Deposition in N. benthamiana Leaves
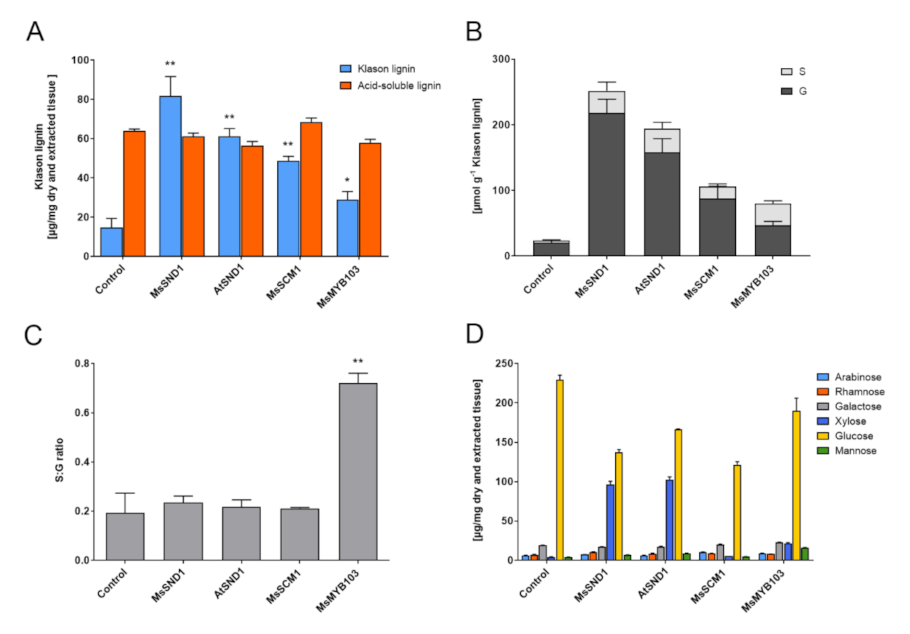
2.3. Expression of MsSND1, MsMYB103, and MsSCM1 Results in Distinct Secondary Cell Wall Composition
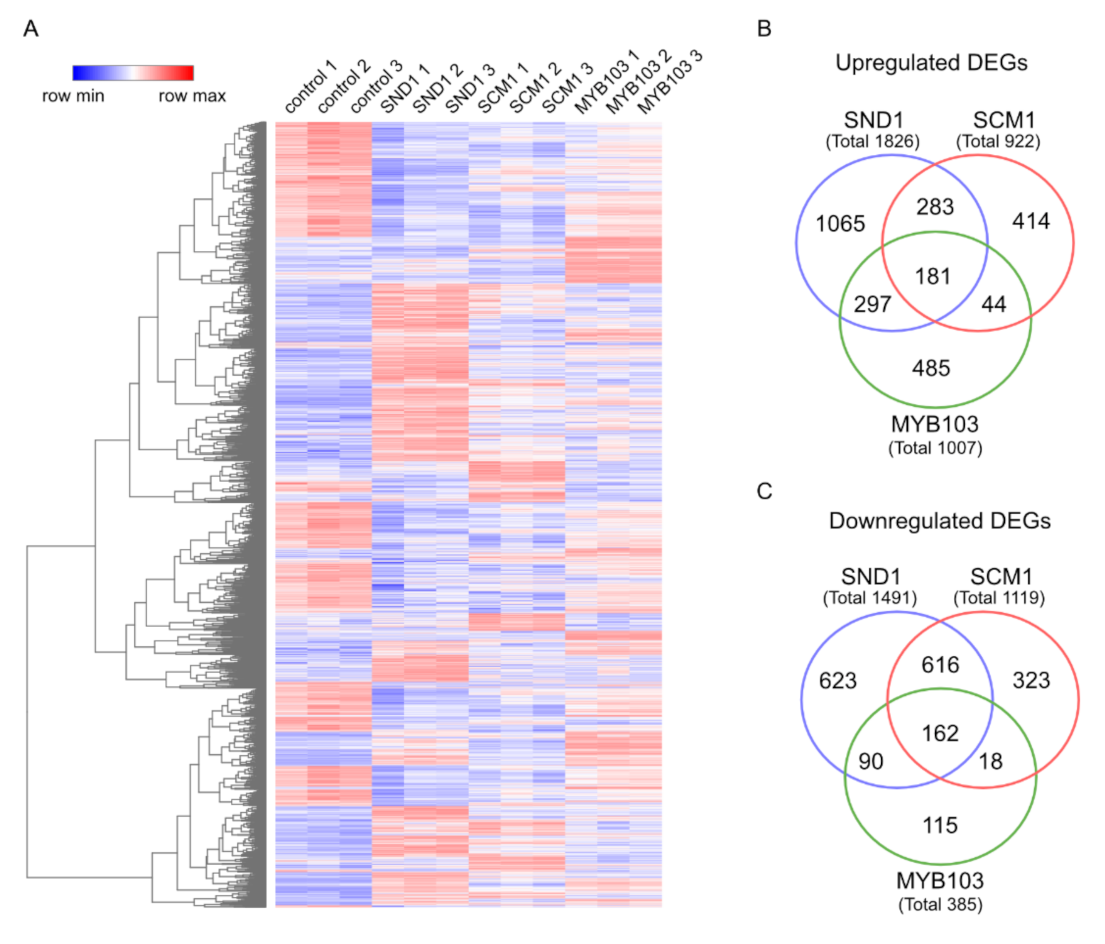
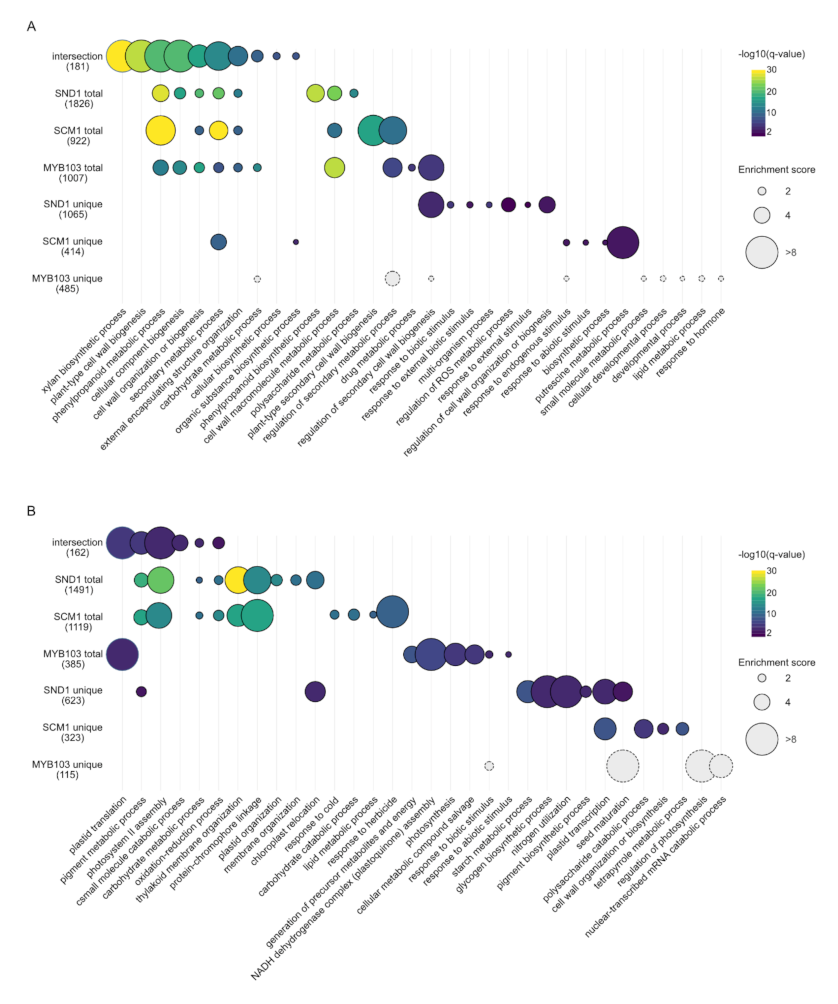
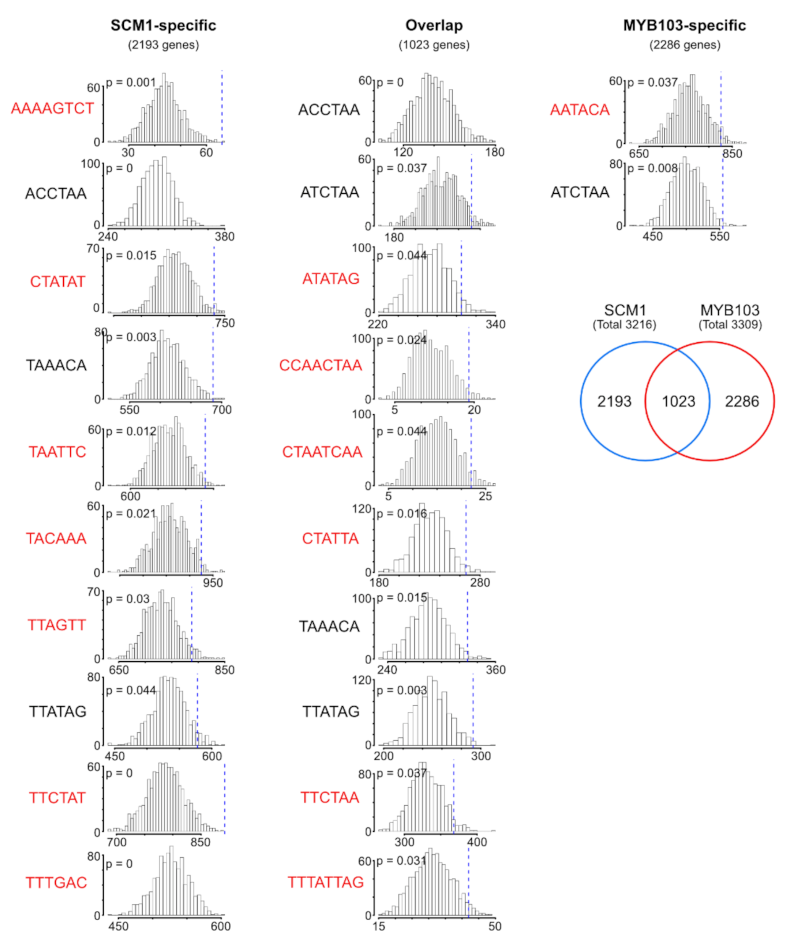
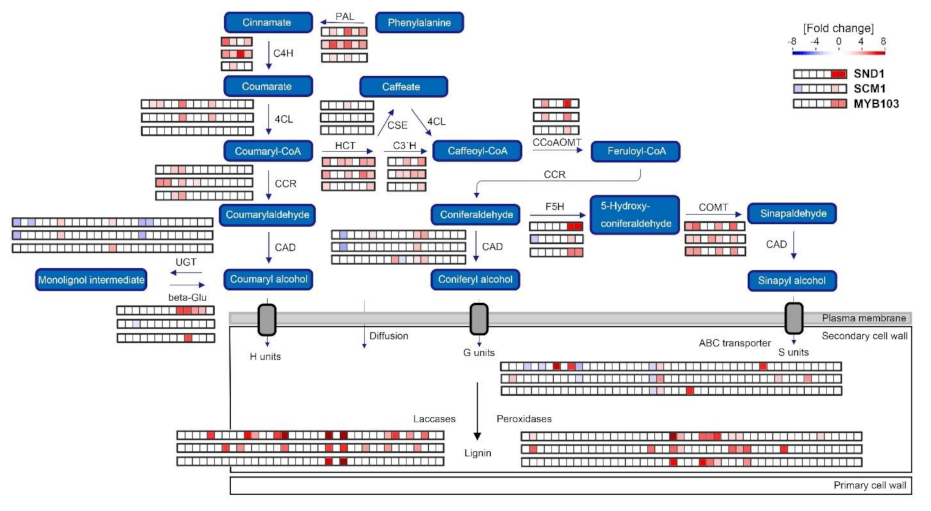
2.4. RNA-Seq Analysis Following Expression of MsSND1, MsSCM1, and MsMYB103 Delineates Diverging Transcriptional Landscapes
2.5. Expression Atlas Highlights Essential Genes of Lignin Biosynthesis
3. Materials and Methods
3.1. Plasmid Construction
3.2. Gene Expression Analysis
3.3. Nicotiana benthamiana Infiltration
3.4. Tissue Staining and Microscopy
3.5. Cell Wall Analysis
3.6. Transcriptome Analysis
3.7. Accession Numbers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pauly, M.; Keegstra, K. Plant Cell Wall Polymers as Precursors for Biofuels. Curr. Opin. Plant Biol. 2010, 13, 304–311. [Google Scholar] [CrossRef]
- Himmel, M.E.; Ding, S.-Y.; Johnson, D.K.; Adney, W.S.; Nimlos, M.R.; Brady, J.W.; Foust, T.D. Biomass Recalcitrance: Engineering Plants and Enzymes for Biofuels Production. Science 2007, 315, 804–807. [Google Scholar] [CrossRef] [PubMed]
- Bonawitz, N.D.; Chapple, C. Can Genetic Engineering of Lignin Deposition Be Accomplished without an Unacceptable Yield Penalty? Curr. Opin. Biotechnol. 2013, 24, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Mottiar, Y.; Vanholme, R.; Boerjan, W.; Ralph, J.; Mansfield, S.D. Designer Lignins: Harnessing the Plasticity of Lignification. Curr. Opin. Biotechnol. 2016, 37, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Clifton-Brown, J.; Hastings, A.; Mos, M.; McCalmont, J.P.; Ashman, C.; Awty-Carroll, D.; Cerazy, J.; Chiang, Y.-C.; Cosentino, S.; Cracroft-Eley, W.; et al. Progress in Upscaling Miscanthus Biomass Production for the European Bio-Economy with Seed-Based Hybrids. GCB Bioenergy 2017, 9, 6–17. [Google Scholar] [CrossRef]
- da Costa, R.M.F.; Pattathil, S.; Avci, U.; Lee, S.J.; Hazen, S.P.; Winters, A.; Hahn, M.G.; Bosch, M. A Cell Wall Reference Profile for Miscanthus Bioenergy Crops Highlights Compositional and Structural Variations Associated with Development and Organ Origin. New Phytol. 2017, 213, 1710–1725. [Google Scholar] [CrossRef] [PubMed]
- Barling, A.; Swaminathan, K.; Mitros, T.; James, B.T.; Morris, J.; Ngamboma, O.; Hall, M.C.; Kirkpatrick, J.; Alabady, M.; Spence, A.K.; et al. A Detailed Gene Expression Study of the Miscanthus Genus Reveals Changes in the Transcriptome Associated with the Rejuvenation of Spring Rhizomes. BMC Genom. 2013, 14, 864. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Ye, Z.-H. Secondary Cell Walls: Biosynthesis, Patterned Deposition and Transcriptional Regulation. Plant Cell Physiol. 2015, 56, 195–214. [Google Scholar] [CrossRef] [PubMed]
- Houston, K.; Tucker, M.R.; Chowdhury, J.; Shirley, N.; Little, A. The Plant Cell Wall: A Complex and Dynamic Structure As Revealed by the Responses of Genes under Stress Conditions. Front. Plant Sci. 2016, 7, 984. [Google Scholar] [CrossRef] [PubMed]
- Taylor-Teeples, M.; Lin, L.; de Lucas, M.; Turco, G.; Toal, T.W.; Gaudinier, A.; Young, N.F.; Trabucco, G.M.; Veling, M.T.; Lamothe, R.; et al. An Arabidopsis Gene Regulatory Network for Secondary Cell Wall Synthesis. Nature 2015, 517, 571–575. [Google Scholar] [CrossRef]
- Nakano, Y.; Yamaguchi, M.; Endo, H.; Rejab, N.A.; Ohtani, M. NAC-MYB-Based Transcriptional Regulation of Secondary Cell Wall Biosynthesis in Land Plants. Front. Plant Sci. 2015, 6, 288. [Google Scholar] [CrossRef]
- Zhong, R.; Demura, T.; Ye, Z.-H. SND1, a NAC Domain Transcription Factor, Is a Key Regulator of Secondary Wall Synthesis in Fibers of Arabidopsis. Plant Cell 2006, 18, 3158–3170. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Richardson, E.A.; Ye, Z.-H. Two NAC Domain Transcription Factors, SND1 and NST1, Function Redundantly in Regulation of Secondary Wall Synthesis in Fibers of Arabidopsis. Planta 2007, 225, 1603–1611. [Google Scholar] [CrossRef]
- Mitsuda, N.; Iwase, A.; Yamamoto, H.; Yoshida, M.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. NAC Transcription Factors, NST1 and NST3, Are Key Regulators of the Formation of Secondary Walls in Woody Tissues of Arabidopsis. Plant Cell 2007, 19, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Kubo, M.; Udagawa, M.; Nishikubo, N.; Horiguchi, G.; Yamaguchi, M.; Ito, J.; Mimura, T.; Fukuda, H.; Demura, T. Transcription Switches for Protoxylem and Metaxylem Vessel Formation. Genes Dev. 2005, 19, 1855–1860. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, R.L.; Zhong, R.; Ye, Z.-H. MYB83 Is a Direct Target of SND1 and Acts Redundantly with MYB46 in the Regulation of Secondary Cell Wall Biosynthesis in Arabidopsis. Plant Cell Physiol. 2009, 50, 1950–1964. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Richardson, E.A.; Ye, Z.-H. The MYB46 Transcription Factor Is a Direct Target of SND1 and Regulates Secondary Wall Biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2776–2792. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Lee, C.; Zhou, J.; McCarthy, R.L.; Ye, Z.-H. A Battery of Transcription Factors Involved in the Regulation of Secondary Cell Wall Biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef]
- Zhou, J.; Lee, C.; Zhong, R.; Ye, Z.-H. MYB58 and MYB63 Are Transcriptional Activators of the Lignin Biosynthetic Pathway during Secondary Cell Wall Formation in Arabidopsis. Plant Cell 2009, 21, 248–266. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Dixon, R. A Transcriptional Networks for Lignin Biosynthesis: More Complex than We Thought? Trends Plant Sci. 2011, 16, 227–233. [Google Scholar] [CrossRef]
- Noda, S.; Koshiba, T.; Hattori, T.; Yamaguchi, M.; Suzuki, S.; Umezawa, T. The Expression of a Rice Secondary Wall-Specific Cellulose Synthase Gene, OsCesA7, Is Directly Regulated by a Rice Transcription Factor, OsMYB58/63. Planta 2015, 242, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Scully, E.D.; Gries, T.; Sarath, G.; Palmer, N.A.; Baird, L.; Serapiglia, M.J.; Dien, B.S.; Boateng, A.A.; Ge, Z.; Funnell-Harris, D.L.; et al. Overexpression of SbMyb60 Impacts Phenylpropanoid Biosynthesis and Alters Secondary Cell Wall Composition in Sorghum Bicolor. Plant J. 2016, 85, 378–395. [Google Scholar] [CrossRef] [PubMed]
- Öhman, D.; Demedts, B.; Kumar, M.; Gerber, L.; Gorzsás, A.; Goeminne, G.; Hedenström, M.; Ellis, B.; Boerjan, W.; Sundberg, B. MYB103 Is Required for FERULATE-5-HYDROXYLASE Expression and Syringyl Lignin Biosynthesis in Arabidopsis Stems. Plant J. 2013, 73, 63–76. [Google Scholar] [CrossRef]
- Yang, C.; Li, D.; Liu, X.; Ji, C.; Hao, L.; Zhao, X.; Li, X.; Chen, C.; Cheng, Z.; Zhu, L. OsMYB103L, an R2R3-MYB Transcription Factor, Influences Leaf Rolling and Mechanical Strength in Rice (Oryza Sativa L.). BMC Plant Biol. 2014, 14, 158. [Google Scholar] [CrossRef]
- Hirano, K.; Kondo, M.; Aya, K.; Miyao, A.; Sato, Y.; Antonio, B.A.; Namiki, N.; Nagamura, Y.; Matsuoka, M. Identification of Transcription Factors Involved in Rice Secondary Cell Wall Formation. Plant Cell Physiol. 2013, 54, 1791–1802. [Google Scholar] [CrossRef]
- Ye, Y.; Liu, B.; Zhao, M.; Wu, K.; Cheng, W.; Chen, X.; Liu, Q.; Liu, Z.; Fu, X.; Wu, Y. CEF1/OsMYB103L Is Involved in GA-Mediated Regulation of Secondary Wall Biosynthesis in Rice. Plant Mol. Biol. 2015, 89, 385–401. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Dixon, R.A. Current Models for Transcriptional Regulation of Secondary Cell Wall Biosynthesis in Grasses. Front. Plant Sci. 2018, 9, 399. [Google Scholar] [CrossRef]
- Rabinowicz, P.D.; Braun, E.L.; Wolfe, A.D.; Bowen, B.; Grotewold, E. Maize R2R3 Myb Genes: Sequence Analysis Reveals Amplification in the Higher Plants. Genetics 1999, 153, 427–444. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Bartley, L.E. Comparative Genomic Analysis of the R2R3 MYB Secondary Cell Wall Regulators of Arabidopsis, Poplar, Rice, Maize, and Switchgrass. BMC Plant Biol. 2014, 14, 135. [Google Scholar] [CrossRef]
- Golfier, P.; Volkert, C.; He, F.; Rausch, T.; Wolf, S. Regulation of Secondary Cell Wall Biosynthesis by a NAC Transcription Factor from Miscanthus. Plant Direct 2017, 1, e00024. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Czedik-Eysenberg, A.; Mertz, R.A.; Si, Y.; Tohge, T.; Nunes-Nesi, A.; Arrivault, S.; Dedow, L.K.; Bryant, D.W.; Zhou, W.; et al. Comparative Analyses of C4 and C3 Photosynthesis in Developing Leaves of Maize and Rice. Nat. Biotechnol. 2014, 32, 1158–1165. [Google Scholar] [CrossRef]
- Li, P.; Ponnala, L.; Gandotra, N.; Wang, L.; Si, Y.; Tausta, S.L.; Kebrom, T.H.; Provart, N.; Patel, R.; Myers, C.R.; et al. The Developmental Dynamics of the Maize Leaf Transcriptome. Nat. Genet. 2010, 42, 1060–1067. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.G.; Owen, T.P.; Koonce, L.T.; Haigler, C.H. Dispersed Lignin in Tracheary Elements Treated with Cellulose Synthesis Inhibitors Provides Evidence That Molecules of the Secondary Cell Wall Mediate Wall Patterning. Plant J. 1992, 2, 959–970. [Google Scholar] [CrossRef]
- Smith, R.A.; Schuetz, M.; Roach, M.; Mansfield, S.D.; Ellis, B.; Samuels, L. Neighboring Parenchyma Cells Contribute to Arabidopsis Xylem Lignification, While Lignification of Interfascicular Fibers Is Cell Autonomous. Plant Cell 2013, 25, 3988–3999. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Chen, X.; Shen, H.; Ma, Q.; Li, G.; Tang, Y.; Pena, M.; York, W.; Frazier, T.P.; Lenaghan, S.; et al. Gene Regulatory Networks for Lignin Biosynthesis in Switchgrass (Panicum Virgatum). Plant Biotechnol. J. 2019, 17, 580–593. [Google Scholar] [CrossRef] [PubMed]
- Bond, D.M.; Albert, N.W.; Lee, R.H.; Gillard, G.B.; Brown, C.M.; Hellens, R.P.; Macknight, R.C. Infiltration-RNAseq: TRANSCRIPTOME Profiling of Agrobacterium-Mediated Infiltration of Transcription Factors to Discover Gene Function and Expression Networks in Plants. Plant Methods 2016, 12, 41. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fernández-Pérez, F.; Pomar, F.; Pedreño, M.A.; Novo-Uzal, E. Suppression of Arabidopsis Peroxidase 72 Alters Cell Wall and Phenylpropanoid Metabolism. Plant Sci. 2015, 239, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Voxeur, A.; Wang, Y.; Sibout, R. Lignification: Different Mechanisms for a Versatile Polymer. Curr. Opin. Plant Biol. 2015, 23, 83–90. [Google Scholar] [CrossRef]
- Berthet, S.; Demont-Caulet, N.; Pollet, B.; Bidzinski, P.; Cezard, L.; Le Bris, P.; Borrega, N.; Herve, J.; Blondet, E.; Balzergue, S.; et al. Disruption of LACCASE4 and 17 Results in Tissue-Specific Alterations to Lignification of Arabidopsis Thaliana Stems. Plant Cell 2011, 23, 1124–1137. [Google Scholar] [CrossRef]
- Kourelis, J.; Kaschani, F.; Grosse-Holz, F.M.; Homma, F.; Kaiser, M.; Hoorn, R.A.L. van der Re-Annotated Nicotiana Benthamiana Gene Models for Enhanced Proteomics and Reverse Genetics. bioRxiv 2018, 9, 373506. [Google Scholar] [CrossRef]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. Mapman: A User-Driven Tool to Display Genomics Data Sets onto Diagrams of Metabolic Pathways and Other Biological Processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- Lampropoulos, A.; Sutikovic, Z.; Wenzl, C.; Maegele, I.; Lohmann, J.U.; Forner, J. GreenGate—A Novel, Versatile, and Efficient Cloning System for Plant Transgenesis. PLoS ONE 2013, 8, e83043. [Google Scholar] [CrossRef]
- Clifton-Brown, J.C.; Lewandowski, I. Screening Miscanthus Genotypes in Field Trials to Optimise Biomass Yield and Quality in Southern Germany. Eur. J. Agron. 2002, 16, 97–110. [Google Scholar] [CrossRef]
- Merai, Z.; Kerenyi, Z.; Molnar, A.; Barta, E.; Valoczi, A.; Bisztray, G.; Havelda, Z.; Burgyan, J.; Silhavy, D. Aureusvirus P14 Is an Efficient RNA Silencing Suppressor That Binds Double-Stranded RNAs without Size Specificity. J. Virol. 2005, 79, 7217–7226. [Google Scholar] [CrossRef]
- Coleman, H.D.; Park, J.-Y.; Nair, R.; Chapple, C.; Mansfield, S.D. RNAi-Mediated Suppression of p-Coumaroyl-CoA 3′-Hydroxylase in Hybrid Poplar Impacts Lignin Deposition and Soluble Secondary Metabolism. Proc. Natl. Acad. Sci. USA 2008, 105, 4501–4506. [Google Scholar] [CrossRef] [PubMed]
- Robinson, A.R.; Mansfield, S.D. Rapid Analysis of Poplar Lignin Monomer Composition by a Streamlined Thioacidolysis Procedure and Near-Infrared Reflectance-Based Prediction Modeling. Plant J. 2009, 58, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Golfier, P.; Ermakova, O.; Unda, F.; Murphy, E.K.; Xie, J.; He, F.; Zhang, W.; Lohmann, J.U.; Mansfield, S.D.; Rausch, T.; et al. Distinct and Overlapping Functions of Miscanthus sinensis MYB Transcription Factors SCM1 and MYB103 in Lignin Biosynthesis. Int. J. Mol. Sci. 2021, 22, 12395. https://doi.org/10.3390/ijms222212395
Golfier P, Ermakova O, Unda F, Murphy EK, Xie J, He F, Zhang W, Lohmann JU, Mansfield SD, Rausch T, et al. Distinct and Overlapping Functions of Miscanthus sinensis MYB Transcription Factors SCM1 and MYB103 in Lignin Biosynthesis. International Journal of Molecular Sciences. 2021; 22(22):12395. https://doi.org/10.3390/ijms222212395
Chicago/Turabian StyleGolfier, Philippe, Olga Ermakova, Faride Unda, Emily K. Murphy, Jianbo Xie, Feng He, Wan Zhang, Jan U. Lohmann, Shawn D. Mansfield, Thomas Rausch, and et al. 2021. "Distinct and Overlapping Functions of Miscanthus sinensis MYB Transcription Factors SCM1 and MYB103 in Lignin Biosynthesis" International Journal of Molecular Sciences 22, no. 22: 12395. https://doi.org/10.3390/ijms222212395
APA StyleGolfier, P., Ermakova, O., Unda, F., Murphy, E. K., Xie, J., He, F., Zhang, W., Lohmann, J. U., Mansfield, S. D., Rausch, T., & Wolf, S. (2021). Distinct and Overlapping Functions of Miscanthus sinensis MYB Transcription Factors SCM1 and MYB103 in Lignin Biosynthesis. International Journal of Molecular Sciences, 22(22), 12395. https://doi.org/10.3390/ijms222212395






