Unravel the Local Complexity of Biological Environments by MALDI Mass Spectrometry Imaging
Abstract
:1. Introduction
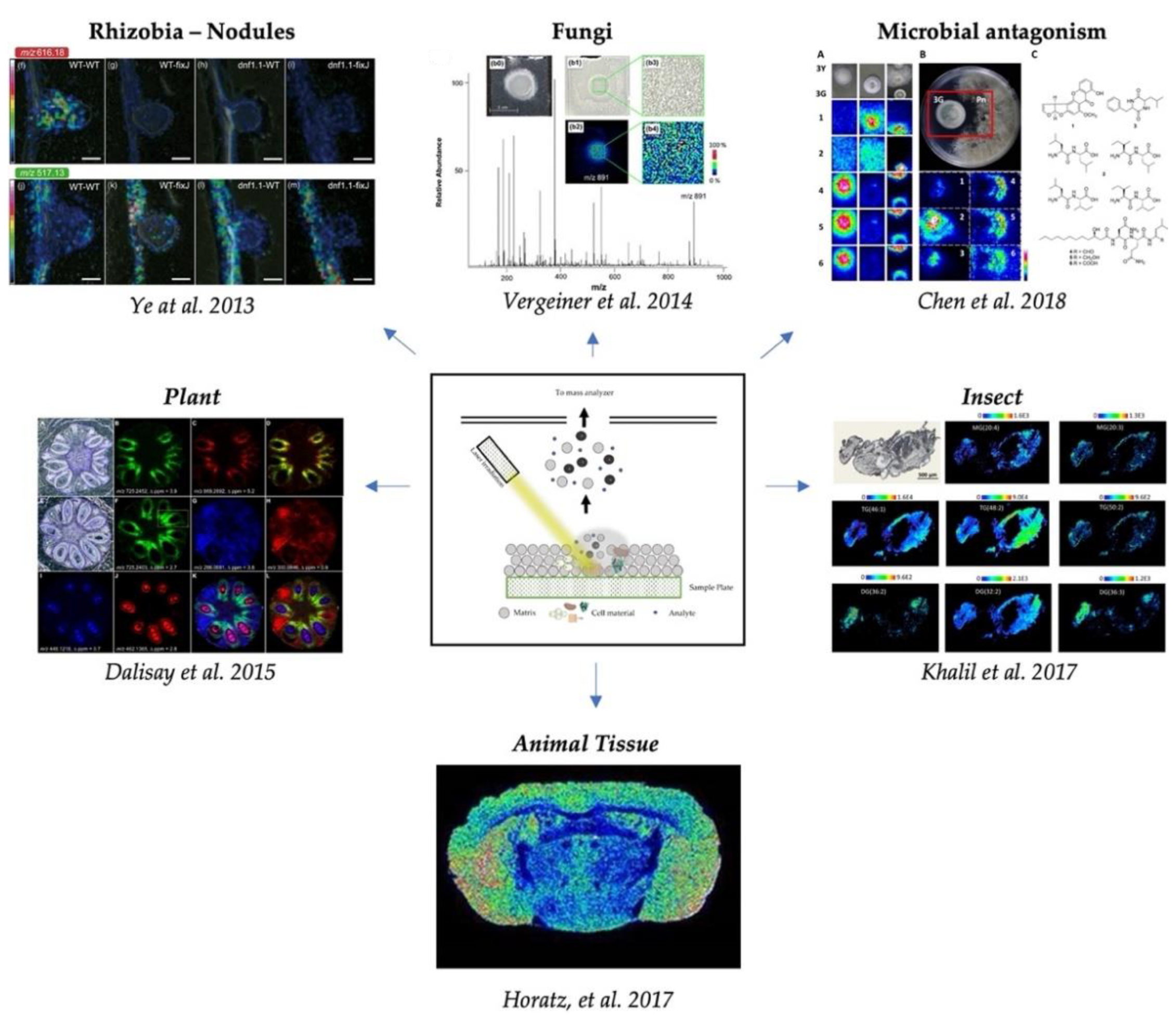
2. Metabolites Associated with Microbial Interactions and Antagonism
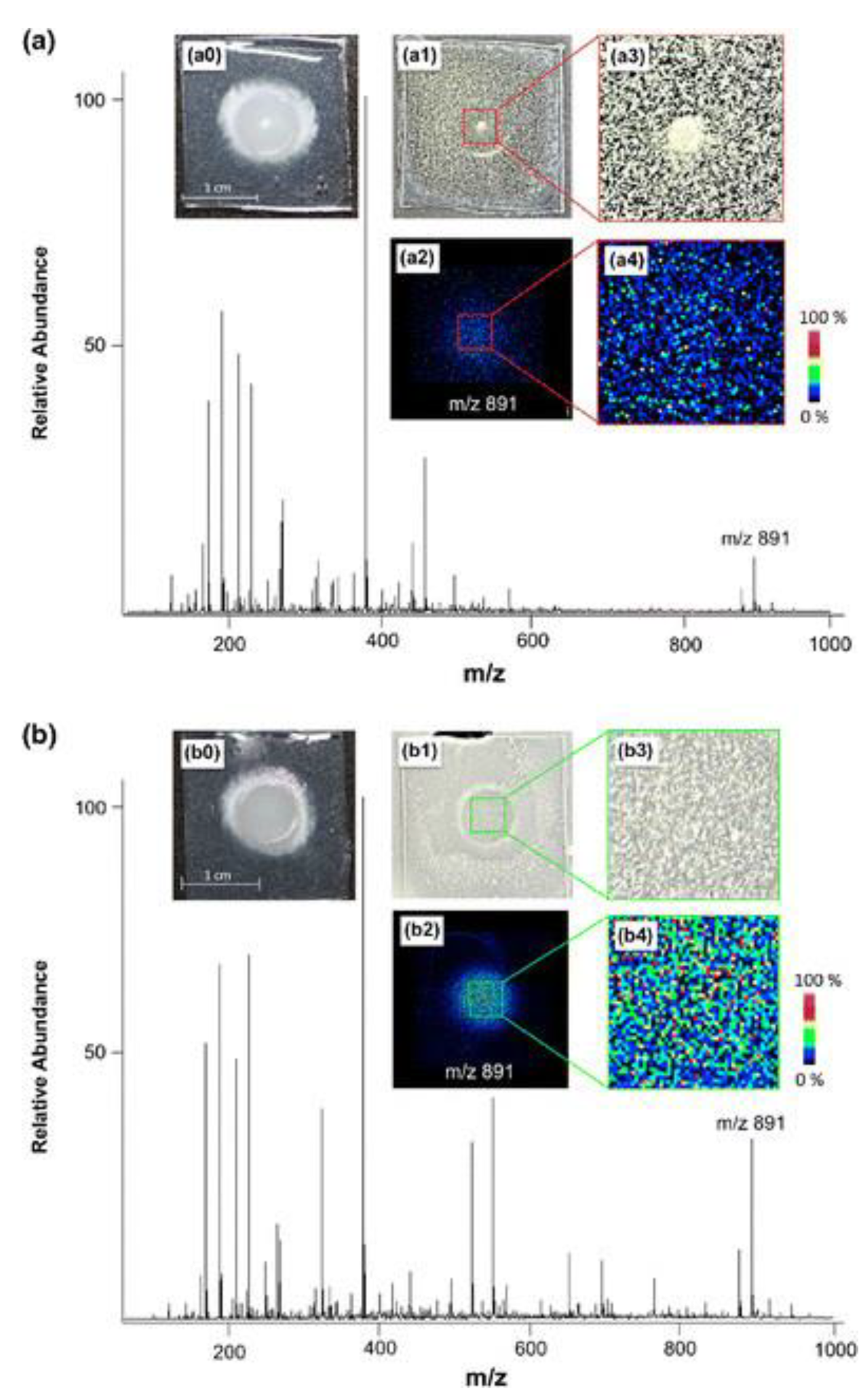
3. Spatial Localization of Fungal Metabolites
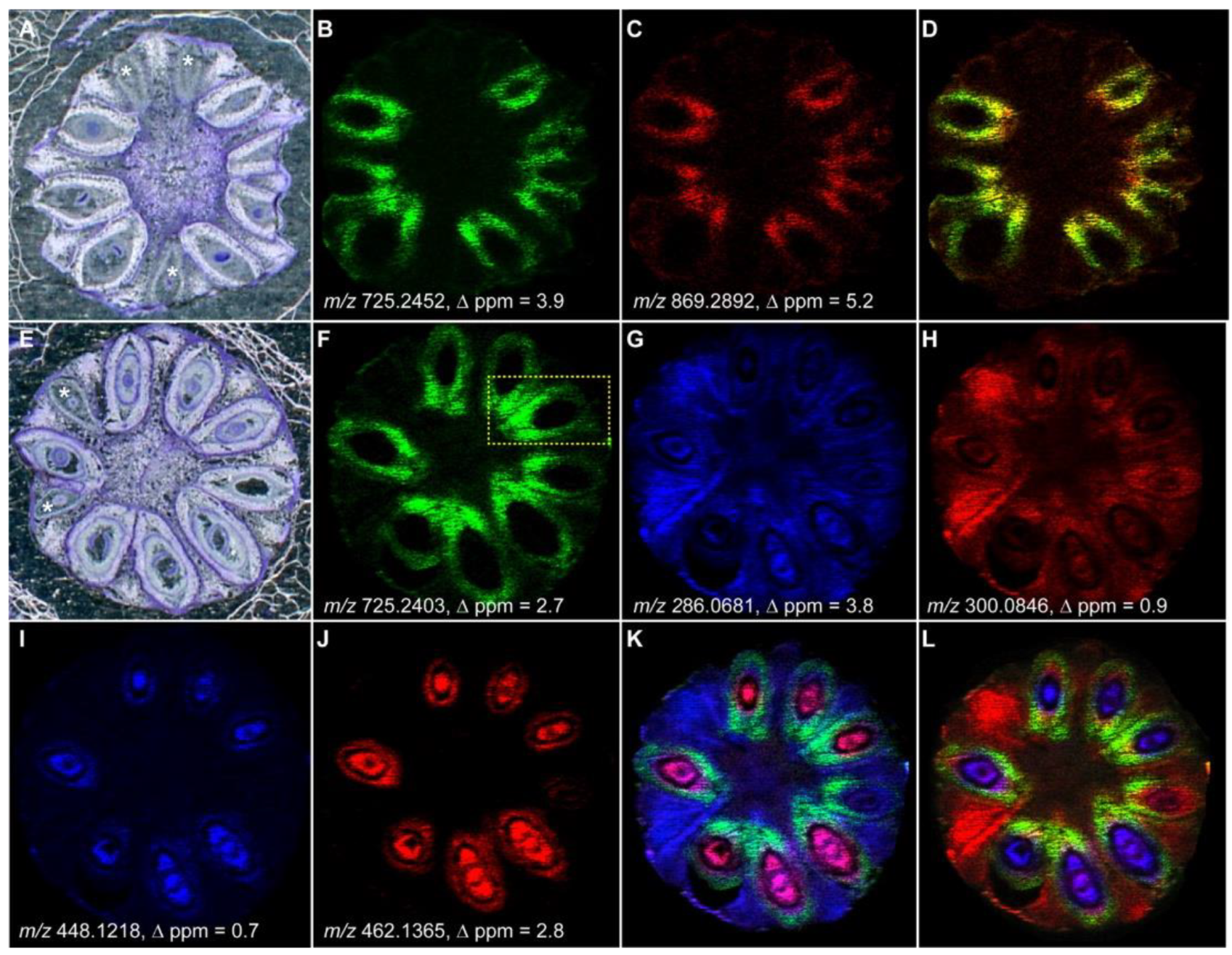
4. Spatial Localization of Plant Metabolites
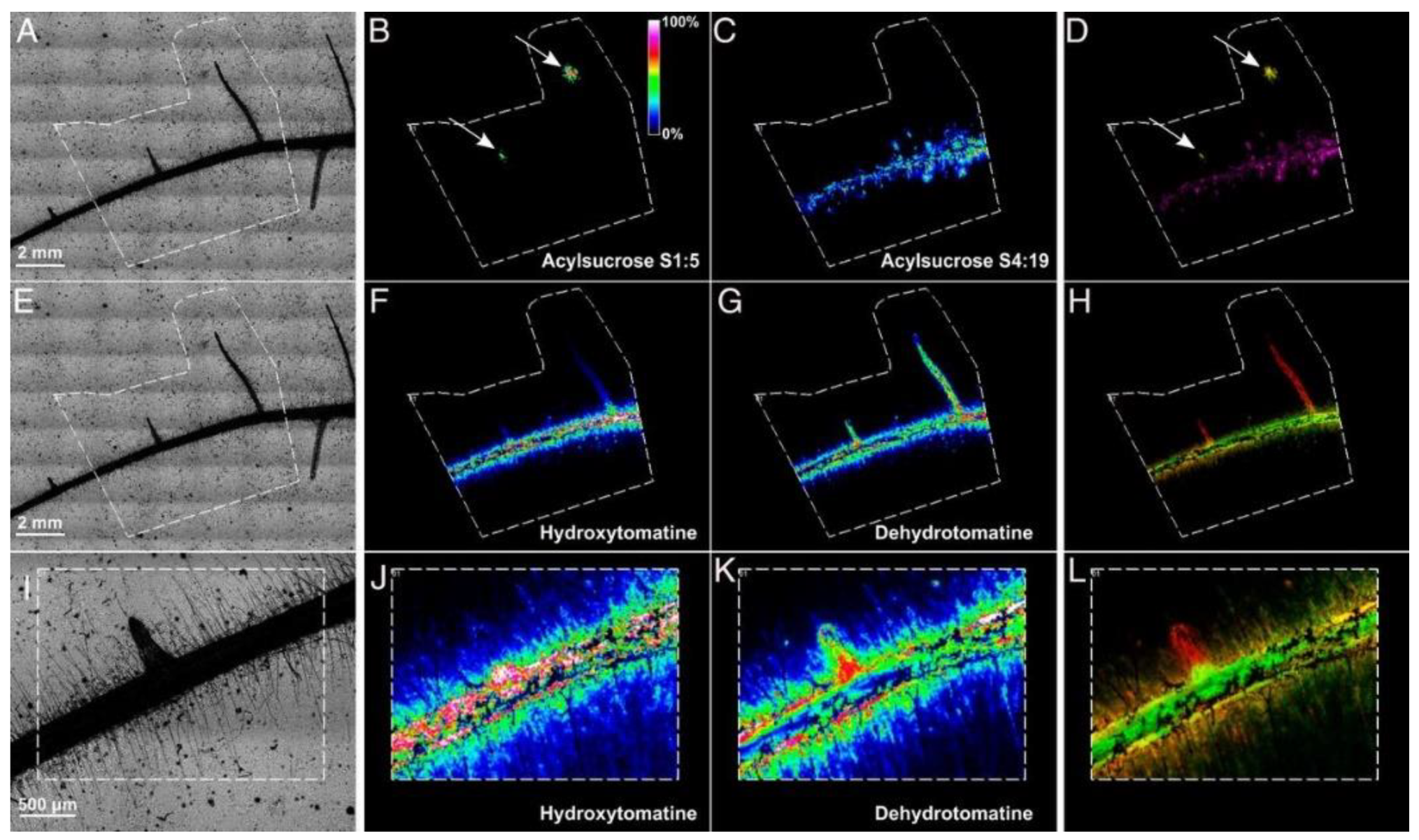
5. The Molecular Complexity of Plant–Microbe Interactions
6. Molecular Imaging in Insects, Nematodes, and Worms
7. Animal Tissues as Exemplary for Molecular Imaging: Brain, Testicles, and Kidney
8. Conclusions
| Sample | Compartment | Class of Compound | Matrix | Technique | Reference |
|---|---|---|---|---|---|
| Dried agar slices | Microbial extracellular interface | Siderophores | DHB/CHCA (1:1) | MALDI TOF MSI and ion mobility | [26,30,37] |
| Piece of medium placed on MALDI plate | Interface of fungal colonies | Indol alkaloids | DHB and DHB/CHCA (1:1) | MALDI TOF MSI | [34] |
| Agar piece placed on MALDI plate | Fungal mycelium | Polyketide | Nanostructured silicon | MALDI TOF MSI on nanosilicone | [25] |
| Ant thorax section on ITO slide | Ant exoskeleton: propleural plate | Ergothioneine | DHB | MALDI Orbitrap MSI | [32] |
| Agar section on MALDI plate | Microbial extracellular interface | Quinolones rhamnolipids | DHB/CHCA (1:1) | MALDI FT ICR MSI | [40,41] |
| Agar section on MALDI plate | Chlamydospores | Cyclo-lipopeptide | DHB/CHCA (1:1) | MALDI MSI | [43] |
| Cryosection of sclerotia | Sclerotia | Alkaloids | DHB | MALDI MSI (Synapt G2 with ion mobility) | [44] |
| Dried colonies on the slide | Inhibitory zone at the microbial interface | Solanapyrones, disaccharides, isoflavones, and isoflavonoids | DHB | MALDI FT ICR MSI | [46] |
| Agar cut on ITO slide | Inhibitory zone | lipopeptaibol | DHB/CHCA (1:1) | MALDI MSI | [48] |
| Surface tissue section on MALDI plate | Pea pods | Aromatic compound | DHB | MALDI MSI with ion mobility | [74] |
| Leaf imprints on porous PTFE sheet | Rice root | Diterpenoids | Fe3O4 nanoparticles | MALDI MSI | [60] |
| Root on MALDI plate with nylon membrane | Root | Organic acids | CHCA | MALDI MSI Synapt G1 | [61] |
| Lyophilized root on slide | Rhizosphere | Acyl sugars and steroidal glycoalkaloids | DHB | MALDI FT ICR MSI | [62] |
| PVDF membrane with roots on MALDI plate | Rhizosphere interface | Nucleobase, amino acid, and derivatives | DHB | MALDI FT ICR MSI | [63] |
| Root-colonized growth on ITO slide | Aerial roots/root biofilm coated | Cyclo-lipopeptides | CHCA | MALDI TOF MSI | [65,66] |
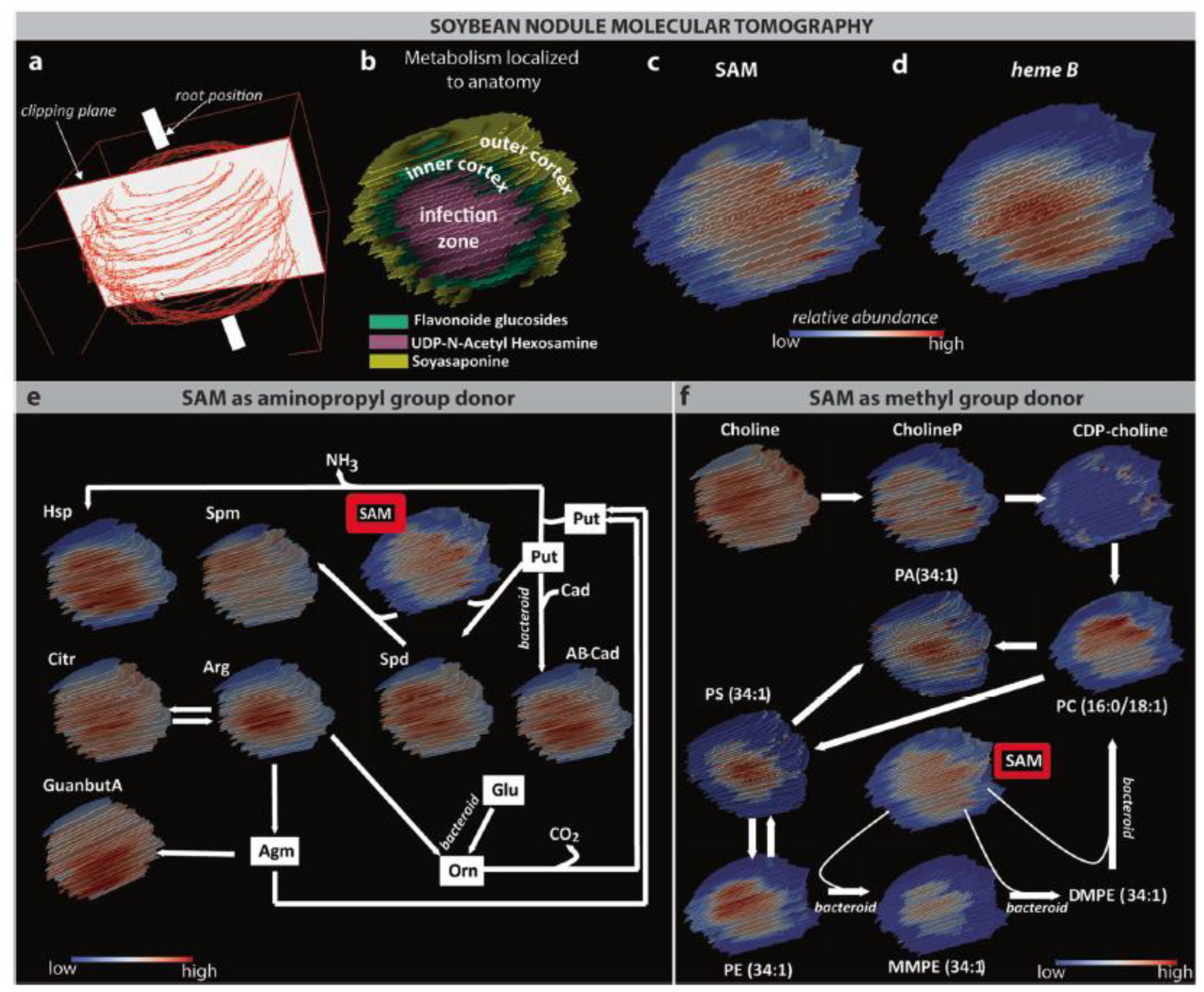
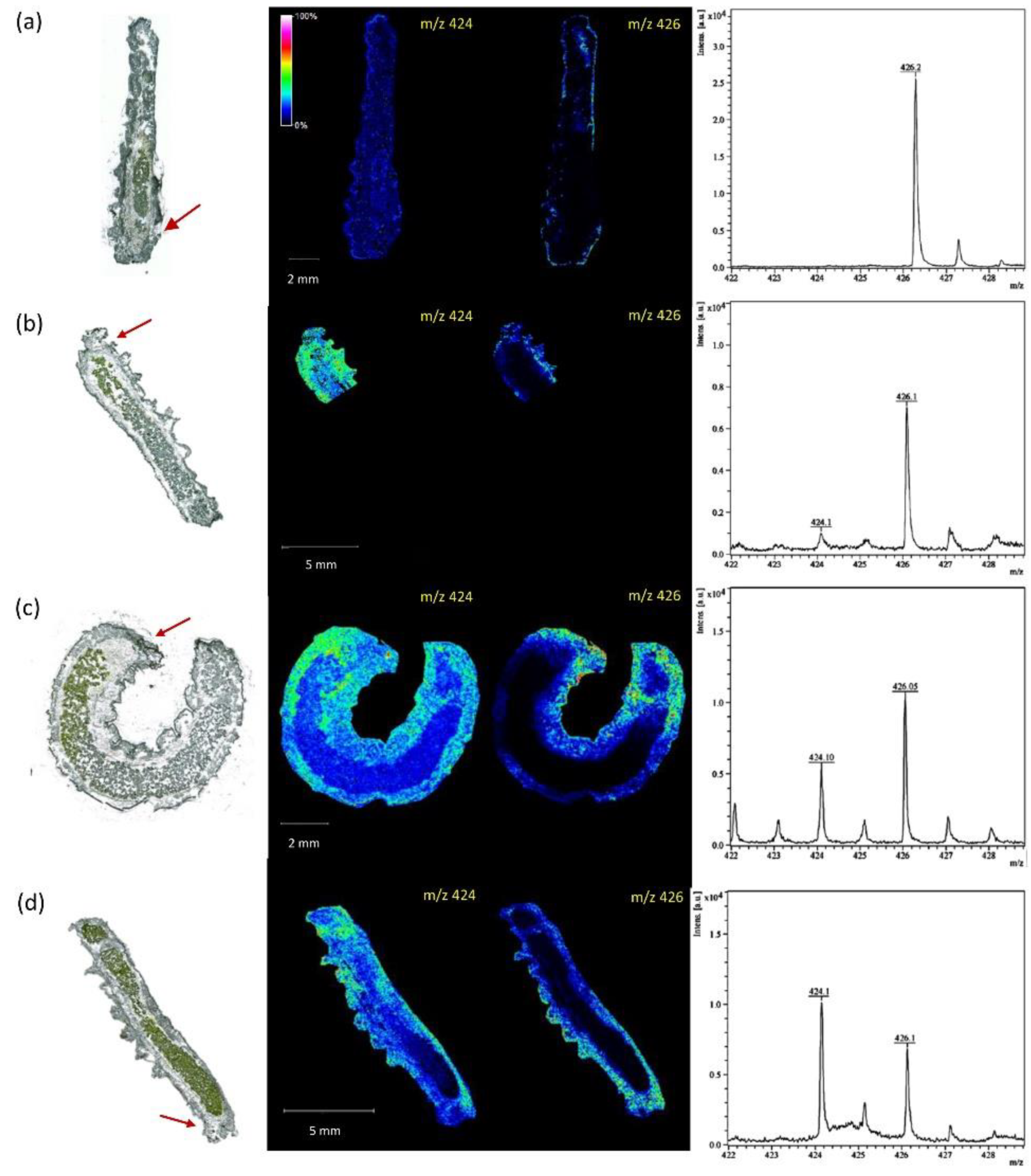
| Cryosection of root on ITO slide | Root nodule | Choline derivative, organic acid, and amino acids | Norharmane | 3D MALDI FTICR MSI | [67] |
| Cryosection of nodules on plain glass microscope slide | Nodule | Amino acid, triterpenoid, phospholipid, disaccharide, nucleobases, vitamins | DHB and CHCA | AP MALDI MSI Orbitrap | [70,71] |
| Tissue covered with aluminum foil and embedded in paraffin. Microtome sections on ITO slide | Stem, leaf | flavonoids | DHB/CHCA (1:1) | MALDI TOF MSI | [75] |
| Cryosection of nodules on glass microscope slide | Nodule | Peptide | DHB and CHCA | MALDI Orbitrap MSI | [69] |
| Cryosections on ITO slides | Leaf epidermises, vascular bundles, roots, petiole collenchyma, root | Cyclotides | DHB | MALDI TOF MSI | [72] |
| Leaf fixed with aluminized tape on MALDI plate | Leaf | Stilbenoid and polyphenol | DHB | MALDI TOF MSI | [73] |
| Cryosections | Brain | Neuropeptide and tryptic peptides | CHCA | MALDI TOF MSI | [76,81] |
| Cryosections on frosted glass slides | Fly genitals | Pheromones and lipids | DHB | MALDI Orbitrap MSI | [27] |
| Cryosection on ITO slides | Hemolymph | Glucosinolate | CHCA | MALDI TOF MSI | [82] |
| Cryosections on ITO slides | Worm | Amino acid, sugars, and polyamine | 2-MBT | MALDI TOF MSI | [89] |
| Cryosections | Rat brain | Phospholipids, neurotransmitters, nucleotides | DHB, nanostructure initiator, DAN, BNDM, and norharmane | MALDI TOF MSI MALDI Orbitrap MSI MALDI Orbitrap with ozonolysis | [92,93,94,95,100,102] |
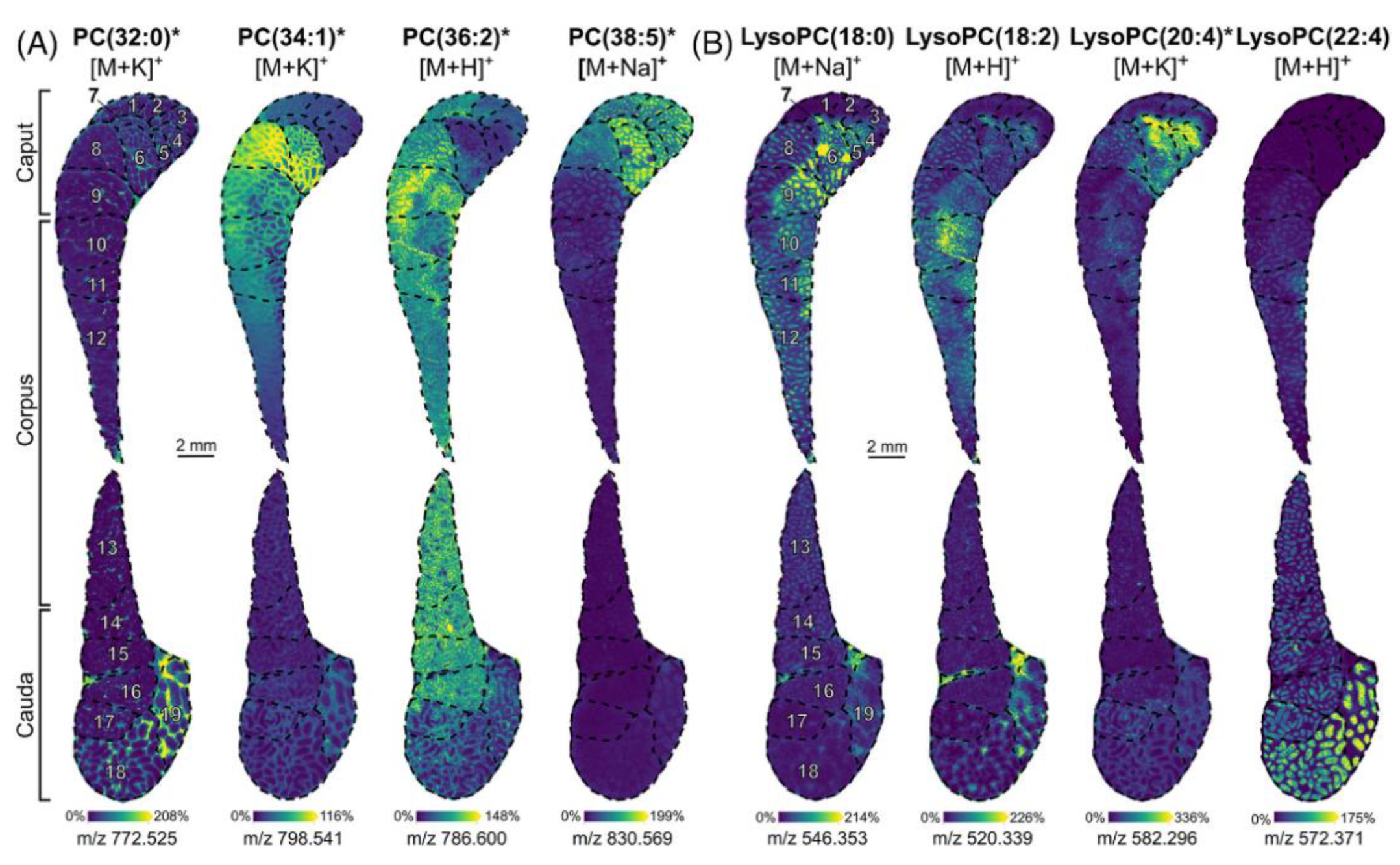
| Cryosections | Rat testicles | Seminolipids, phospholipids, and steroids | CHCA, DHB, and AA. Derivatization for steroids. | MALDI TOF MSI MALDI FT ICR MSI iMScope QT | [105,106,108,109,110] |
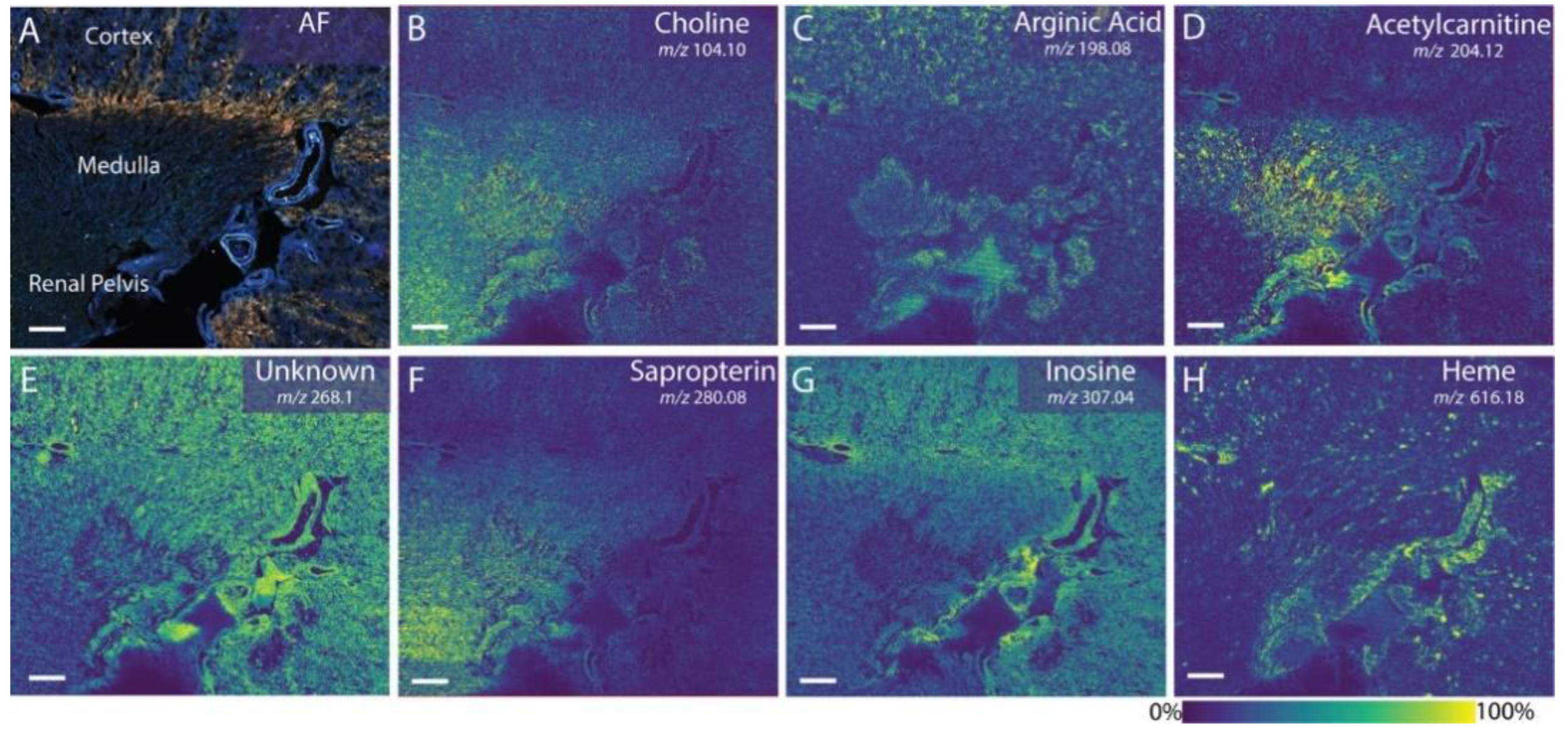
| Cryosections | Rat kidney | Phospholipids and small-molecular-weight metabolites | DHB, benzoylpyridine CHCA, and DAN | AP-MALDI Orbitrap MSI MALDI timsTOF MSI MALDI TOF MSI | [112,113,114] |
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Megason, S.G.; Fraser, S.E. Imaging in Systems Biology. Cell 2007, 130, 784–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kherlopian, A.R.; Song, T.; Duan, Q.; Neimark, M.A.; Po, M.J.; Gohagan, J.K.; Laine, A.F. A review of imaging techniques for systems biology. BMC Syst. Biol. 2008, 2, 74. [Google Scholar] [CrossRef] [Green Version]
- Kamweru, P.K. Imaging from anatomic to molecular and atomic resolution scales: A review. Afr. J. Biotechnol. 2021, 20, 266–274. [Google Scholar]
- Spraker, J.E.; Luu, G.T.; Sanchez, L.M. Imaging mass spectrometry for natural products discovery: A review of ionization methods. Nat. Prod. Rep. 2020, 37, 150–162. [Google Scholar] [CrossRef]
- Dilillo, M.; Pellegrini, D.; Ait-Belkacem, R.; de Graaf, E.L.; Caleo, M.; McDonnell, L.A. Mass Spectrometry Imaging, Laser Capture Microdissection, and LC-MS/MS of the Same Tissue Section. J. Proteome Res. 2017, 16, 2993–3001. [Google Scholar] [CrossRef] [PubMed]
- Breuker, K.; Knochenmuss, R.; Zenobi, R. Gas-phase basicities of deprotonated matrix-assisted laser desorption/ionization matrix molecules. Int. J. Mass Spectrom. 1999, 184, 25–38. [Google Scholar] [CrossRef]
- Knochenmuss, R. Ion formation mechanisms in UV-MALDI. Analyst 2006, 131, 966–986. [Google Scholar] [CrossRef]
- Karas, M.; Glückmann, M.; Schäfer, J. Ionization in Matrix-Assisted Laser Desorption/Ionization: Singly Charged Molecular Ions Are the Lucky Survivors. J. Mass Spectrom. 2000, 35, 1–12. [Google Scholar] [CrossRef]
- Knochenmuss, R. The Coupled Chemical and Physical Dynamics Model of MALDI. Annu. Rev. Anal. Chem. 2016, 9, 365–385. [Google Scholar] [CrossRef]
- McEwen, C.N.; Inutan, E.D.; Moreno-Pedraza, A.; Lu, I.C.; Hoang, K.; Pophristic, M.; Trimpin, S. Sublimation Driven Ionization for Use in Mass Spectrometry: Mechanistic Implications. J. Am. Soc. Mass Spectrom. 2021, 32, 114–123. [Google Scholar] [CrossRef]
- Giraldo-Dávila, D.; Chacón-Patiño, M.L.; Ramirez-Pradilla, J.S.; Blanco-Tirado, C.; Combariza, M.Y. Selective Ionization by Electron-Transfer MALDI-MS of Vanadyl Porphyrins from Crude Oils. Fuel 2018, 226, 103–111. [Google Scholar] [CrossRef]
- Knochenmuss, R.; Stortelder, A.; Breuker, K.; Zenobi, R. Secondary ion-molecule reactions in matrix-assisted laser desorption/ionization. J. Mass Spectrom. 2000, 35, 1237–1245. [Google Scholar] [CrossRef]
- Zhou, Q.; Fülöp, A.; Hopf, C. Recent developments of novel matrices and on-tissue chemical derivatization reagents for MALDI-MSI. Anal. Bioanal. Chem. 2020, 413, 2599–2617. [Google Scholar] [CrossRef] [PubMed]
- Corinti, D.; Crestoni, M.E.; Fornarini, S.; Pieper, M.; Niehaus, K.; Giampà, M. An integrated approach to study novel properties of a MALDI matrix (4-maleicanhydridoproton sponge) for MS imaging analyses. Anal. Bioanal. Chem. 2019, 411, 953–964. [Google Scholar] [CrossRef] [Green Version]
- Horatz, K.; Giampà, M.; Qiao, Z.; Moestue, S.A.; Lissel, F. Polymerization as a Strategy to Improve Small Organic Matrices for Low-Molecular-Weight Compound Analytics with MALDI MS and MALDI MS Imaging. ACS Appl. Polym. Mater. 2021, 3, 4234–4244. [Google Scholar] [CrossRef]
- Spengler, B.; Hubert, M. Scanning microprobe matrix-assisted laser desorption ionization (SMALDI) mass spectrometry: Instrumentation for sub-micrometer resolved LDI and MALDI surface analysis. J. Am. Soc. Mass Spectrom. 2002, 13, 735–748. [Google Scholar] [CrossRef] [Green Version]
- McDonnell, L.A.; Heeren, R.M.A. Imaging mass spectrometry. Mass Spectrom. Rev. 2007, 26, 606–643. [Google Scholar] [CrossRef] [Green Version]
- Ščupáková, K.; Balluff, B.; Tressler, C.; Adelaja, T.; Heeren, R.M.A.; Glunde, K.; Ertaylan, G. Cellular resolution in clinical MALDI mass spectrometry imaging: The latest advancements and current challenges. Clin. Chem. Lab. Med. 2020, 58, 914–929. [Google Scholar] [CrossRef]
- Horatz, K.; Ditte, K.; Prenveille, T.; Zhang, K.-N.; Jehnichen, D.; Kiriy, A.; Voit, B.; Lissel, F. Cover Feature: Amorphous Conjugated Polymers as Efficient Dual-Mode MALDI Matrices for Low-Molecular-Weight Analytes. ChemPlusChem 2019, 84, 1171. [Google Scholar] [CrossRef] [Green Version]
- Giampà, M.; Lissel, M.B.; Patschkowski, T.; Fuchser, J.; Hans, V.H.; Gembruch, O.; Bednarz, H.; Niehaus, K. Maleic Anhydride Proton Sponge as a Novel MALDI Matrix for the Visualization of Small Molecules (<250 m/z) in Brain Tumors by Routine MALDI ToF Imaging Mass Spectrometry. Chem. Commun. 2016, 52, 9801–9804. [Google Scholar] [CrossRef]
- Yuan, M.; Breitkopf, S.B.; Yang, X.; Asara, J.M. A Positive/Negative Ion–Switching, Targeted Mass Spectrometry–Based Metabolomics Platform for Bodily Fluids, Cells, and Fresh and Fixed Tissue. Nat. Protoc. 2012, 7, 872–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buszewska-Forajta, M.; Patejko, M.; Macioszek, S.; Sigorski, D.; Iżycka-Świeszewska, E.; Markuszewski, M.J. Paraffin-Embedded Tissue as a Novel Matrix in Metabolomics Study: Optimization of Metabolite Extraction Method. Chromatographia 2019, 82, 1501–1513. [Google Scholar] [CrossRef] [Green Version]
- Ye, H.; Gemperline, E.; Venkateshwaran, M.; Chen, R.; Delaux, P.-M.; Howes-Podoll, M.; Ané, J.-M.; Li, L. MALDI mass spectrometry-assisted molecular imaging of metabolites during nitrogen fixation in the Medicago truncatula–Sinorhizobium meliloti symbiosis. Plant J. 2013, 75, 130–145. [Google Scholar] [CrossRef] [PubMed]
- Dalisay, D.S.; Kim, K.W.; Lee, C.; Yang, H.; Rübel, O.; Bowen, B.P.; Davin, L.B.; Lewis, N.G. Dirigent Protein-Mediated Lignan and Cyanogenic Glucoside Formation in Flax Seed: Integrated Omics and MALDI Mass Spectrometry Imaging. J. Nat. Prod. 2015, 78, 1231–1242. [Google Scholar] [CrossRef]
- Chen, P.Y.; Hsieh, C.Y.; Shih, C.J.; Lin, Y.J.; Tsao, C.W.; Yang, Y.L. Exploration of Fungal Metabolic Interactions Using Imaging Mass Spectrometry on Nanostructured Silicon. J. Nat. Prod. 2018, 81, 1527–1533. [Google Scholar] [CrossRef]
- Vergeiner, S.; Schafferer, L.; Haas, H.; Müller, T. Improved MALDI-TOF microbial mass spectrometry imaging by application of a dispersed solid matrix. J. Am. Soc. Mass Spectrom. 2014, 25, 1498–1501. [Google Scholar] [CrossRef] [PubMed]
- Khalil, S.M.; Pretzel, J.; Becker, K.; Spengler, B. High-resolution AP-SMALDI mass spectrometry imaging of Drosophila melanogaster. Int. J. Mass Spectrom. 2017, 416, 1–19. [Google Scholar] [CrossRef]
- Horatz, K.; Giampà, M.; Karpov, Y.; Sahre, K.; Bednarz, H.; Kiriy, A.; Voit, B.; Niehaus, K.; Hadjichristidis, N.; Michels, D.L.; et al. Conjugated Polymers as a New Class of Dual-Mode Matrices for MALDI Mass Spectrometry and Imaging. J. Am. Chem. Soc. 2018, 140, 11416–11423. [Google Scholar] [CrossRef] [Green Version]
- Feichtmayer, J.; Deng, L.; Griebler, C. Antagonistic Microbial Interactions: Contributions and Potential Applications for Controlling Pathogens in the Aquatic Systems. Front. Microbiol. 2017, 8, 2192. [Google Scholar] [CrossRef]
- Boya, P.R.A.; Martin, H.C.; Fernandez-Marin, H.; Gutierrez, M. Fungus-Growing Ant’s Microbial Interaction of Streptomyces sp. and Escovopsis sp. through Molecular Networking and MALDI Imaging. Nat. Prod. Commun. 2019, 14, 63–66. [Google Scholar]
- Cheah, I.K.; Halliwell, B. Ergothioneine; antioxidant potential, physiological function and role in disease. Biochim. Biophys. Acta 2012, 1822, 784–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gemperline, E.; Horn, H.A.; DeLaney, K.; Currie, C.R.; Li, L. Imaging with Mass Spectrometry of Bacteria on the Exoskeleton of Fungus-Growing Ants. ACS Chem. Biol. 2017, 12, 1980–1985. [Google Scholar] [CrossRef]
- Torres, M.J.; Brandan, C.P.; Petroselli, G.; Erra-Balsells, R.; Audisio, M.C. Antagonistic effects of Bacillus subtilis subsp. subtilis and B. amyloliquefaciens against Macrophomina phaseolina: SEM study of fungal changes and UV-MALDI-TOF MS analysis of their bioactive compounds. Microbiol. Res. 2016, 182, 31–39. [Google Scholar] [CrossRef]
- Bai, J.; Zhang, P.; Bao, G.; Gu, J.-G.; Han, L.; Zhang, L.-W.; Xu, Y. Imaging mass spectrometry-guided fast identification of antifungal secondary metabolites from Penicillium polonicum. Appl. Microbiol. Biotechnol. 2018, 102, 8493–8500. [Google Scholar] [CrossRef]
- Chung, C.L.; Huang, S.Y.; Huang, Y.C.; Tzean, S.S.; Ann, P.J.; Tsai, J.N.; Yang, C.C.; Lee, H.H.; Huang, T.W.; Huang, H.Y.; et al. The Genetic Structure of Phellinus noxius and Dissemination Pattern of Brown Root Rot Disease in Taiwan. PLoS ONE 2015, 10, e0139445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raines, D.J.; Sanderson, T.J.; Wilde, E.J.; Duhme-Klair, A.K. Siderophores. In Reference Module in Chemistry, Molecular Sciences and Chemical Engineering; Elsevier: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Ho, Y.N.; Hoo, S.Y.; Wang, B.W.; Hsieh, C.T.; Lin, C.C.; Sun, C.H.; Peng, C.C.; Lin, C.; Yang, Y.L. Specific inactivation of an antifungal bacterial siderophore by a fungal plant pathogen. ISME J. 2021, 15, 1858–1861. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Perdian, D.C.; Song, Z.; Yeung, E.S.; Nikolau, B.J. Use of mass spectrometry for imaging metabolites in plants. Plant J. 2012, 70, 81–95. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, T.; Dorrestein, P.C. Homogeneous Matrix Deposition on Dried Agar for MALDI Imaging Mass Spectrometry of Microbial Cultures. J. Am. Soc. Mass Spectrom. 2015, 26, 1959–1962. [Google Scholar] [CrossRef] [Green Version]
- Moree, W.J.; Phelan, V.V.; Wu, C.-H.; Bandeira, N.; Cornett, D.S.; Duggan, B.M.; Dorrestein, P.C. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc. Natl. Acad. Sci. USA 2012, 109, 13811–13816. [Google Scholar] [CrossRef] [Green Version]
- Moree, W.J.; Yang, J.Y.; Zhao, X.; Liu, W.T.; Aparicio, M.; Atencio, L.; Ballesteros, J.; Sánchez, J.; Gavilán, R.G.; Gutiérrez, M.; et al. Imaging mass spectrometry of a coral microbe interaction with fungi. J. Chem. Ecol. 2013, 39, 1045–1054. [Google Scholar] [CrossRef] [Green Version]
- Soberón-Chávez, G.; Lépine, F.; Déziel, E. Production of rhamnolipids by Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2005, 68, 718–725. [Google Scholar] [CrossRef]
- Spraker, J.E.; Sanchez, L.M.; Lowe, T.M.; Dorrestein, P.C.; Keller, N.P. Ralstonia solanacearum lipopeptide induces chlamydospore development in fungi and facilitates bacterial entry into fungal tissues. ISME J. 2016, 10, 2317–2330. [Google Scholar] [CrossRef] [Green Version]
- Dopstadt, J.; Vens-Cappell, S.; Neubauer, L.; Tudzynski, P.; Cramer, B.; Dreisewerd, K.; Humpf, H.-U. Localization of ergot alkaloids in sclerotia of Claviceps purpurea by matrix-assisted laser desorption/ionization mass spectrometry imaging. Anal. Bioanal. Chem. 2017, 409, 1221–1230. [Google Scholar] [CrossRef]
- Kasahara, K.; Miyamoto, T.; Fujimoto, T.; Oguri, H.; Tokiwano, T.; Oikawa, H.; Ebizuka, Y.; Fujii, I. Solanapyrone synthase, a possible Diels-Alderase and iterative type I polyketide synthase encoded in a biosynthetic gene cluster from Alternaria solani. ChemBioChem 2010, 11, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Park, J.-J.; Dugan, F.M.; Peever, T.L.; Gang, D.R.; Vandemark, G.; Chen, W. Production of the antibiotic secondary metabolite solanapyrone A by the fungal plant pathogen Ascochyta rabiei during fruiting body formation in saprobic growth. Environ. Microbiol. 2017, 19, 1822–1835. [Google Scholar] [CrossRef] [PubMed]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of Action of Microbial Biological Control Agents Against Plant Diseases: Relevance Beyond Efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Khan, R.A.A.; Yue, Q.; Jiao, Y.; Yang, Y.; Li, Y.; Xie, B. Discovery of a new antifungal lipopeptaibol from Purpureocillium lilacinum using MALDI-TOF-IMS. Biochem. Biophys. Res. Commun. 2020, 527, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Han, Y.; Sheng, Y.; Wang, Y.; Pan, Q.; Nie, H. Mass spectrometry imaging for direct visualization of components in plants tissues. J. Sep. Sci. 2021, 44, 3462–3476. [Google Scholar] [CrossRef]
- Bednarz, H.; Roloff, N.; Niehaus, K. Mass Spectrometry Imaging of the Spatial and Temporal Localization of Alkaloids in Nightshades. J. Agric. Food. Chem. 2019, 67, 13470–13477. [Google Scholar] [CrossRef]
- Deng, Y.; He, M.; Feng, F.; Feng, X.; Zhang, Y.; Zhang, F. The distribution and changes of glycoalkaloids in potato tubers under different storage time based on MALDI-TOF mass spectrometry imaging. Talanta 2021, 221, 121453. [Google Scholar] [CrossRef]
- Kamjijam, B.; Suwannaporn, P.; Bednarz, H.; Jom, K.N.; Niehaus, K. Elevation of gamma-aminobutyric acid (GABA) and essential amino acids in vacuum impregnation mediated germinated rice traced by MALDI imaging. Food Chem. 2021, 365, 130399. [Google Scholar] [CrossRef]
- Zhang, Y.-X.; Zhao, X.-B.; Ha, W.; Zhang, Y.-D.; Shi, Y.-P. Spatial distribution analysis of phospholipids in rice by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry imaging. J. Chromatogr. A 2021, 1651, 462302. [Google Scholar] [CrossRef]
- Horn, P.J.; Silva, J.E.; Anderson, D.; Fuchs, J.; Borisjuk, L.; Nazarenus, T.J.; Shulaev, V.; Cahoon, E.B.; Chapman, K.D. Imaging heterogeneity of membrane and storage lipids in transgenic Camelina sativa seeds with altered fatty acid profiles. Plant J. 2013, 76, 138–150. [Google Scholar] [PubMed]
- Seaman, C.; Flinders, B.; Eijkel, G.; Heeren, R.M.; Bricklebank, N.; Clench, M.R. “Afterlife experiment”: Use of MALDI-MS and SIMS imaging for the study of the nitrogen cycle within plants. Anal. Chem. 2014, 86, 10071–10077. [Google Scholar] [CrossRef] [PubMed]
- Dueñas, M.E.; Larson, E.A.; Lee, Y.J. Toward Mass Spectrometry Imaging in the Metabolomics Scale: Increasing Metabolic Coverage Through Multiple On-Tissue Chemical Modifications. Front. Plant Sci. 2019, 10, 860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feenstra, A.D.; Dueñas, M.E.; Lee, Y.J. Five Micron High Resolution MALDI Mass Spectrometry Imaging with Simple, Interchangeable, Multi-Resolution Optical System. J. Am. Soc. Mass Spectrom. 2017, 28, 434–442. [Google Scholar] [CrossRef]
- Shiono, K.; Taira, S. Imaging of Multiple Plant Hormones in Roots of Rice (Oryza sativa) Using Nanoparticle-Assisted Laser Desorption/Ionization Mass Spectrometry. J. Agric. Food Chem. 2020, 68, 6770–6775. [Google Scholar] [CrossRef]
- Shiono, K.; Hashizaki, R.; Nakanishi, T.; Sakai, T.; Yamamoto, T.; Ogata, K.; Harada, K.I.; Ohtani, H.; Katano, H.; Taira, S. Multi-imaging of Cytokinin and Abscisic Acid on the Roots of Rice (Oryza sativa) Using Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. J. Agric. Food Chem. 2017, 65, 7624–7628. [Google Scholar] [CrossRef] [Green Version]
- Klein, A.T.; Yagnik, G.B.; Hohenstein, J.D.; Ji, Z.; Zi, J.; Reichert, M.D.; MacIntosh, G.C.; Yang, B.; Peters, R.J.; Vela, J.; et al. Investigation of the Chemical Interface in the Soybean-Aphid and Rice-Bacteria Interactions Using MALDI-Mass Spectrometry Imaging. Anal. Chem. 2015, 87, 5294–5301. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Zepeda, D.; Frausto, M.; Nájera-González, H.-R.; Herrera-Estrella, L.; Ordaz-Ortiz, J.-J. Mass Spectrometry-Based Quantification and Spatial Localization of Small Organic Acid Exudates in Plant Roots under Phosphorus Deficiency and Aluminum Toxicity. Plant J. 2021, 106, 1791–1806. [Google Scholar] [CrossRef]
- Korenblum, E.; Dong, Y.; Szymanski, J.; Panda, S.; Jozwiak, A.; Massalha, H.; Meir, S.; Rogachev, I.; Aharoni, A. Rhizosphere Microbiome Mediates Systemic Root Metabolite Exudation by Root-To-Root Signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 3874–3883. [Google Scholar] [CrossRef]
- Veličković, D.; Lin, V.S.; Rivas, A.; Anderton, C.R.; Moran, J.J. An approach for broad molecular imaging of the root-soil interface via indirect matrix-assisted laser desorption/ionization mass spectrometry. Soil Biol. Biochem. 2020, 146, 107804. [Google Scholar] [CrossRef]
- Rajendran, G.; Sing, F.; Desai, A.J.; Archana, G. Enhanced growth and nodulation of pigeon pea by co-inoculation of Bacillus strains with Rhizobium spp. Bioresour. Technol. 2008, 99, 4544–4550. [Google Scholar] [CrossRef] [PubMed]
- Pathak, K.V.; Keharia, H. Characterization of Fungal Antagonistic Bacilli Isolated from Aerial Roots of Banyan (Ficus Benghalensis) Using Intact-Cell MALDI-TOF Mass Spectrometry (ICMS). J. Appl. Microbiol. 2013, 114, 1300–1310. [Google Scholar] [CrossRef]
- Debois, D.; Jourdan, E.; Smargiasso, N.; Thonart, P.; De Pauw, E.; Ongena, M. Spatiotemporal monitoring of the antibiome secreted by Bacillus biofilms on plant roots using MALDI mass spectrometry imaging. Anal. Chem. 2014, 86, 4431–4438. [Google Scholar] [CrossRef] [PubMed]
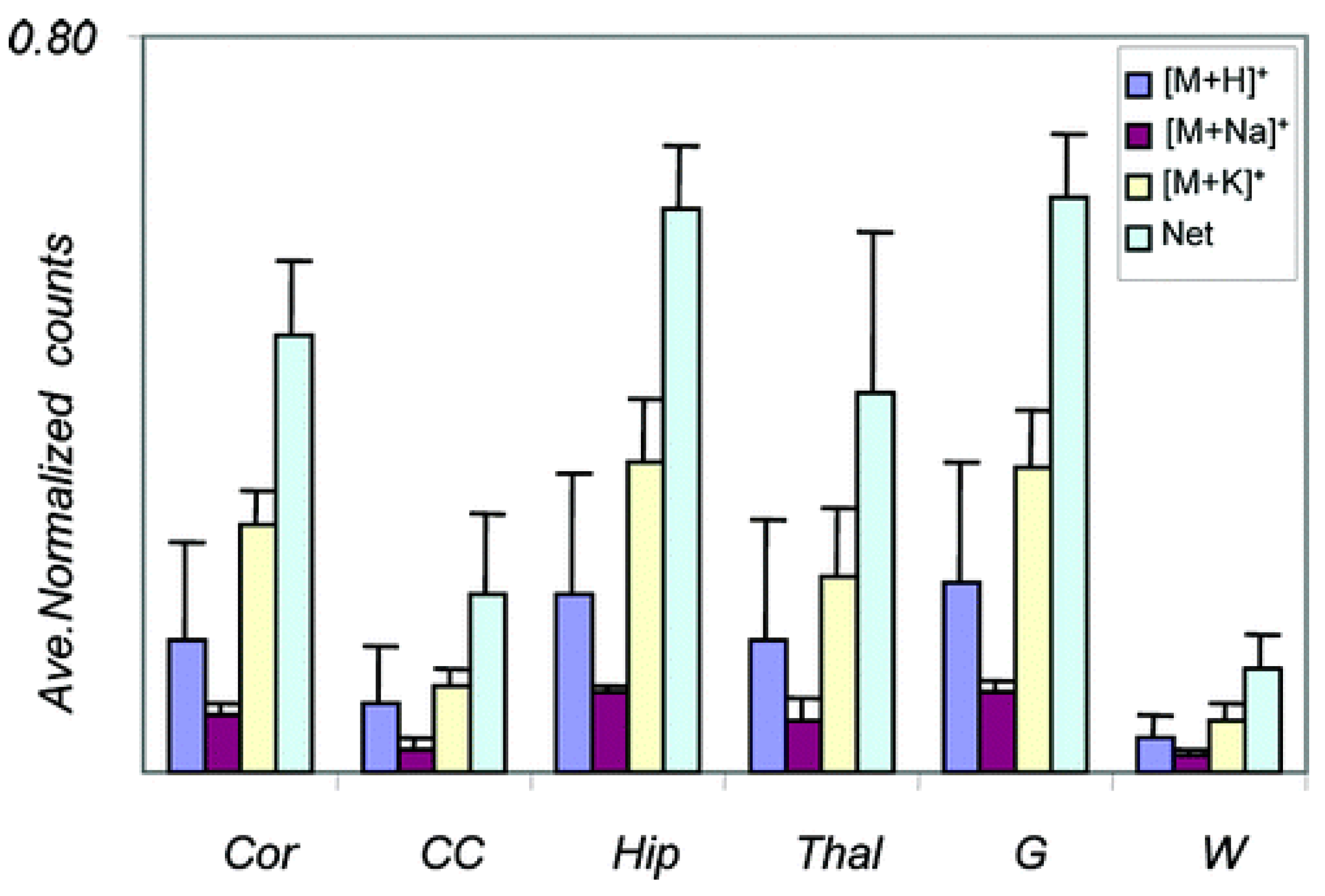
- Veličković, D.; Agtuca, B.J.; Stopka, S.A.; Vertes, A.; Koppenaal, D.W.; Paša-Tolić, L.; Stacey, G.; Anderton, C.R. Observed Metabolic Asymmetry within Soybean Root Nodules Reflects Unexpected Complexity in Rhizobacteria-Legume Metabolite Exchange. ISME J. 2018, 12, 2335–2338. [Google Scholar] [CrossRef] [Green Version]
- Gemperline, E.; Jayaraman, D.; Maeda, J.; Ané, J.M.; Li, L. Multifaceted investigation of metabolites during nitrogen fixation in Medicago via high resolution MALDI-MS imaging and ESI-MS. J. Am. Soc. Mass Spectrom. 2015, 26, 149–158. [Google Scholar] [CrossRef] [Green Version]
- Gemperline, E.; Keller, C.; Jayaraman, D.; Maeda, J.; Sussman, M.R.; Ané, J.M.; Li, L. Examination of Endogenous Peptides in Medicago truncatula Using Mass Spectrometry Imaging. J. Proteome Res. 2016, 15, 4403–4411. [Google Scholar] [CrossRef]
- Keller, C.; Maeda, J.; Jayaraman, D.; Chakraborty, S.; Sussman, M.R.; Harris, J.M.; Ané, J.-M.; Li, L. Comparison of Vacuum MALDI and AP-MALDI Platforms for the Mass Spectrometry Imaging of Metabolites Involved in Salt Stress in Medicago truncatula. Front. Plant Sci. 2018, 9, 1238. [Google Scholar] [CrossRef]
- Dokwal, D.; Romsdahl, T.B.; Kunz, D.A.; Alonso, A.P.; Dickstein, R. Phosphorus deprivation affects composition and spatial distribution of membrane lipids in legume nodules. Plant Physiol. 2021, 185, 1847–1859. [Google Scholar] [CrossRef]
- Slazak, B.; Kapusta, M.; Strömstedt, A.A.; Słomka, A.; Krychowiak, M.; Shariatgorji, M.; Andrén, P.E.; Bohdanowicz, J.; Kuta, E.; Göransson, U. How Does the Sweet Violet (Viola odorata L.) Fight Pathogens and Pests—Cyclotides as a Comprehensive Plant Host Defense System. Front. Plant Sci. 2018, 9, 1296. [Google Scholar] [CrossRef] [Green Version]
- Becker, L.; Carré, V.; Poutaraud, A.; Merdinoglu, D.; Chaimbault, P. MALDI mass spectrometry imaging for the simultaneous location of resveratrol, pterostilbene and viniferins on grapevine leaves. Molecules 2014, 19, 10587–10600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seneviratne, H.K.; Dalisay, D.S.; Kim, K.W.; Moinuddin, S.G.; Yang, H.; Hartshorn, C.M.; Davin, L.B.; Lewis, N.G. Non-host disease resistance response in pea (Pisum sativum) pods: Biochemical function of DRR206 and phytoalexin pathway localization. Phytochemistry 2015, 113, 140–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soares, M.S.; da Silva, D.F.; Forim, M.R.; da Silva, M.F.; Fernandes, J.B.; Vieira, P.C.; Silva, D.B.; Lopes, N.P.; de Carvalho, S.A.; de Souza, A.A.; et al. Quantification and localization of hesperidin and rutin in Citrus sinensis grafted on C. limonia after Xylella fastidiosa infection by HPLC-UV and MALDI imaging mass spectrometry. Phytochemistry 2015, 115, 161–170. [Google Scholar] [CrossRef]
- Catae, A.F.; da Silva Menegasso, A.R.; Pratavieira, M.; Palma, M.S.; Malaspina, O.; Roat, T.C. MALDI-imaging analyses of honeybee brains exposed to a neonicotinoid insecticide. Pest Manag. Sci. 2019, 75, 607–615. [Google Scholar] [CrossRef]
- Ly, A.; Ragionieri, L.; Liessem, S.; Becker, M.; Deininger, S.-O.; Neupert, S.; Predel, R. Enhanced Coverage of Insect Neuropeptides in Tissue Sections by an Optimized Mass-Spectrometry-Imaging Protocol. Anal. Chem. 2019, 91, 1980–1988. [Google Scholar] [CrossRef] [Green Version]
- Habenstein, J.; Schmitt, F.; Liessem, S.; Ly, A.; Trede, D.; Wegener, C.; Predel, R.; Rössler, W.; Neupert, S. Transcriptomic, peptidomic, and mass spectrometry imaging analysis of the brain in the ant Cataglyphis nodus. J. Neurochem. 2021, 158, 391–412. [Google Scholar] [CrossRef] [PubMed]
- Khalil, S.M.; Römpp, A.; Pretzel, J.; Becker, K.; Spengler, B. Phospholipid Topography of Whole-Body Sections of the Anopheles stephensi Mosquito, Characterized by High-Resolution Atmospheric-Pressure Scanning Microprobe Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging. Anal. Chem. 2015, 87, 11309–11316. [Google Scholar] [CrossRef] [Green Version]
- Yang, E.; Gamberi, C.; Chaurand, P. Mapping the fly Malpighian tubule lipidome by imaging mass spectrometry. J. Mass Spectrom. 2019, 54, 557–566. [Google Scholar] [CrossRef]
- Pratavieira, M.; da Silva Menegasso, A.R.; Garcia, A.M.C.; dos Santos, D.S.; Gomes, P.C.; Malaspina, O.; Palma, M.S. MALDI Imaging Analysis of Neuropeptides in the Africanized Honeybee (Apis mellifera) Brain: Effect of Ontogeny. J. Proteome Res. 2014, 13, 3054–3064. [Google Scholar] [CrossRef]
- Abdalsamee, M.K.; Giampà, M.; Niehaus, K.; Müller, C. Rapid incorporation of glucosinolates as a strategy used by a herbivore to prevent activation by myrosinases. Insect Biochem. Mol. Biol. 2014, 52, 115–123. [Google Scholar] [CrossRef]
- Kompauer, M.; Heiles, S.; Spengler, B. Autofocusing MALDI mass spectrometry imaging of tissue sections and 3D chemical topography of nonflat surfaces. Nat. Methods 2017, 14, 1156–1158. [Google Scholar] [CrossRef] [PubMed]
- Paine, M.R.L.; Ellis, S.R.; Maloney, D.; Heeren, R.M.A.; Verhaert, P. Digestion-Free Analysis of Peptides from 30-Year-Old Formalin-Fixed, Paraffin-Embedded Tissue by Mass Spectrometry Imaging. Anal. Chem. 2018, 90, 9272–9280. [Google Scholar] [CrossRef] [Green Version]
- Kaftan, F.; Vrkoslav, V.; Kynast, P.; Kulkarni, P.; Böcker, S.; Cvačka, J.; Knaden, M.; Svatoš, A. Mass spectrometry imaging of surface lipids on intact Drosophila melanogaster flies. J. Mass Spectrom. 2014, 49, 223–232. [Google Scholar] [CrossRef]
- Robert, F.M.; Chaevien, S.C.; Louis, A.S.; Arthur, S.E.; Richard, A.Y. MALDI Mass Spectrometric Imaging of the Nematode Caenorhabditis elegans. Curr. Metabolomics 2015, 3, 130–137. [Google Scholar]
- Hameed, S.; Ikegami, K.; Sugiyama, E.; Matsushita, S.; Kimura, Y.; Hayasaka, T.; Sugiura, Y.; Masaki, N.; Waki, M.; Ohta, I.; et al. Direct Profiling of the Phospholipid Composition of Adult Caenorhabditis Elegans Using Whole-Body Imaging Mass Spectrometry. Anal. Bioanal. Chem. 2015, 407, 7589–7602. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, E.A.; Bonfim, M.F.; Bloch, C.; Engler, G.; Rocha, T.; de Almeida Engler, J. Imaging Mass Spectrometry of Endogenous Polypeptides and Secondary Metabolites from Galls Induced by Root-Knot Nematodes in Tomato Roots. Mol. Plant Microbe Interact. 2018, 31, 1048–1059. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Qin, L.; Sun, J.; Chen, L.; Jia, L.; Zhao, J.; Yang, H.; Xue, K.; Wang, X.; Sang, W. Metabolite Changes Associated with Earthworms (Eisenia Fetida) Graphene Exposure Revealed by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging. Ecotoxicol. Environ. Saf. 2020, 205, 111102. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Sun, R.; Gordon, A.; Ge, J.; Zhang, Y.; Li, P.; Yang, H. 3-Aminophthalhydrazide (Luminol) as a Novel Matrix for Dual-Polarity MALDI MS Imaging. Anal. Chem. 2019, 91, 8221–8228. [Google Scholar] [CrossRef] [PubMed]
- Belov, M.E.; Ellis, S.R.; Dilillo, M.; Paine, M.R.L.; Danielson, W.F.; Anderson, G.A.; de Graaf, E.L.; Eijkel, G.B.; Heeren, R.M.A.; McDonnell, L.A. Design and Performance of a Novel Interface for Combined Matrix-Assisted Laser Desorption Ionization at Elevated Pressure and Electrospray Ionization with Orbitrap Mass Spectrometry. Anal. Chem. 2017, 89, 7493–7501. [Google Scholar] [CrossRef]
- Hankin, J.A.; Murphy, R.C. Relationship between MALDI IMS Intensity and Measured Quantity of Selected Phospholipids in Rat Brain Sections. Anal. Chem. 2010, 82, 8476–8484. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Liu, W.; Mu, Y.; Wang, X. 1,1′-binaphthyl-2,2′-diamine as a novel MALDI matrix to enhance the in situ imaging of metabolic heterogeneity in lung cancer. Talanta 2020, 209, 120557. [Google Scholar] [CrossRef]
- Ivanisevic, J.; Epstein, A.A.; Kurczy, M.E.; Benton, P.H.; Uritboonthai, W.; Fox, H.S.; Boska, M.D.; Gendelman, H.E.; Siuzdak, G. Brain Region Mapping Using Global Metabolomics. Chem. Biol. 2014, 21, 1575–1584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angel, P.M.; Spraggins, J.M.; Baldwin, H.S.; Caprioli, R. Enhanced Sensitivity for High Spatial Resolution Lipid Analysis by Negative Ion Mode Matrix Assisted Laser Desorption Ionization Imaging Mass Spectrometry. Anal. Chem. 2012, 84, 1557–1564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.-Y.J.; Post, S.N.J.J.; Woods, A.S. A minimalist approach to MALDI imaging of glycerophospholipids and sphingolipids in rat brain sections. Int. J. Mass Spectrom. 2008, 278, 143–149. [Google Scholar] [CrossRef] [Green Version]
- Murphy, R.C.; Hankin, J.A.; Barkley, R.M. Imaging of lipid species by MALDI mass spectrometry. J. Lipid Res. 2009, 50, S317–S322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruce, K.D.; Zsombok, A.; Eckel, R.H. Lipid Processing in the Brain: A Key Regulator of Systemic Metabolism. Front. Endocrinol. 2017, 8, 60. [Google Scholar] [CrossRef] [Green Version]
- Harris, A.; Roseborough, A.; Mor, R.; Yeung, K.K.C.; Whitehead, S.N. Ganglioside Detection from Formalin-Fixed Human Brain Tissue Utilizing MALDI Imaging Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2020, 31, 479–487. [Google Scholar] [CrossRef]
- Ye, H.; Wang, J.; Greer, T.; Strupat, K.; Li, L. Visualizing Neurotransmitters and Metabolites in the Central Nervous System by High Resolution and High Accuracy Mass Spectrometric Imaging. ACS Chem. Neurosci. 2013, 4, 1049–1056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeisel, S.H.; da Costa, K.-A. Choline: An essential nutrient for public health. Nutr. Rev. 2009, 67, 615–623. [Google Scholar] [CrossRef] [Green Version]
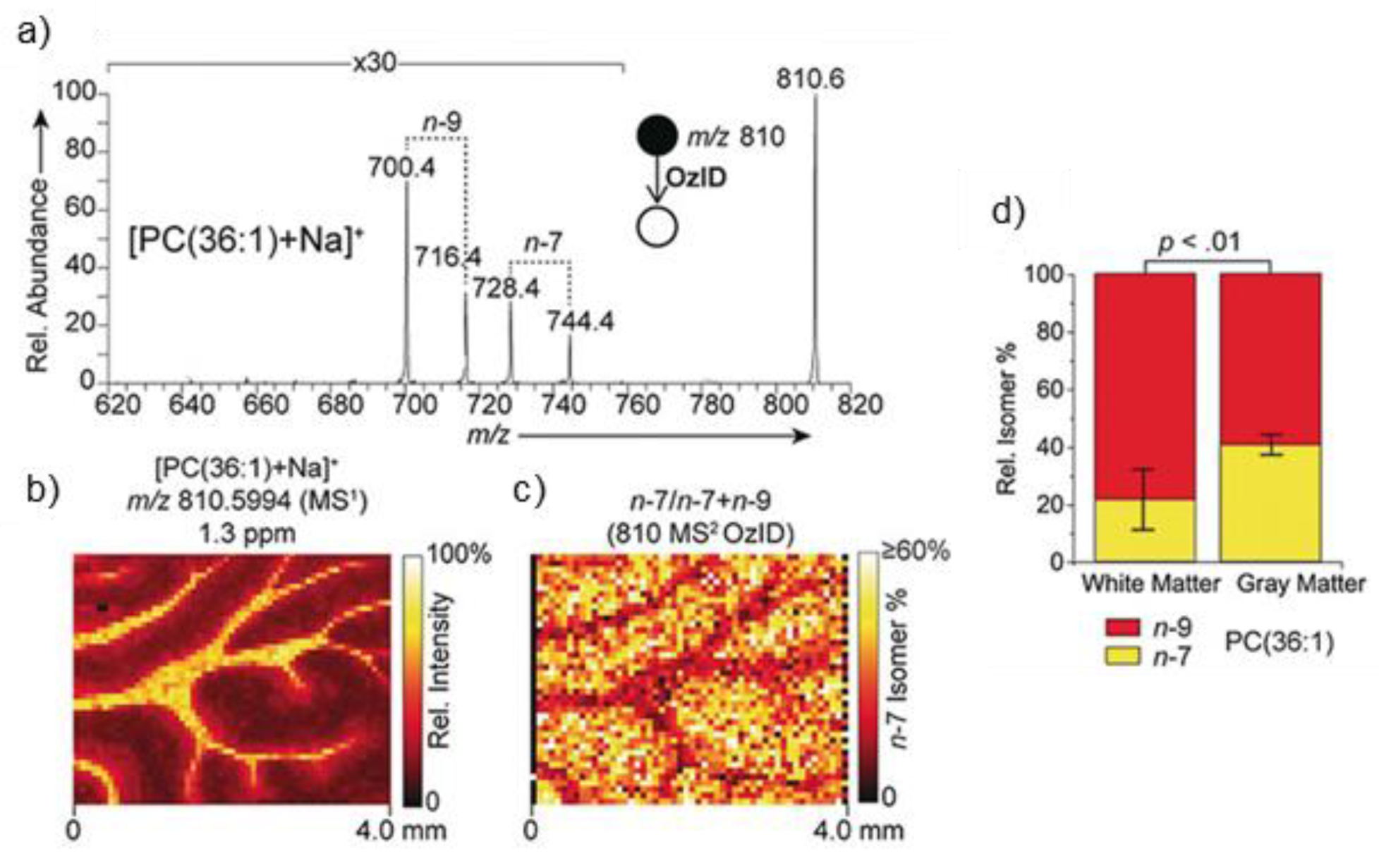
- Paine, M.R.L.; Poad, B.L.J.; Eijkel, G.B.; Marshall, D.L.; Blanksby, S.J.; Heeren, R.M.A.; Ellis, S.R. Mass Spectrometry Imaging with Isomeric Resolution Enabled by Ozone-Induced Dissociation. Angew. Chem. Int. Ed. 2018, 57, 10530–10534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Claes, B.S.R.; Bowman, A.P.; Poad, B.L.J.; Young, R.S.E.; Heeren, R.M.A.; Blanksby, S.J.; Ellis, S.R. Mass Spectrometry Imaging of Lipids with Isomer Resolution Using High-Pressure Ozone-Induced Dissociation. Anal. Chem. 2021, 93, 9826–9834. [Google Scholar] [CrossRef]
- Brown, S.H.J.; Mitchell, T.W.; Blanksby, S.J. Analysis of Unsaturated Lipids by Ozone-Induced Dissociation. Biochim. Biophys. Acta 2011, 1811, 807–817. [Google Scholar] [CrossRef] [PubMed]
- Goto-Inoue, N.; Hayasaka, T.; Zaima, N.; Setou, M. Imaging mass spectrometry reveals changes of metabolites distribution in mouse testis during testicular maturation. Surf. Interface Anal. 2012, 44, 749–754. [Google Scholar] [CrossRef]
- Goto-Inoue, N.; Hayasaka, T.; Zaima, N.; Setou, M. The specific localization of seminolipid molecular species on mouse testis during testicular maturation revealed by imaging mass spectrometry. Glycobiology 2009, 19, 950–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, A.; Phapale, P.; Chernyavsky, I.; Lavigne, R.; Fay, D.; Tarasov, A.; Kovalev, V.; Fuchser, J.; Nikolenko, S.; Pineau, C.; et al. FDR-Controlled Metabolite Annotation for High-Resolution Imaging Mass Spectrometry. Nat. Methods 2017, 14, 57–60. [Google Scholar] [CrossRef]
- Lagarrigue, M.; Lavigne, R.; Guével, B.; Palmer, A.; Rondel, K.; Guillot, L.; Kobarg, J.H.; Trede, D.; Pineau, C. Spatial segmentation and metabolite annotation involved in sperm maturation in the rat epididymis by MALDI imaging mass spectrometry. J. Mass Spectrom. 2020, 55, e4633. [Google Scholar] [CrossRef]
- Shimma, S.; Kumada, H.-O.; Taniguchi, H.; Konno, A.; Yao, I.; Furuta, K.; Matsuda, T.; Ito, S. Microscopic visualization of testosterone in mouse testis by use of imaging mass spectrometry. Anal. Bioanal. Chem. 2016, 408, 7607–7615. [Google Scholar] [CrossRef] [PubMed]
- Cobice, D.F.; Livingstone, D.E.W.; Mackay, C.L.; Goodwin, R.J.A.; Smith, L.B.; Walker, B.R.; Andrew, R. Spatial Localization and Quantitation of Androgens in Mouse Testis by Mass Spectrometry Imaging. Anal. Chem. 2016, 88, 10362–10367. [Google Scholar] [CrossRef] [Green Version]
- Bowman, A.P.; Bogie, J.F.J.; Hendriks, J.J.A.; Haidar, M.; Belov, M.; Heeren, R.M.A.; Ellis, S.R. Evaluation of Lipid Coverage and High Spatial Resolution MALDI-Imaging Capabilities of Oversampling Combined with Laser Post-Ionisation. Anal. Bioanal. Chem. 2020, 412, 2277–2289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wäldchen, F.; Mohr, F.; Wagner, A.H.; Heiles, S. Multifunctional Reactive MALDI Matrix Enabling High-Lateral Resolution Dual Polarity MS Imaging and Lipid C=C Position-Resolved MS2 Imaging. Anal. Chem. 2020, 92, 14130–14138. [Google Scholar] [CrossRef]
- Neumann, E.K.; Migas, L.G.; Allen, J.L.; Caprioli, R.M.; Van de Plas, R.; Spraggins, J.M. Spatial Metabolomics of the Human Kidney using MALDI Trapped Ion Mobility Imaging Mass Spectrometry. Anal. Chem. 2020, 92, 13084–13091. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Liu, W.; Geng, Y.; Wang, X. On-Tissue Derivatization Strategy for Mass Spectrometry Imaging of Carboxyl-Containing Metabolites in Biological Tissues. Anal. Chem. 2020, 92, 12126–12131. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sgobba, E.; Daguerre, Y.; Giampà, M. Unravel the Local Complexity of Biological Environments by MALDI Mass Spectrometry Imaging. Int. J. Mol. Sci. 2021, 22, 12393. https://doi.org/10.3390/ijms222212393
Sgobba E, Daguerre Y, Giampà M. Unravel the Local Complexity of Biological Environments by MALDI Mass Spectrometry Imaging. International Journal of Molecular Sciences. 2021; 22(22):12393. https://doi.org/10.3390/ijms222212393
Chicago/Turabian StyleSgobba, Elvira, Yohann Daguerre, and Marco Giampà. 2021. "Unravel the Local Complexity of Biological Environments by MALDI Mass Spectrometry Imaging" International Journal of Molecular Sciences 22, no. 22: 12393. https://doi.org/10.3390/ijms222212393








