Olfactory Receptor OR7A17 Expression Correlates with All-Trans Retinoic Acid (ATRA)-Induced Suppression of Proliferation in Human Keratinocyte Cells
Abstract
:1. Introduction
2. Results
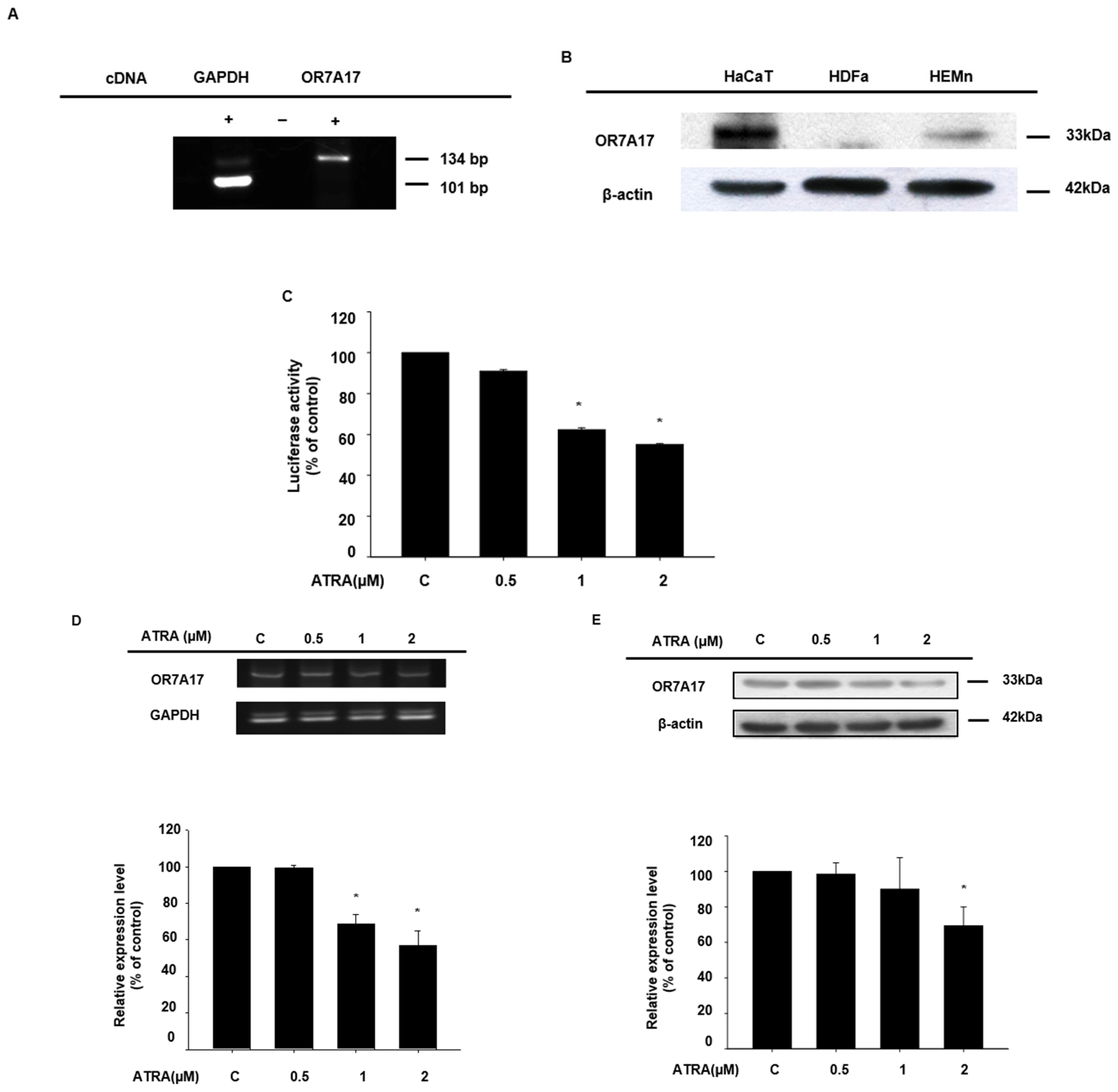
2.1. ATRA Downregulated the Expression of OR7A17 in Human Keratinocytes
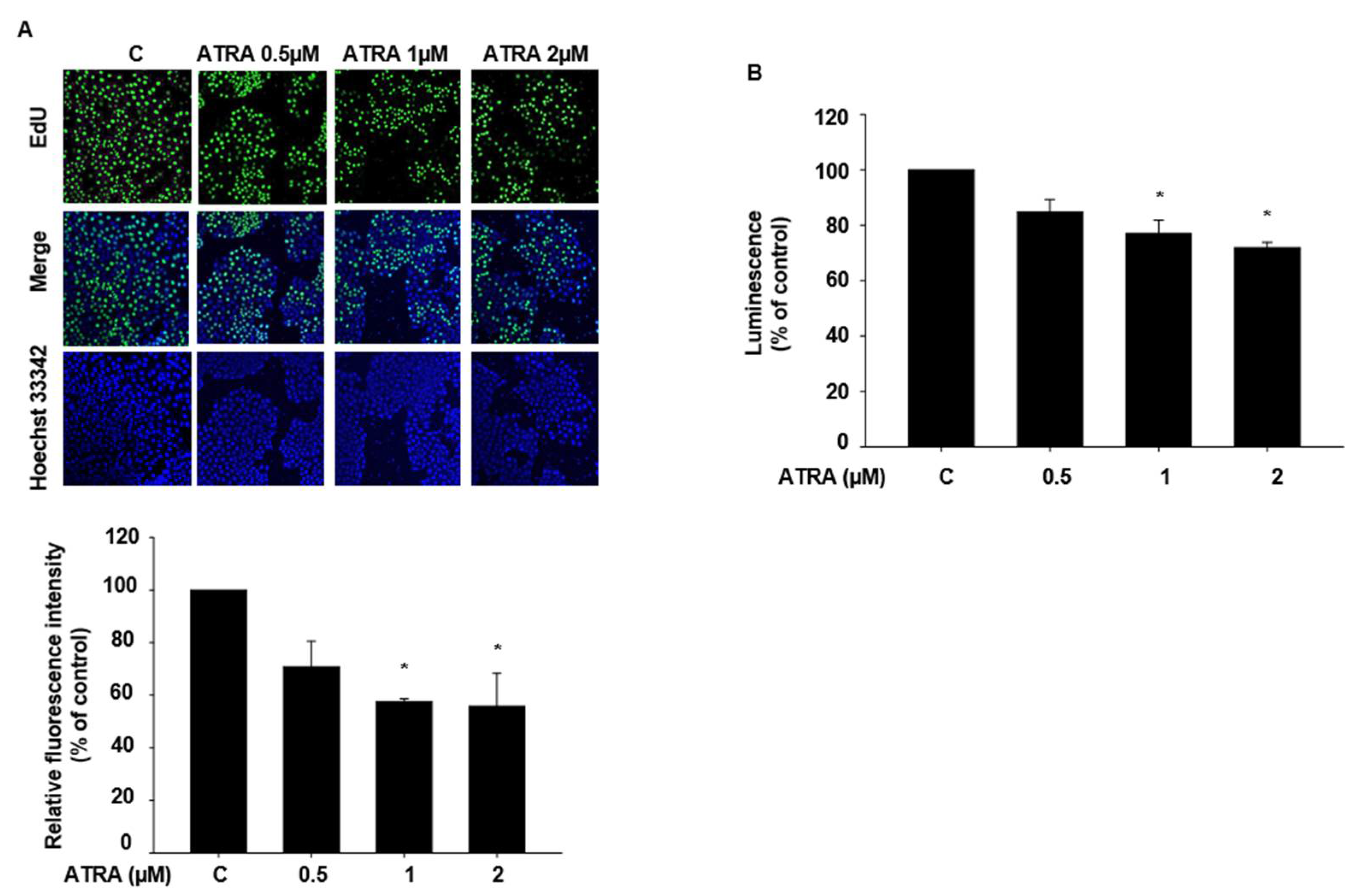
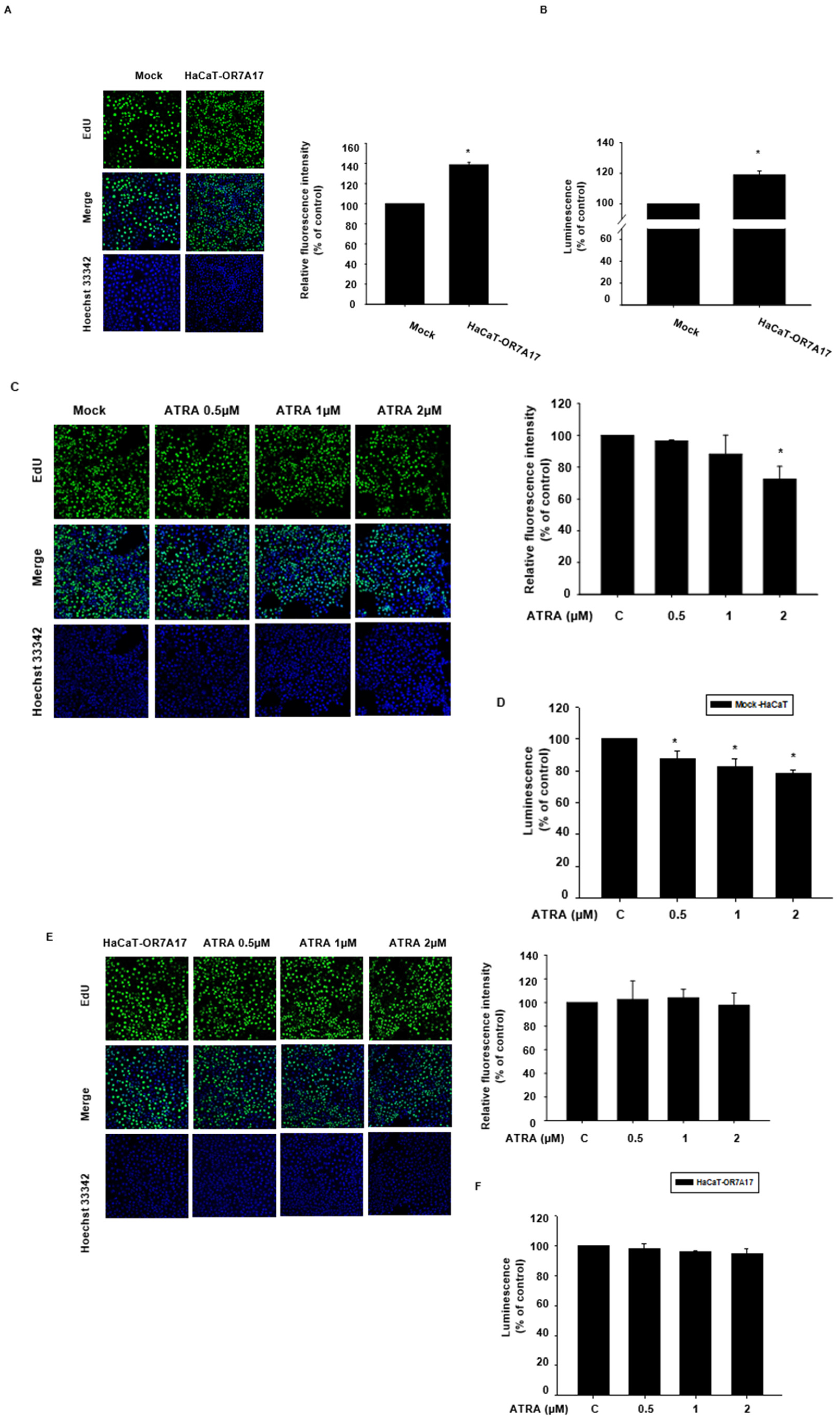
2.2. ATRA Suppresses the Proliferation of HaCaT Cells
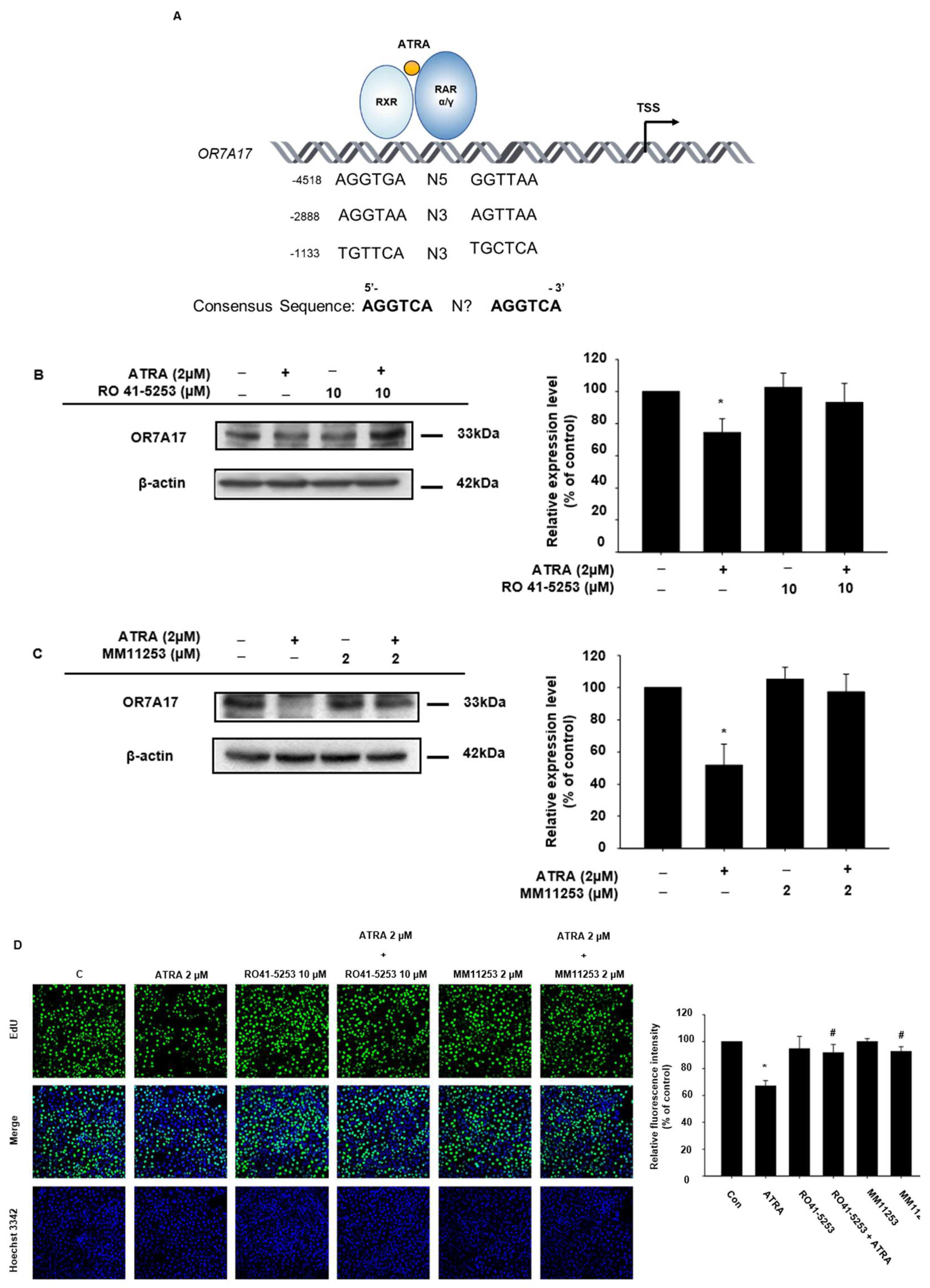
2.3. OR7A17 Expression Is Regulated via RAR Signaling
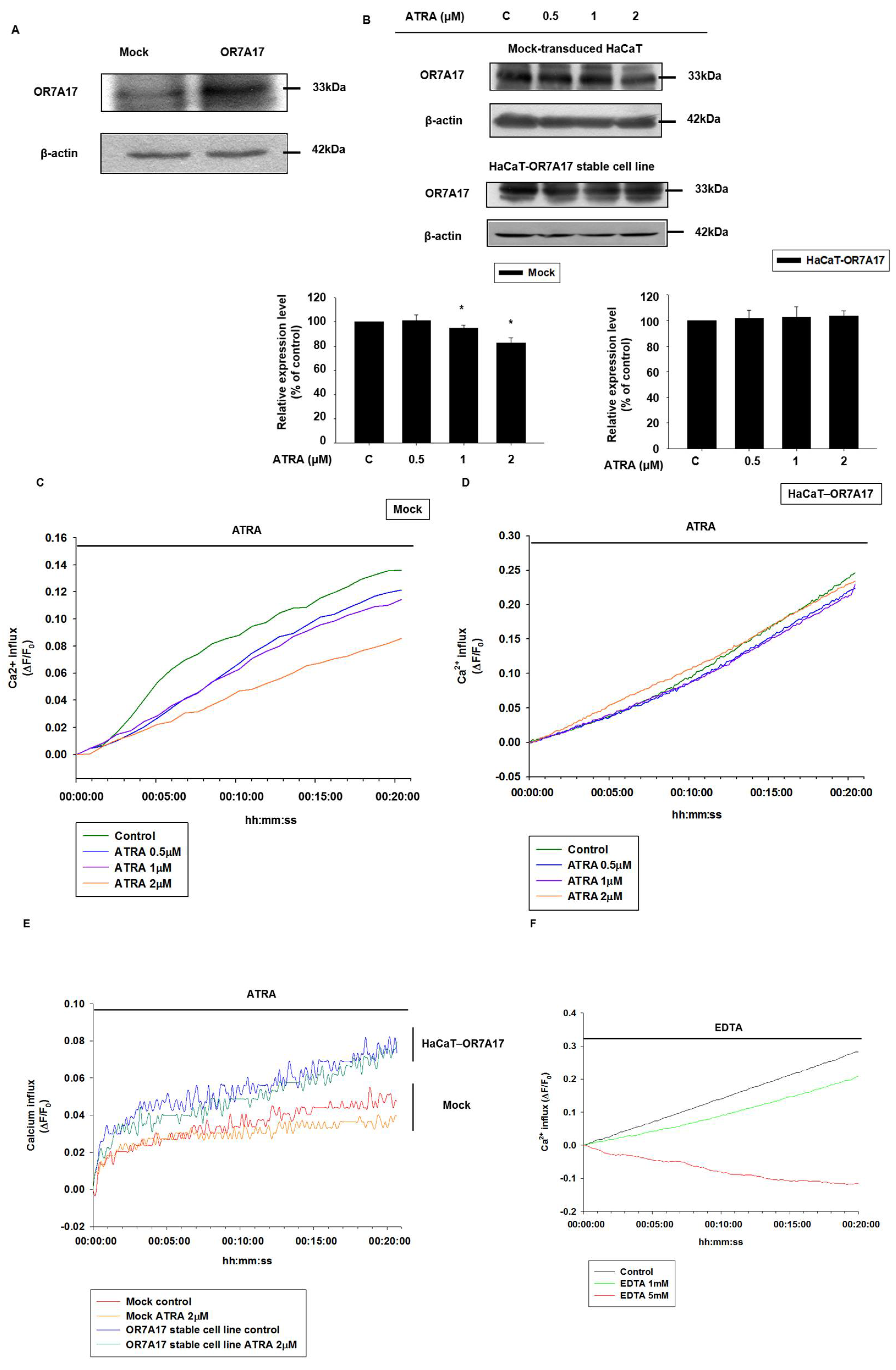
2.4. Overexpression of OR7A17 Interferes with the Suppressive Effect of ATRA
2.5. OR7A17 Overexpression Induces Proliferation of HaCaT Cells
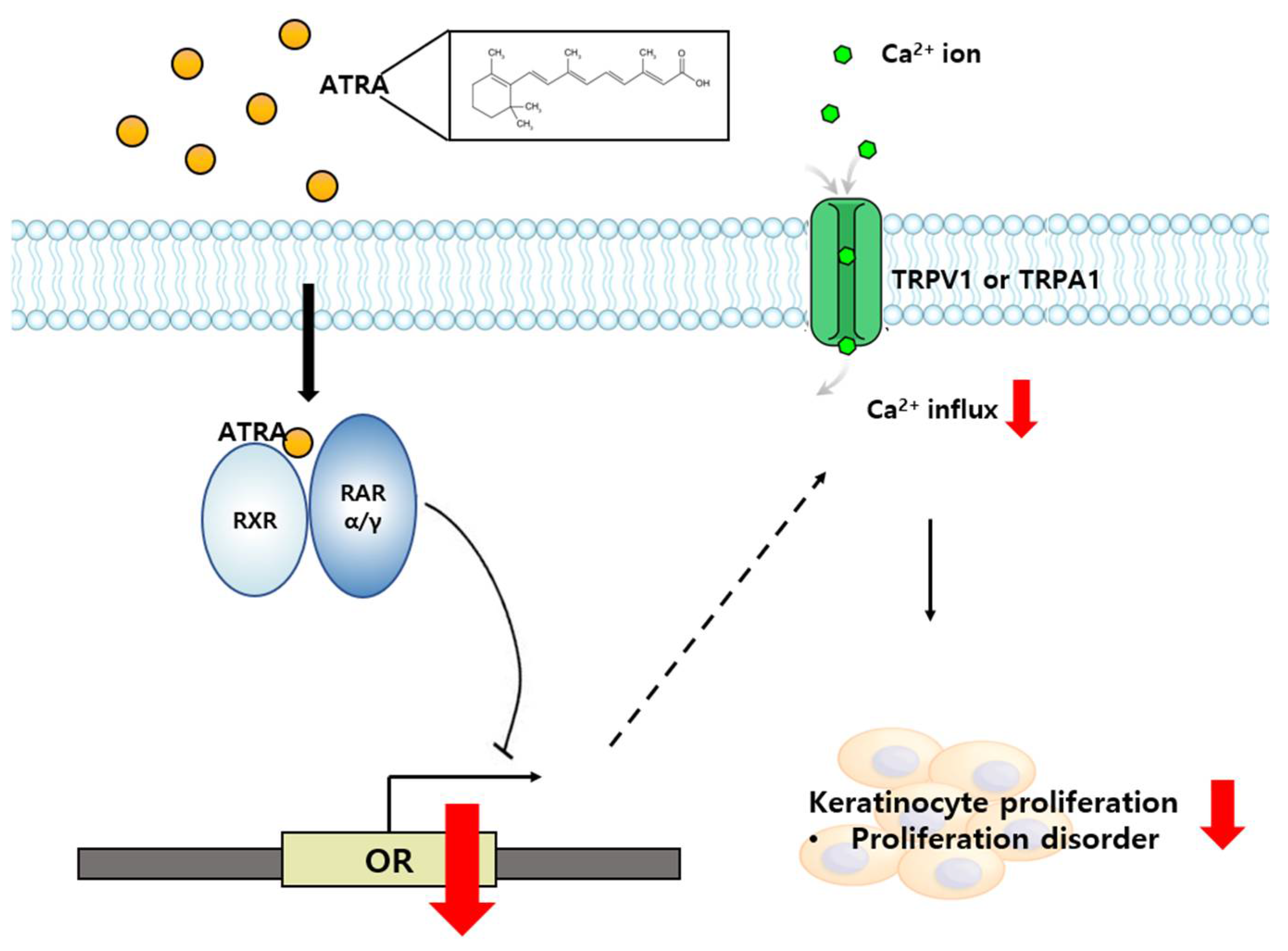
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. ATRA Treatment
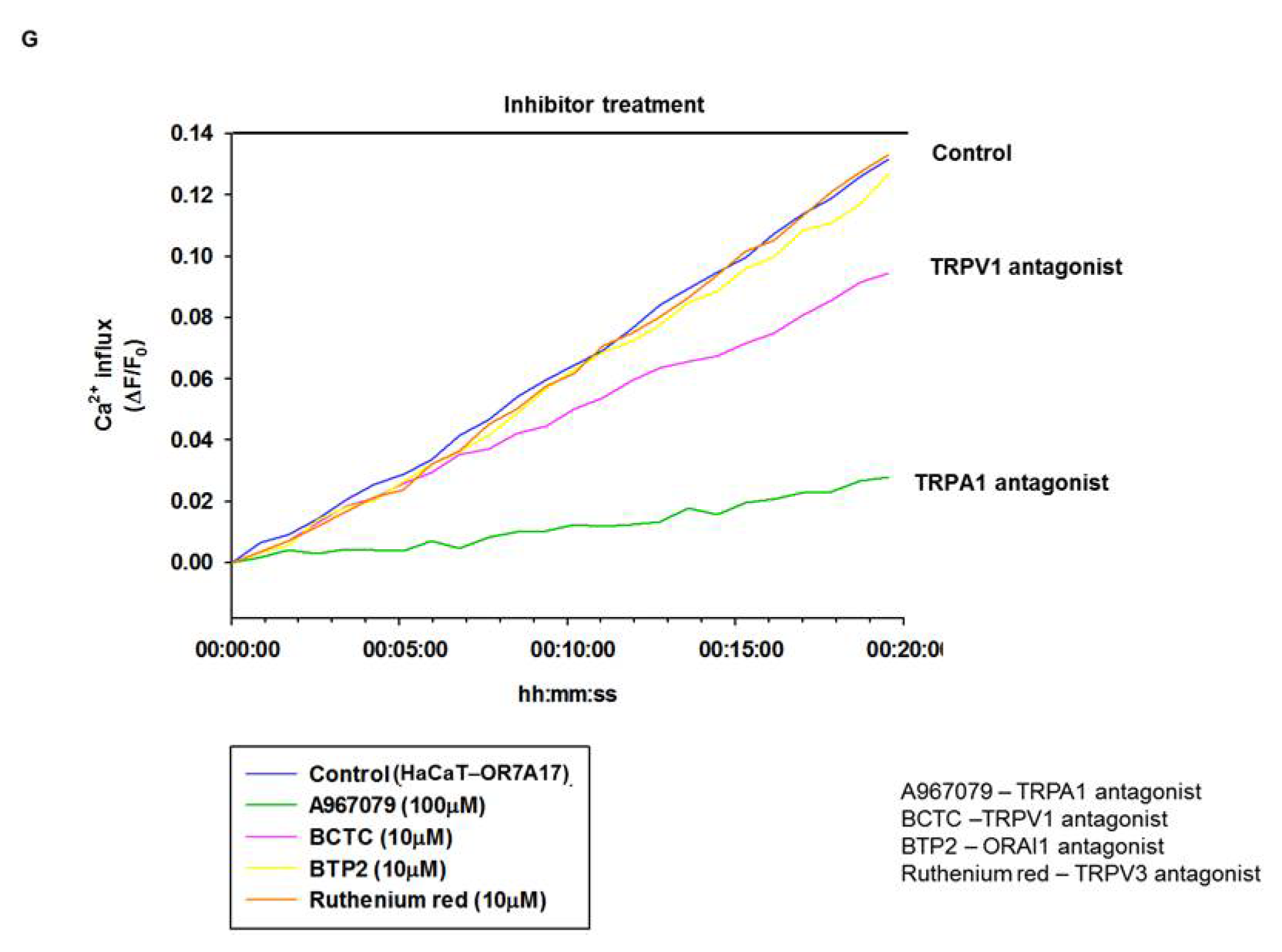
4.3. Fluo-4 Ca2+ Influx Assay
4.4. Luciferase Reporter Assay and β-Galactosidase Activity Assay
4.5. Western Blotting Analysis
4.6. Antibodies
4.7. Reverse Transcription qPCR (RT-qPCR)
4.8. EdU Incorporation Assay
4.9. CellTiter Glo® 2.0 Assay for Cell Proliferation Analysis
4.10. Lentiviral Transduction of HaCaT Cells
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Antunes, G.; de Souza, F.M.S. Olfactory receptor signaling. Methods Cell Biol. 2016, 132, 127–145. [Google Scholar] [CrossRef]
- Flegel, C.; Manteniotis, S.; Osthold, S.; Hatt, H.; Gisselmann, G. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS ONE 2013, 8, e55368. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.J.; Depoortere, I.; Hatt, H. Therapeutic potential of ectopic olfactory and taste receptors. Nat. Rev. Drug Discov. 2019, 18, 116–138. [Google Scholar] [CrossRef]
- Busse, D.; Kudella, P.; Gruning, N.M.; Gisselmann, G.; Stander, S.; Luger, T.; Jacobsen, F.; Steinstrasser, L.; Paus, R.; Gkogkolou, P.; et al. A synthetic sandalwood odorant induces wound-healing processes in human keratinocytes via the olfactory receptor OR2AT4. J. Investig. Dermatol. 2014, 134, 2823–2832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, T.; Veitinger, S.; Peek, I.; Busse, D.; Eckardt, J.; Vladimirova, D.; Jovancevic, N.; Wojcik, S.; Gisselmann, G.; Altmuller, J.; et al. Two olfactory receptors-OR2A4/7 and OR51B5-differentially affect epidermal proliferation and differentiation. Exp. Dermatol. 2017, 26, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Gelis, L.; Jovancevic, N.; Veitinger, S.; Mandal, B.; Arndt, H.D.; Neuhaus, E.M.; Hatt, H. Functional Characterization of the Odorant Receptor 51E2 in Human Melanocytes. J. Biol. Chem. 2016, 291, 17772–17786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tham, E.H.; Dyjack, N.; Kim, B.E.; Rios, C.; Seibold, M.A.; Leung, D.Y.M.; Goleva, E. Expression and function of the ectopic olfactory receptor OR10G7 in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2019, 143, 1838–1848.e1834. [Google Scholar] [CrossRef]
- Thielitz, A.; Gollnick, H. Topical retinoids in acne vulgaris: Update on efficacy and safety. Am. J. Clin. Dermatol. 2008, 9, 369–381. [Google Scholar] [CrossRef]
- Torma, H.; Bergstrom, A.; Ghiasifarahani, G.; Berne, B. The effect of two endogenous retinoids on the mRNA expression profile in human primary keratinocytes, focusing on genes causing autosomal recessive congenital ichthyosis. Arch. Dermatol. Res 2014, 306, 739–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.L.; Tao, Y.; Zhou, W.Q.; Ma, P.C.; Cao, Y.P.; He, C.D.; Wei, J.; Li, L.J. All-trans retinoic acid induces cell-cycle arrest in human cutaneous squamous carcinoma cells by inhibiting the mitogen-activated protein kinase-activated protein 1 pathway. Clin. Exp. Dermatol. 2014, 39, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Ipek, Y.; Hulya, D.; Melih, A. Disseminated exfoliative dermatitis associated with all-transretinoic Acid in the treatment of acute promyelocytic leukemia. Case Rep. Med. 2012, 2012, 236174. [Google Scholar] [CrossRef] [Green Version]
- Ale, S.I.; Laugier, J.P.; Maibach, H.I. Differential irritant skin responses to tandem application of topical retinoic acid and sodium lauryl sulphate: II. Effect of time between first and second exposure. Br. J. Dermatol. 1997, 137, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, Q.; Geng, S. Alltrans retinoic acid alters the expression of the tight junction proteins Claudin1 and 4 and epidermal barrier functionassociated genes in the epidermis. Int. J. Mol. Med. 2019, 43, 1789–1805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duong, V.; Rochette-Egly, C. The molecular physiology of nuclear retinoic acid receptors. From health to disease. Biochim. Biophys. Acta 2011, 1812, 1023–1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tasseff, R.; Jensen, H.A.; Congleton, J.; Dai, D.; Rogers, K.V.; Sagar, A.; Bunaciu, R.P.; Yen, A.; Varner, J.D. An Effective Model of the Retinoic Acid Induced HL-60 Differentiation Program. Sci. Rep. 2017, 7, 14327. [Google Scholar] [CrossRef] [Green Version]
- Jung, H.; Chae, Y.C.; Kim, J.Y.; Jeong, O.S.; Kook, H.; Seo, S.B. Author Correction: Regulatory role of G9a and LSD1 in the Transcription of Olfactory Receptors during Leukaemia Cell Differentiation. Sci. Rep. 2018, 8, 17075. [Google Scholar] [CrossRef]
- Memezawa, A.; Takada, I.; Takeyama, K.; Igarashi, M.; Ito, S.; Aiba, S.; Kato, S.; Kouzmenko, A.P. Id2 gene-targeted crosstalk between Wnt and retinoid signaling regulates proliferation in human keratinocytes. Oncogene 2007, 26, 5038–5045. [Google Scholar] [CrossRef] [Green Version]
- Zouboulis, C.C.; Seltmann, H.; Sass, J.O.; Ruhl, R.; Plum, C.; Hettmannsperger, U.; Blume-Peytavi, U.; Nau, H.; Orfanos, C.E. Retinoid signaling by all-trans retinoic acid and all-trans retinoyl-beta-D-glucuronide is attenuated by simultaneous exposure of human keratinocytes to retinol. J. Investig. Dermatol. 1999, 112, 157–164. [Google Scholar] [CrossRef] [Green Version]
- Chang Ge, C.; Wu, M.; Chen, G.; Yu, G.; Ji, D.; Wang, S. Identification of molecular characteristics induced by radiotherapy in rectal cancer based on microarray data. Oncol. Lett. 2017, 13, 2777–2783. [Google Scholar] [CrossRef]
- Butty, A.M.; Chud, T.C.S.; Cardoso, D.F.; Lopes, L.S.F.; Miglior, F.; Schenkel, F.S.; Canovas, A.; Hafliger, I.M.; Drogemuller, C.; Stothard, P.; et al. Genome-wide association study between copy number variants and hoof health traits in Holstein dairy cattle. J. Dairy Sci. 2021, 104, 8050–8061. [Google Scholar] [CrossRef]
- Li, J.V.; Ng, C.A.; Cheng, D.; Zhou, Z.; Yao, M.; Guo, Y.; Cox, C.D. Modified N-linked glycosylation status predicts trafficking defective human Piezo1 channel mutations. Commun. Biol. 2021, 4, 1–17. [Google Scholar] [CrossRef]
- Crump, N.T.; Hazzalin, C.A.; Bowers, E.M.; Alani, R.M.; Cole, P.A.; Mahadevan, L.C. Dynamic acetylation of all lysine-4 trimethylated histone H3 is evolutionarily conserved and mediated by p300/CBP. PNAS 2011, 108, 7814–7819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Assi, M.; Pirlot, B.; Stroobant, V.; Thissen, J.P.; Jacquemin, P. A Novel KRAS Antibody Highlights a Regulation Mechanism of Post-Translational Modifications of KRAS during Tumorigenesis. Int. J. Mol. Sci. 2020, 21, 6361. [Google Scholar] [CrossRef]
- Ghezzi, A.; Liebeskind, B.J.; Thompson, A.; Atkinson, N.S.; ZakonFront, H.H. Ancient association between cation leak channels and Mid1 proteins is conserved in fungi and animals. Mol. Neurosci. 2014, 7, 15. [Google Scholar] [CrossRef] [Green Version]
- Spehr, J.; Gelis, L.; Osterloh, M.; Oberland, S.; Hatt, H.; Spehr, M.; Neuhaus, E.M. G protein-coupled receptor signaling via Src kinase induces endogenous human transient receptor potential vanilloid type 6 (TRPV6) channel activation. J. Biol. Chem. 2011, 286, 13184–13192. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, T.; Gold, G.H. A cyclic nucleotide-gated conductance in olfactory receptor cilia. Nature 1987, 325, 442–444. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Reed, R.R. Golf: An olfactory neuron specific-G protein involved in odorant signal transduction. Science 1989, 244, 790–795. [Google Scholar] [CrossRef]
- Parmentier, M.; Libert, F.; Schurmans, S.; Schiffmann, S.; Lefort, A.; Eggerickx, D.; Ledent, C.; Mollereau, C.; Gerard, C.; Perret, J.; et al. Expression of members of the putative olfactory receptor gene family in mammalian germ cells. Nature 1992, 355, 453–455. [Google Scholar] [CrossRef]
- Spehr, M.; Munger, S.D. Olfactory receptors: G protein-coupled receptors and beyond. J. Neurochem. 2009, 109, 1570–1583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denda, M. Newly discovered olfactory receptors in epidermal keratinocytes are associated with proliferation, migration, and re-epithelialization of keratinocytes. J. Investig. Dermatol. 2014, 134, 2677–2679. [Google Scholar] [CrossRef] [Green Version]
- Clagett-Dame, M.; Knutson, D. Vitamin A in reproduction and development. Nutrients 2011, 3, 385–428. [Google Scholar] [CrossRef] [Green Version]
- Kowanetz, M.; Valcourt, U.; Bergstrom, R.; Heldin, C.H.; Moustakas, A. Id2 and Id3 define the potency of cell proliferation and differentiation responses to transforming growth factor beta and bone morphogenetic protein. Mol. Cell. Biol. 2004, 24, 4241–4254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norton, J.D.; Deed, R.W.; Craggs, G.; Sablitzky, F. Id helix-loop-helix proteins in cell growth and differentiation. Trends Cell Biol. 1998, 8, 58–65. [Google Scholar]
- Cunningham, T.J.; Duester, G. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat. Rev. Mol. Cell Biol. 2015, 16, 110–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elias, P.; Ahn, S.; Brown, B.; Crumrine, D.; Feingold, K.R. Origin of the epidermal calcium gradient: Regulation by barrier status and role of active vs passive mechanisms. J. Investig. Dermatol. 2002, 119, 1269–1274. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.E.; Lee, S.H. Skin Barrier and Calcium. Ann. Dermatol. 2018, 30, 265–275. [Google Scholar] [CrossRef]
- Lee, S.H.; Elias, P.M.; Proksch, E.; Menon, G.K.; Mao-Quiang, M.; Feingold, K.R. Calcium and potassium are important regulators of barrier homeostasis in murine epidermis. J. Clin. Investig. 1992, 89, 530–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menon, G.K.; Elias, P.M.; Lee, S.H.; Feingold, K.R. Localization of calcium in murine epidermis following disruption and repair of the permeability barrier. Cell Tissue Res. 1992, 270, 503–512. [Google Scholar] [CrossRef]
- Tsutsumi, M.; Denda, S.; Inoue, K.; Ikeyama, K.; Denda, M. Calcium ion gradients and dynamics in cultured skin slices of rat hindpaw in response to stimulation with ATP. J. Investig. Dermatol. 2009, 129, 584–589. [Google Scholar] [CrossRef] [Green Version]
- Kahl, C.R.; Means, A.R. Regulation of cell cycle progression by calcium/calmodulin-dependent pathways. Endocr. Rev. 2003, 24, 719–736. [Google Scholar] [CrossRef] [Green Version]
- Bonnefond, M.L.; Florent, R.; Lenoir, S.; Lambert, B.; Abeilard, E.; Giffard, F.; Louis, M.H.; Elie, N.; Briand, M.; Vivien, D.; et al. Inhibition of store-operated channels by carboxyamidotriazole sensitizes ovarian carcinoma cells to anti-BclxL strategies through Mcl-1 down-regulation. Oncotarget 2018, 9, 33896–33911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munaron, L.; Antoniotti, S.; Lovisolo, D. Intracellular calcium signals and control of cell proliferation: How many mechanisms? J. Cell. Mol. Med. 2004, 8, 161–168. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Park, S.-H.; Oh, S.W.; Kwon, K.; Park, S.J.; Yu, E.; Yang, S.; Park, J.Y.; Choi, S.; Yang, S.; et al. Olfactory Receptor OR7A17 Expression Correlates with All-Trans Retinoic Acid (ATRA)-Induced Suppression of Proliferation in Human Keratinocyte Cells. Int. J. Mol. Sci. 2021, 22, 12304. https://doi.org/10.3390/ijms222212304
Kim H, Park S-H, Oh SW, Kwon K, Park SJ, Yu E, Yang S, Park JY, Choi S, Yang S, et al. Olfactory Receptor OR7A17 Expression Correlates with All-Trans Retinoic Acid (ATRA)-Induced Suppression of Proliferation in Human Keratinocyte Cells. International Journal of Molecular Sciences. 2021; 22(22):12304. https://doi.org/10.3390/ijms222212304
Chicago/Turabian StyleKim, Hyeyoun, See-Hyoung Park, Sae Woong Oh, Kitae Kwon, Se Jung Park, Eunbi Yu, Seyoung Yang, Jung Yoen Park, Seoyoung Choi, Seoyoun Yang, and et al. 2021. "Olfactory Receptor OR7A17 Expression Correlates with All-Trans Retinoic Acid (ATRA)-Induced Suppression of Proliferation in Human Keratinocyte Cells" International Journal of Molecular Sciences 22, no. 22: 12304. https://doi.org/10.3390/ijms222212304
APA StyleKim, H., Park, S.-H., Oh, S. W., Kwon, K., Park, S. J., Yu, E., Yang, S., Park, J. Y., Choi, S., Yang, S., Han, S. B., Song, M., Cho, J. Y., & Lee, J. (2021). Olfactory Receptor OR7A17 Expression Correlates with All-Trans Retinoic Acid (ATRA)-Induced Suppression of Proliferation in Human Keratinocyte Cells. International Journal of Molecular Sciences, 22(22), 12304. https://doi.org/10.3390/ijms222212304








