Dissection of the Regulatory Elements of the Complex Expression Pattern of Puckered, a Dual-Specificity JNK Phosphatase
Abstract
:1. Introduction
2. Results
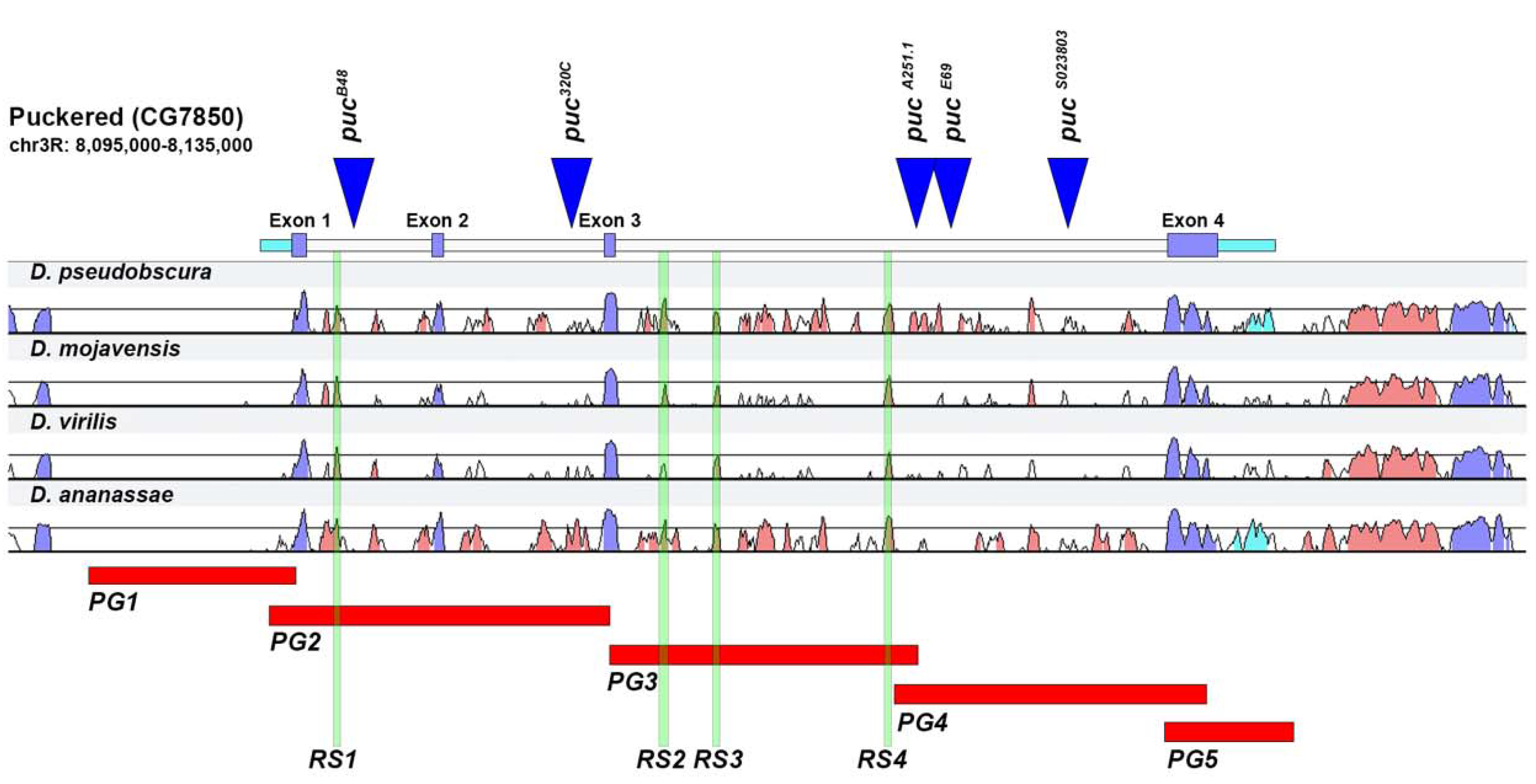
2.1. puc Genomic Domain Analysis
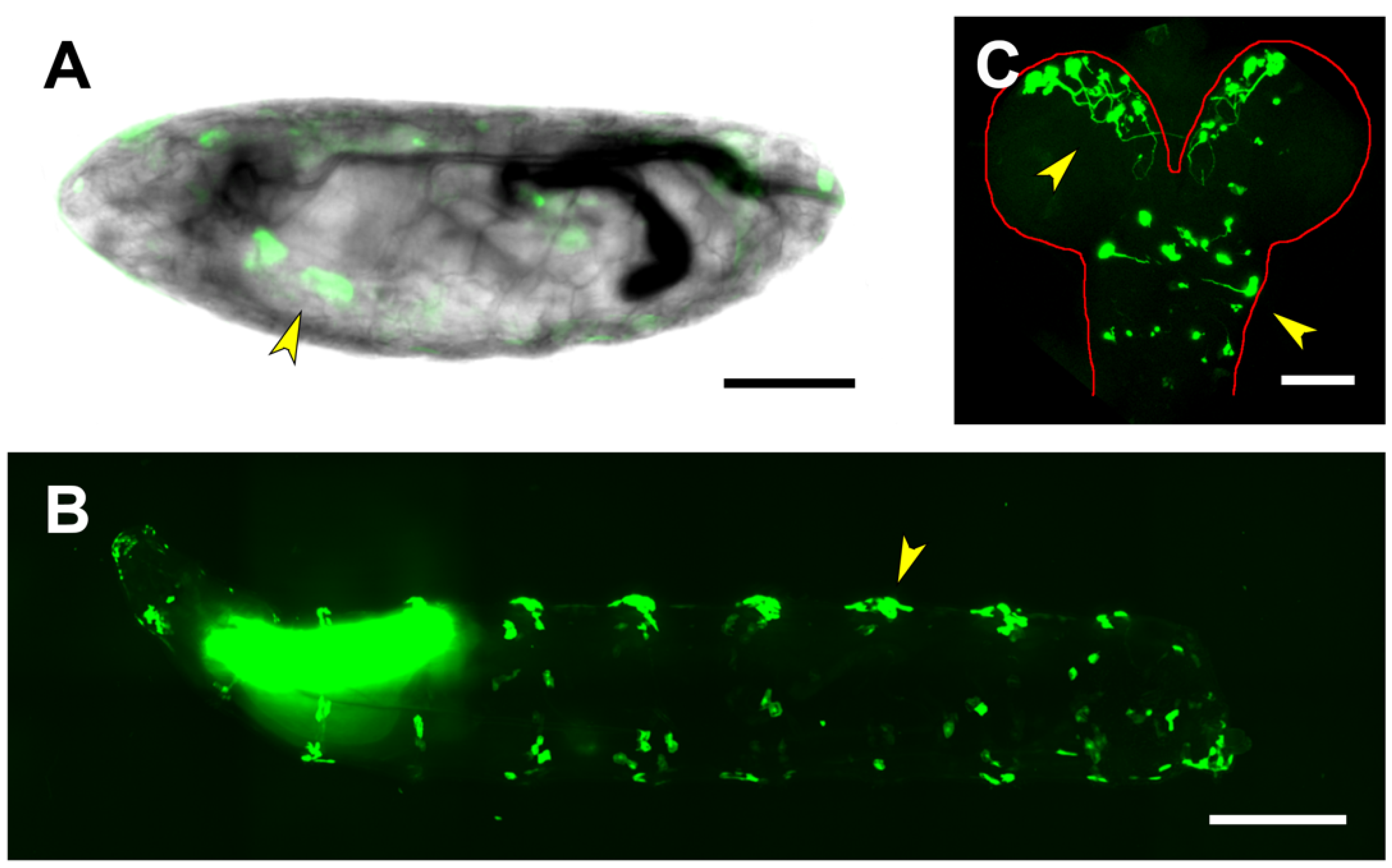
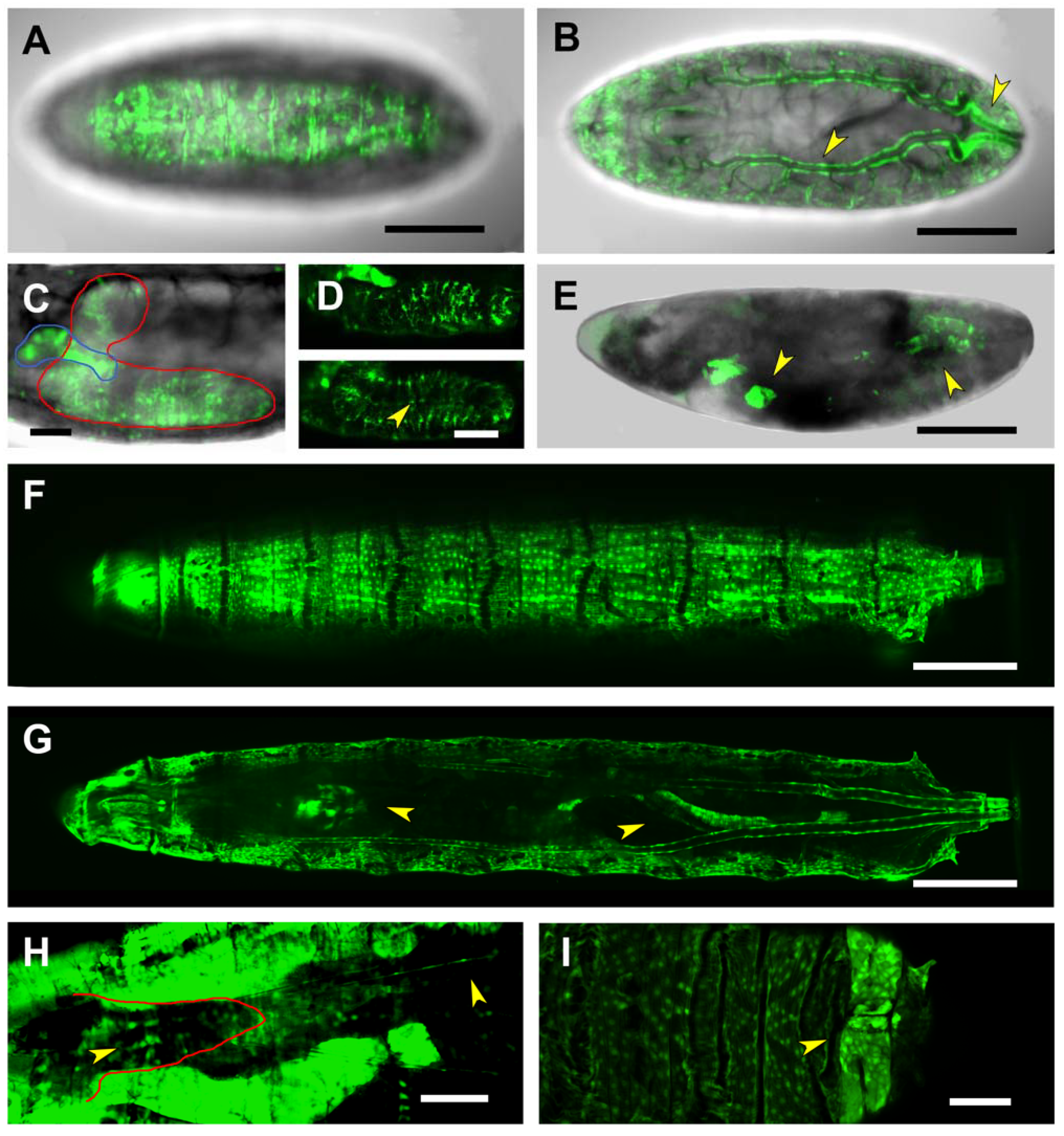
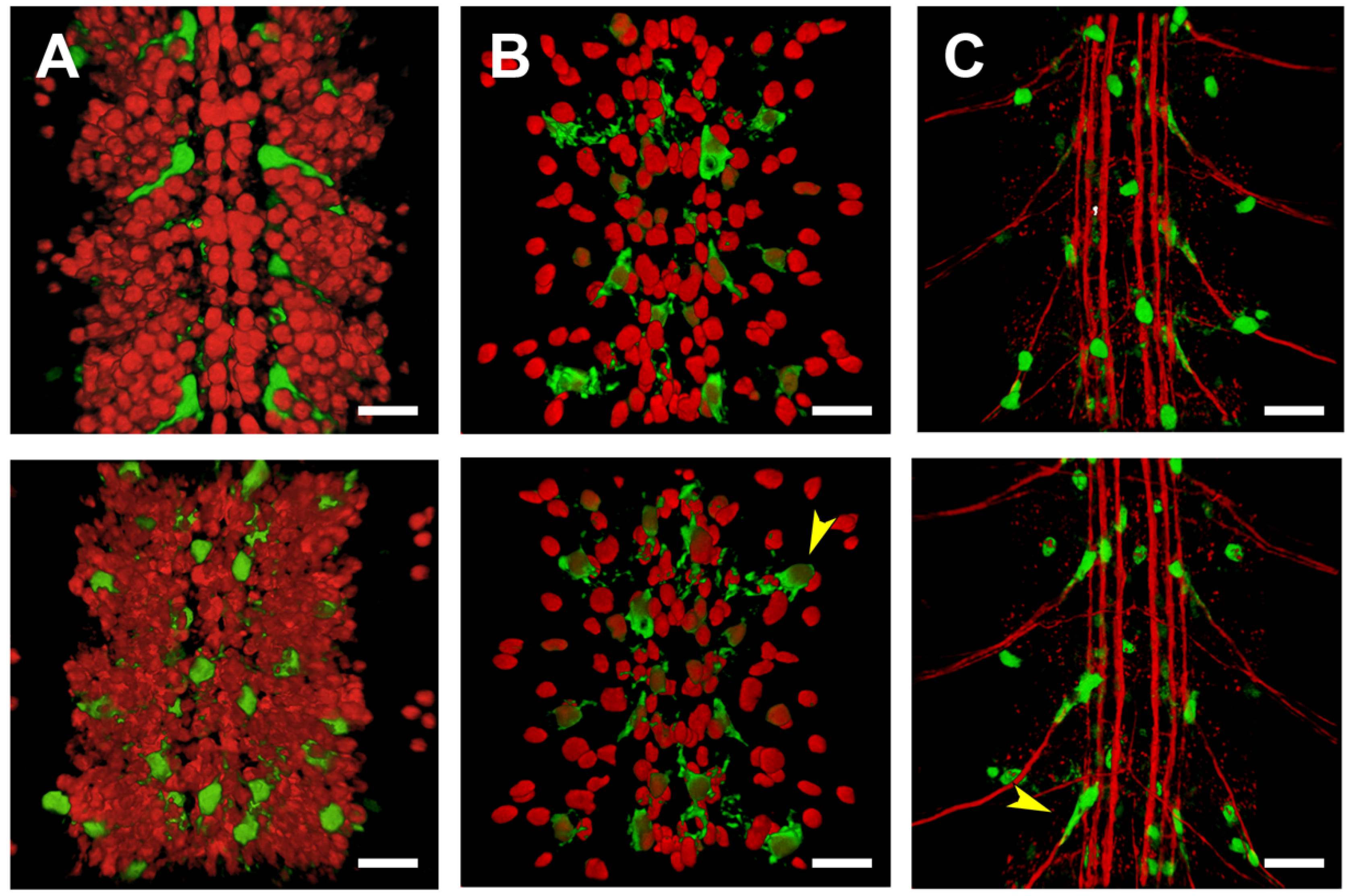
2.2. Analysis of Lines Carrying puc Regions In Vivo
2.3. Upstream Sequences (PG1)
2.4. First and Second Intron (PG2, RS1)
2.5. Third Intron (PG3, PG4, RS2–4)
2.6. Downstream Sequences (PG5)
2.7. Regulatory Control of puc Expression by the JNK Signaling Cascade
3. Discussion
3.1. puc Expression Regulatory Domains
3.2. puc Expression Regulatory Factors
4. Methods
4.1. Cloning of puc Regulatory Sequences
4.2. Bioinformatic Analyses
4.3. Drosophila Strains and Genetics
4.4. Drosophila Embryo Fixation and Dissections
4.5. Immunohistochemistry
4.6. Image Acquisition
4.7. Larvae Live Imaging
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lemmon, M.A.; Freed, D.M.; Schlessinger, J.; Kiyatkin, A. The Dark Side of Cell Signaling: Positive Roles for Negative Regulators. Cell 2016, 164, 1172–1184. [Google Scholar] [CrossRef] [Green Version]
- Johnson, G.L.; Lapadat, R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 2002, 298, 1911–1912. [Google Scholar] [CrossRef] [Green Version]
- Hibi, M.; Lin, A.; Smeal, T.; Minden, A.; Karin, M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993, 7, 2135–2148. [Google Scholar] [CrossRef] [Green Version]
- Lazarus, P.; Calcagnotto, A. Identification and partial characterization of the c-jun oncogene in Xenopus laevis. Cancer Lett. 1994, 86, 201–208. [Google Scholar] [CrossRef]
- Ip, Y.T.; Davis, R.J. Signal transduction by the c-Jun N-terminal kinase (JNK)—From inflammation to development. Curr. Opin. Cell Biol. 1998, 10, 205–219. [Google Scholar] [CrossRef]
- Martin-Blanco, E.; Pastor-Pareja, J.C.; Garcia-Bellido, A. JNK and decapentaplegic signaling control adhesiveness and cytoskeleton dynamics during thorax closure in Drosophila. Proc. Natl. Acad. Sci. USA 2000, 97, 7888–7893. [Google Scholar] [CrossRef] [Green Version]
- Llense, F.; Martin-Blanco, E. JNK signaling controls border cell cluster integrity and collective cell migration. Curr. Biol. 2008, 18, 538–544. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Karin, M. The control of cell motility and epithelial morphogenesis by Jun kinases. Trends Cell Biol. 2004, 14, 94–101. [Google Scholar] [CrossRef]
- Nikoloudaki, G.; Brooks, S.; Peidl, A.P.; Tinney, D.; Hamilton, D.W. JNK Signaling as a Key Modulator of Soft Connective Tissue Physiology, Pathology, and Healing. Int. J. Mol. Sci. 2020, 21, 1015. [Google Scholar] [CrossRef] [Green Version]
- Craige, S.M.; Chen, K.; Blanton, R.M.; Keaney, J.F., Jr.; Kant, S. JNK and cardiometabolic dysfunction. Biosci. Rep. 2019, 39, 1921. [Google Scholar] [CrossRef] [Green Version]
- De Los Reyes Corrales, T.; Losada-Perez, M.; Casas-Tinto, S. JNK Pathway in CNS Pathologies. Int. J. Mol. Sci. 2021, 22, 3883. [Google Scholar] [CrossRef]
- Pereira, A.M.; Tudor, C.; Kanger, J.S.; Subramaniam, V.; Martin-Blanco, E. Integrin-dependent activation of the JNK signaling pathway by mechanical stress. PLoS ONE 2011, 6, e26182. [Google Scholar] [CrossRef] [Green Version]
- Pereira, A.M.; Tudor, C.; Pouille, P.A.; Shekhar, S.; Kanger, J.S.; Subramaniam, V.; Martin-Blanco, E. Plasticity of the MAPK signaling network in response to mechanical stress. PLoS ONE 2014, 9, e101963. [Google Scholar] [CrossRef] [Green Version]
- Kyriakis, J.M.; Avruch, J. Protein kinase cascades activated by stress and inflammatory cytokines. Bioessays 1996, 18, 567–577. [Google Scholar] [CrossRef]
- Davis, R.J. Signal transduction by the JNK group of MAP kinases. Cell 2000, 103, 239–252. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.H.; Giraud, J.; Davis, R.J.; White, M.F. c-Jun N-terminal kinase (JNK) mediates feedback inhibition of the insulin signaling cascade. J. Biol. Chem. 2003, 278, 2896–2902. [Google Scholar] [CrossRef] [Green Version]
- Glise, B.; Bourbon, H.; Noselli, S. hemipterous encodes a novel Drosophila MAP kinase kinase, required for epithelial cell sheet movement. Cell 1995, 83, 451–461. [Google Scholar] [CrossRef] [Green Version]
- Riesgo-Escovar, J.R.; Jenni, M.; Fritz, A.; Hafen, E. The Drosophila Jun-N-terminal kinase is required for cell morphogenesis but not for DJun-dependent cell fate specification in the eye. Genes Dev. 1996, 10, 2759–2768. [Google Scholar] [CrossRef] [Green Version]
- Harden, N. Signaling pathways directing the movement and fusion of epithelial sheets: Lessons from dorsal closure in Drosophila. Differentiation 2002, 70, 181–203. [Google Scholar] [CrossRef]
- Homsy, J.G.; Jasper, H.; Peralta, X.G.; Wu, H.; Kiehart, D.P.; Bohmann, D. JNK signaling coordinates integrin and actin functions during Drosophila embryogenesis. Dev. Dyn. 2006, 235, 427–434. [Google Scholar] [CrossRef]
- Martin-Blanco, E.; Gampel, A.; Ring, J.; Virdee, K.; Kirov, N.; Tolkovsky, A.M.; Martinez-Arias, A. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 1998, 12, 557–570. [Google Scholar] [CrossRef] [Green Version]
- Martin-Blanco, E. Regulatory control of signal transduction during morphogenesis in Drosophila. Int. J. Dev. Biol. 1998, 42, 363–368. [Google Scholar]
- Ring, J.M.; Martinez Arias, A. puckered, a gene involved in position-specific cell differentiation in the dorsal epidermis of the Drosophila larva. Dev. Suppl. 1993, 251–259. [Google Scholar] [CrossRef]
- Agnes, F.; Suzanne, M.; Noselli, S. The Drosophila JNK pathway controls the morphogenesis of imaginal discs during metamorphosis. Development 1999, 126, 5453–5462. [Google Scholar] [CrossRef]
- Dobens, L.L.; Martin-Blanco, E.; Martinez-Arias, A.; Kafatos, F.C.; Raftery, L.A. Drosophila puckered regulates Fos/Jun levels during follicle cell morphogenesis. Development 2001, 128, 1845–1856. [Google Scholar] [CrossRef]
- Shklover, J.; Mishnaevski, K.; Levy-Adam, F.; Kurant, E. JNK pathway activation is able to synchronize neuronal death and glial phagocytosis in Drosophila. Cell Death Dis. 2015, 6, e1649. [Google Scholar] [CrossRef] [Green Version]
- Reed, B.H.; Wilk, R.; Lipshitz, H.D. Downregulation of Jun kinase signaling in the amnioserosa is essential for dorsal closure of the Drosophila embryo. Curr. Biol. 2001, 11, 1098–1108. [Google Scholar] [CrossRef] [Green Version]
- Ohayon, D.; Pattyn, A.; Venteo, S.; Valmier, J.; Carroll, P.; Garces, A. Zfh1 promotes survival of a peripheral glia subtype by antagonizing a Jun N-terminal kinase-dependent apoptotic pathway. EMBO J. 2009, 28, 3228–3243. [Google Scholar] [CrossRef] [Green Version]
- Zeitlinger, J.; Bohmann, D. Thorax closure in Drosophila: Involvement of Fos and the JNK pathway. Development 1999, 126, 3947–3956. [Google Scholar] [CrossRef]
- Weber, U.; Paricio, N.; Mlodzik, M. Jun mediates Frizzled-induced R3/R4 cell fate distinction and planar polarity determination in the Drosophila eye. Development 2000, 127, 3619–3629. [Google Scholar] [CrossRef]
- Fanto, M.; Weber, U.; Strutt, D.I.; Mlodzik, M. Nuclear signaling by Rac and Rho GTPases is required in the establishment of epithelial planar polarity in the Drosophila eye. Curr. Biol. 2000, 10, 979–988. [Google Scholar] [CrossRef] [Green Version]
- Martin-Blanco, E. Regulation of cell differentiation by the Drosophila Jun kinase cascade. Curr. Opin. Genet. Dev. 1997, 7, 666–671. [Google Scholar] [CrossRef]
- Pastor-Pareja, J.C.; Wu, M.; Xu, T. An innate immune response of blood cells to tumors and tissue damage in Drosophila. Dis. Model. Mech. 2008, 1, 144–154. [Google Scholar] [CrossRef] [Green Version]
- Bosch, M.; Serras, F.; Martin-Blanco, E.; Baguna, J. JNK signaling pathway required for wound healing in regenerating Drosophila wing imaginal discs. Dev. Biol. 2005, 280, 73–86. [Google Scholar] [CrossRef] [Green Version]
- Galko, M.J.; Krasnow, M.A. Cellular and genetic analysis of wound healing in Drosophila larvae. PLoS Biol. 2004, 2, E239. [Google Scholar] [CrossRef] [Green Version]
- Perez-Garijo, A.; Fuchs, Y.; Steller, H. Apoptotic cells can induce non-autonomous apoptosis through the TNF pathway. Elife 2013, 2, e01004. [Google Scholar] [CrossRef]
- Wu, H.; Wang, M.C.; Bohmann, D. JNK protects Drosophila from oxidative stress by trancriptionally activating autophagy. Mech. Dev. 2009, 126, 624–637. [Google Scholar] [CrossRef]
- Srahna, M.; Leyssen, M.; Choi, C.M.; Fradkin, L.G.; Noordermeer, J.N.; Hassan, B.A. A signaling network for patterning of neuronal connectivity in the Drosophila brain. PLoS Biol. 2006, 4, e348. [Google Scholar] [CrossRef]
- Pfeiffer, B.D.; Ngo, T.T.; Hibbard, K.L.; Murphy, C.; Jenett, A.; Truman, J.W.; Rubin, G.M. Refinement of tools for targeted gene expression in Drosophila. Genetics 2010, 186, 735–755. [Google Scholar] [CrossRef] [Green Version]
- Alessi, D.R.; Smythe, C.; Keyse, S.M. The human CL100 gene encodes a Tyr/Thr-protein phosphatase which potently and specifically inactivates MAP kinase and suppresses its activation by oncogenic ras in Xenopus oocyte extracts. Oncogene 1993, 8, 2015–2020. [Google Scholar]
- Adachi-Yamada, T.; Fujimura-Kamada, K.; Nishida, Y.; Matsumoto, K. Distortion of proximodistal information causes JNK-dependent apoptosis in Drosophila wing. Nature 1999, 400, 166–169. [Google Scholar] [CrossRef]
- Kelso, R.J.; Buszczak, M.; Quinones, A.T.; Castiblanco, C.; Mazzalupo, S.; Cooley, L. Flytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster. Nucleic Acids Res. 2004, 32, D418–D420. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Huang, J.; Yang, L.; Yang, Y.; Li, W.; Xue, L. NOPO modulates Egr-induced JNK-independent cell death in Drosophila. Cell Res. 2012, 22, 425–431. [Google Scholar] [CrossRef] [Green Version]
- Weston, C.R.; Davis, R.J. The JNK signal transduction pathway. Curr. Opin. Cell Biol. 2007, 19, 142–149. [Google Scholar] [CrossRef]
- Sancho, R.; Nateri, A.S.; de Vinuesa, A.G.; Aguilera, C.; Nye, E.; Spencer-Dene, B.; Behrens, A. JNK signalling modulates intestinal homeostasis and tumourigenesis in mice. EMBO J. 2009, 28, 1843–1854. [Google Scholar] [CrossRef] [Green Version]
- Cellurale, C.; Sabio, G.; Kennedy, N.J.; Das, M.; Barlow, M.; Sandy, P.; Jacks, T.; Davis, R.J. Requirement of c-Jun NH(2)-terminal kinase for Ras-initiated tumor formation. Mol. Cell Biol. 2011, 31, 1565–1576. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Chen, Y.; Xu, W.; Wu, N.; Li, M.; Cao, Y.; Wu, S.; Li, Q.; Xue, L. Impaired Hippo signaling promotes Rho1-JNK-dependent growth. Proc. Natl. Acad. Sci. USA 2015, 112, 1065–1070. [Google Scholar] [CrossRef] [Green Version]
- Carrasco-Rando, M.; Tutor, A.S.; Prieto-Sanchez, S.; Gonzalez-Perez, E.; Barrios, N.; Letizia, A.; Martin, P.; Campuzano, S.; Ruiz-Gomez, M. Drosophila araucan and caupolican integrate intrinsic and signalling inputs for the acquisition by muscle progenitors of the lateral transverse fate. PLoS Genet. 2011, 7, e1002186. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Silverman, N. Fighting infection fly-style. Fly 2007, 1, 106–109. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Wu, C.; Ding, X.; Li, W.; Xue, L. Toll signaling promotes JNK-dependent apoptosis in Drosophila. Cell Div. 2020, 15, 7. [Google Scholar] [CrossRef]
- Wagner, C.; Uliczka, K.; Bossen, J.; Niu, X.; Fink, C.; Thiedmann, M.; Knop, M.; Vock, C.; Abdelsadik, A.; Zissler, U.M.; et al. Constitutive immune activity promotes JNK- and FoxO-dependent remodeling of Drosophila airways. Cell Rep. 2021, 35, 108956. [Google Scholar] [CrossRef]
- McEwen, D.G.; Cox, R.T.; Peifer, M. The canonical Wg and JNK signaling cascades collaborate to promote both dorsal closure and ventral patterning. Development 2000, 127, 3607–3617. [Google Scholar] [CrossRef]
- Ryoo, H.D.; Gorenc, T.; Steller, H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev. Cell 2004, 7, 491–501. [Google Scholar] [CrossRef] [Green Version]
- Shokri, L.; Inukai, S.; Hafner, A.; Weinand, K.; Hens, K.; Vedenko, A.; Gisselbrecht, S.S.; Dainese, R.; Bischof, J.; Furger, E.; et al. A Comprehensive Drosophila melanogaster Transcription Factor Interactome. Cell Rep. 2019, 27, 955–970. [Google Scholar] [CrossRef] [Green Version]
- Abrams, E.W.; Andrew, D.J. CrebA regulates secretory activity in the Drosophila salivary gland and epidermis. Development 2005, 132, 2743–2758. [Google Scholar] [CrossRef] [Green Version]
- Tubon, T.C., Jr.; Zhang, J.; Friedman, E.L.; Jin, H.; Gonzales, E.D.; Zhou, H.; Drier, D.; Gerstner, J.R.; Paulson, E.A.; Fropf, R.; et al. dCREB2-mediated enhancement of memory formation. J. Neurosci. 2013, 33, 7475–7487. [Google Scholar] [CrossRef] [Green Version]
- Ramet, M.; Lanot, R.; Zachary, D.; Manfruelli, P. JNK signaling pathway is required for efficient wound healing in Drosophila. Dev. Biol. 2002, 241, 145–156. [Google Scholar] [CrossRef] [Green Version]
- Igaki, T.; Miura, M. The Drosophila TNF ortholog Eiger: Emerging physiological roles and evolution of the TNF system. Semin. Immunol. 2014, 26, 267–274. [Google Scholar] [CrossRef] [Green Version]
- Nam, H.J.; Jang, I.H.; You, H.; Lee, K.A.; Lee, W.J. Genetic evidence of a redox-dependent systemic wound response via Hayan protease-phenoloxidase system in Drosophila. EMBO J. 2012, 31, 1253–1265. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Zhang, K.; Sandoval, H.; Yamamoto, S.; Jaiswal, M.; Sanz, E.; Li, Z.; Hui, J.; Graham, B.H.; Quintana, A.; et al. Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell 2015, 160, 177–190. [Google Scholar] [CrossRef] [Green Version]
- Karkali, K.; Saunders, T.E.; Vernon, S.W.; Baines, R.A.; Panayotou, G.; Martín-Blanco, E. JNK signaling in pioneer neurons directs the architectural organization of the CNS and coordinates the motor activity of the Drosophila embryo. bioRxiv 2020, 092486. [Google Scholar] [CrossRef] [Green Version]
- Lacin, H.; Zhu, Y.; Wilson, B.A.; Skeath, J.B. dbx mediates neuronal specification and differentiation through cross-repressive, lineage-specific interactions with eve and hb9. Development 2009, 136, 3257–3266. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.A.; Su, M.T. Excessive Dpp signaling induces cardial apoptosis through dTAK1 and dJNK during late embryogenesis of Drosophila. J. Biomed. Sci. 2011, 18, 85. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Boutros, M. JNK-dependent intestinal barrier failure disrupts host-microbe homeostasis during tumorigenesis. Proc. Natl. Acad. Sci. USA 2020, 117, 9401–9412. [Google Scholar] [CrossRef] [Green Version]
- Cohen, S.M.; Jurgens, G. Proximal-distal pattern formation in Drosophila: Cell autonomous requirement for Distal-less gene activity in limb development. EMBO J. 1989, 8, 2045–2055. [Google Scholar] [CrossRef]
- Parrish, J.Z.; Kim, M.D.; Jan, L.Y.; Jan, Y.N. Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes Dev. 2006, 20, 820–835. [Google Scholar] [CrossRef]
- Lacin, H.; Truman, J.W. Lineage mapping identifies molecular and architectural similarities between the larval and adult Drosophila central nervous system. Elife 2016, 5, e13399. [Google Scholar] [CrossRef]
- Broihier, H.T.; Skeath, J.B. Drosophila homeodomain protein dHb9 directs neuronal fate via crossrepressive and cell-nonautonomous mechanisms. Neuron 2002, 35, 39–50. [Google Scholar] [CrossRef] [Green Version]
- Odden, J.P.; Holbrook, S.; Doe, C.Q. Drosophila HB9 is expressed in a subset of motoneurons and interneurons, where it regulates gene expression and axon pathfinding. J. Neurosci. 2002, 22, 9143–9149. [Google Scholar] [CrossRef] [Green Version]
- Sharma, Y.; Cheung, U.; Larsen, E.W.; Eberl, D.F. PPTGAL, a convenient Gal4 P-element vector for testing expression of enhancer fragments in Drosophila. Genesis 2002, 34, 115–118. [Google Scholar] [CrossRef]
- Larkin, A.; Marygold, S.J.; Antonazzo, G.; Attrill, H.; Dos Santos, G.; Garapati, P.V.; Goodman, J.L.; Gramates, L.S.; Millburn, G.; Strelets, V.B.; et al. FlyBase: Updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 2021, 49, D899–D907. [Google Scholar] [CrossRef]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef]
- Kel, A.E.; Gossling, E.; Reuter, I.; Cheremushkin, E.; Kel-Margoulis, O.V.; Wingender, E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003, 31, 3576–3579. [Google Scholar] [CrossRef] [Green Version]
- Fornes, O.; Castro-Mondragon, J.A.; Khan, A.; van der Lee, R.; Zhang, X.; Richmond, P.A.; Modi, B.P.; Correard, S.; Gheorghe, M.; Baranasic, D.; et al. JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020, 48, D87–D92. [Google Scholar] [CrossRef]
- Patel, N.H. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Methods Cell Biol. 1994, 44, 445–487. [Google Scholar] [CrossRef]
- Landgraf, M.; Bossing, T.; Technau, G.M.; Bate, M. The origin, location, and projections of the embryonic abdominal motorneurons of Drosophila. J. Neurosci. 1997, 17, 9642–9655. [Google Scholar] [CrossRef] [Green Version]

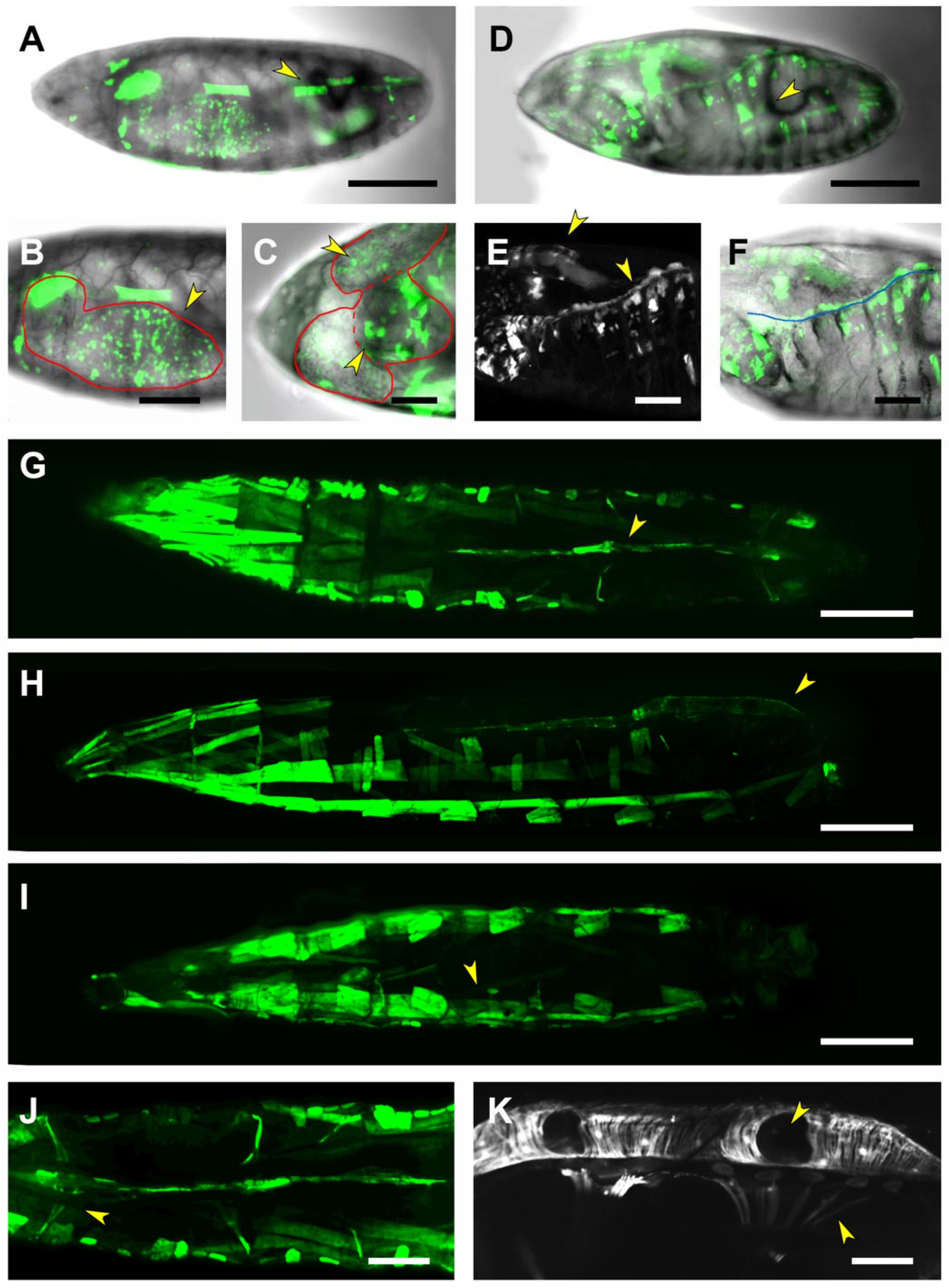
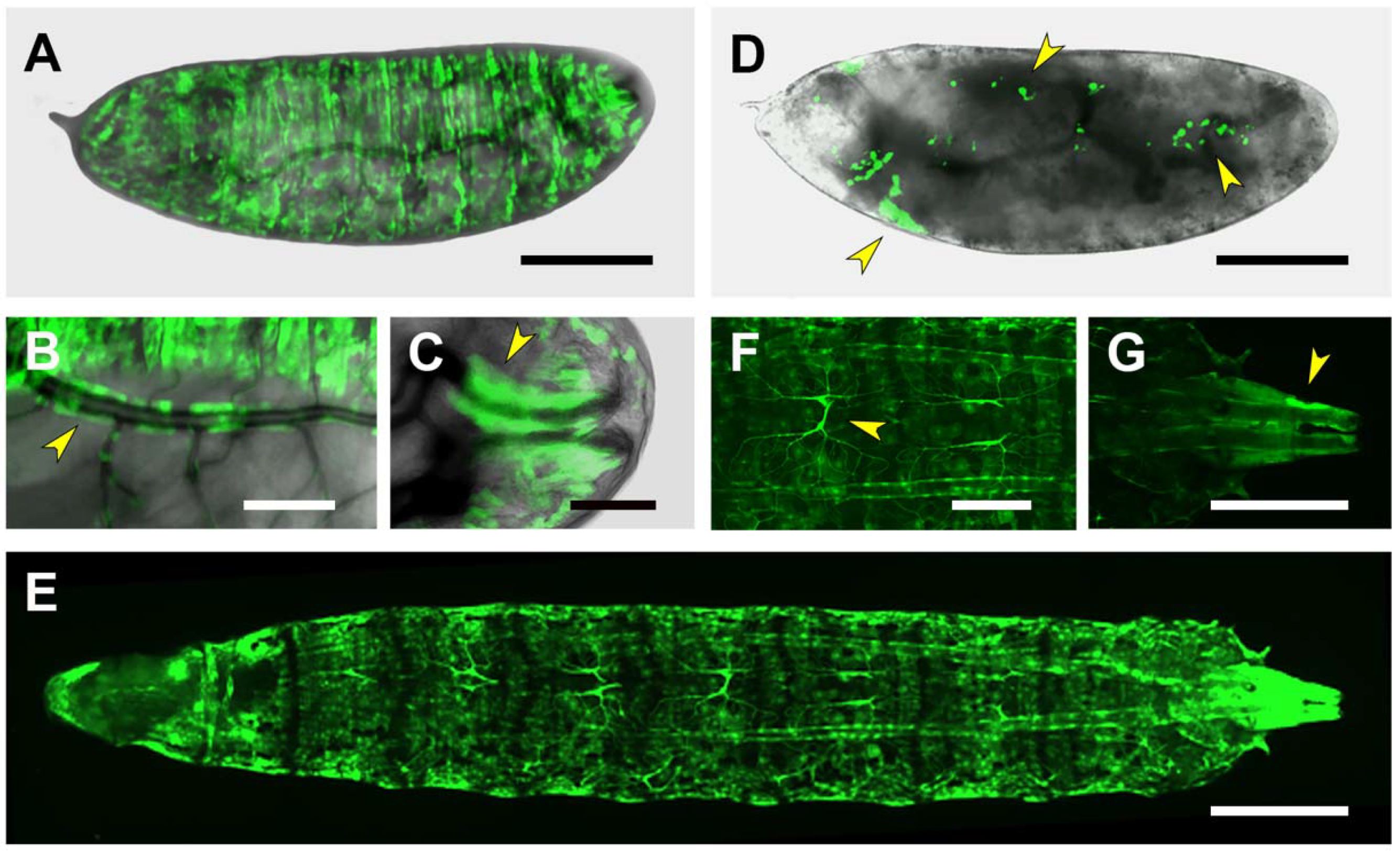
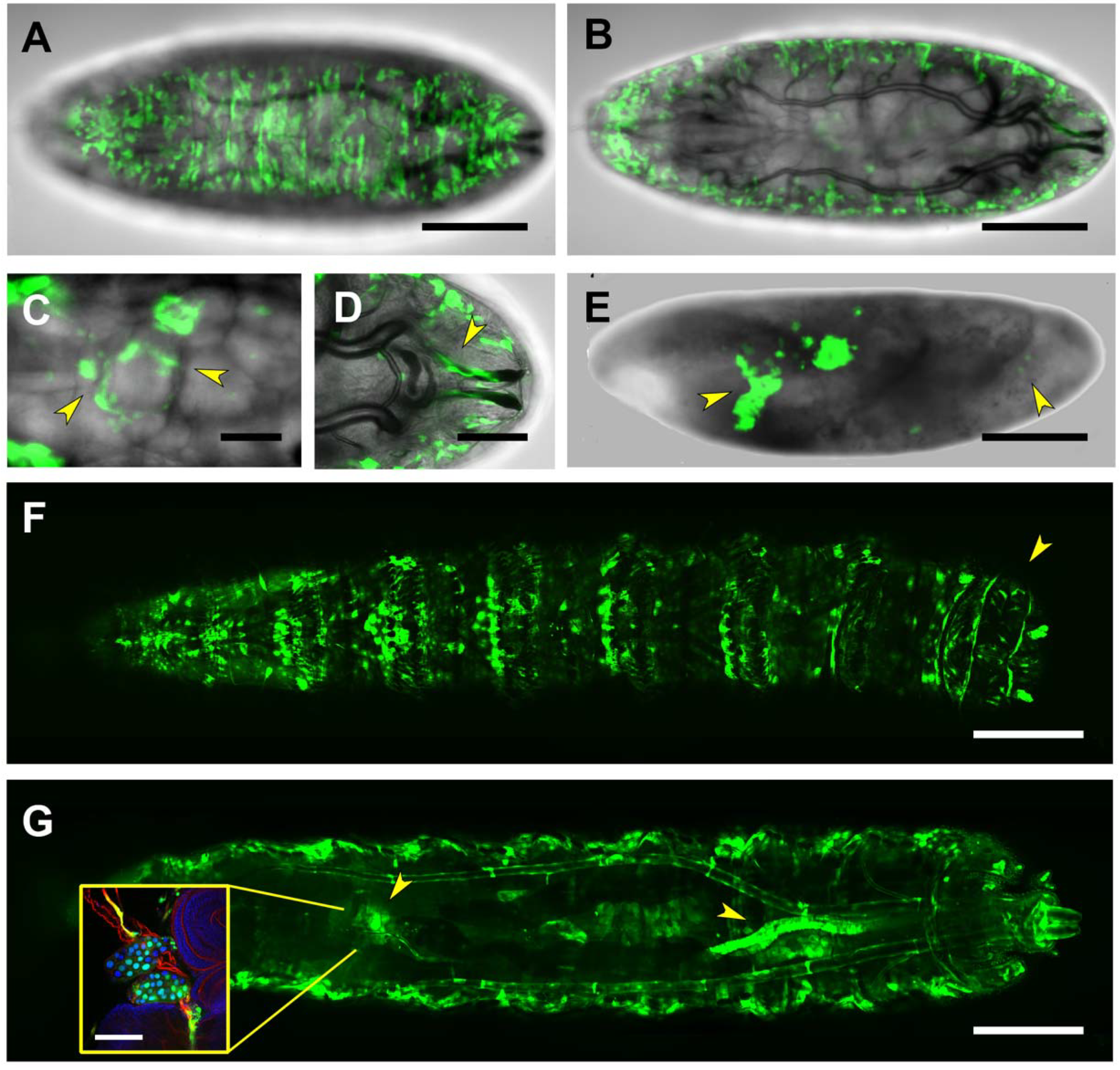
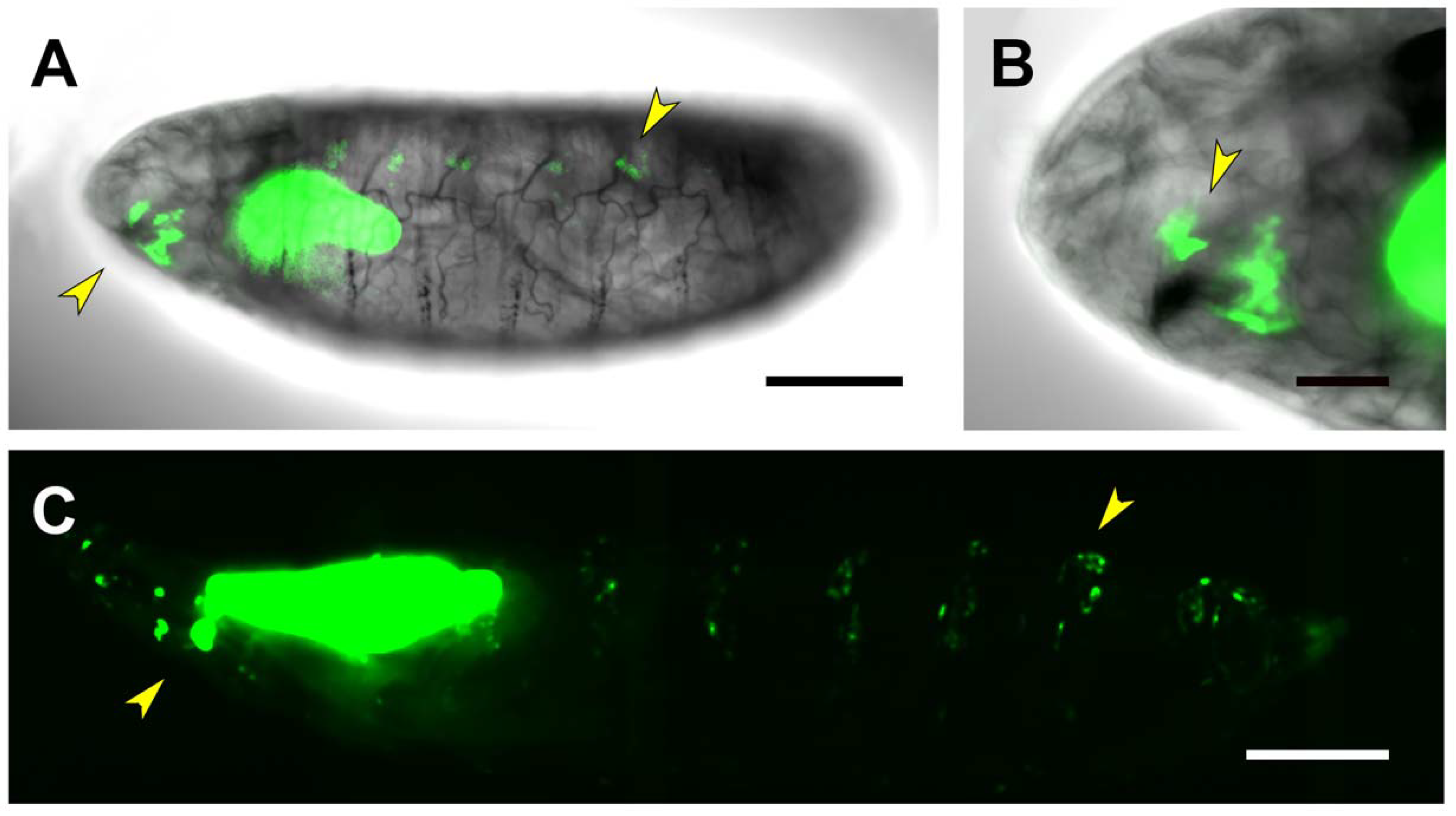
| Embryo | CNS Embryo/1st Instar Larvae | 3rd Instar Larvae | |
|---|---|---|---|
| PG1 | Salivary Glands | Salivary Glands | |
| Posterior Spiracles | Posterior Spiracles | ||
| Brain lobes Neurons | Denticles precursors | ||
| Thoracic ganglion Neurons | |||
| PG2 | Posterior Spiracles | Posterior Spiracles | |
| CNS | CNS Glia | CNS | |
| Epidermis (patchy) | · Perineural/Subperineural | · VNC subset | |
| Tracheal System | · Ensheating | · Glia intersegmental nerve | |
| Foregut | Epidermis | ||
| Tracheal System | |||
| Foregut | |||
| Posterior midgut | |||
| Anal pad | |||
| RS1 | Salivary Glands | ||
| Posterior Spiracles | Posterior Spiracles | ||
| Hindgut | Epidermis (patchy) | ||
| Maxillary primordia | Tracheal System (patchy) | ||
| Epidermis | Maxillary primordia | ||
| PG3 | Salivary Glands | Salivary Glands | |
| CNS | CNS Glia | Midgut/Hindgut | |
| Midgut (weak) | · Repo-negative midline | Body wall muscles | |
| Body wall muscles | · Surface | Heart | |
| Pharynx/ring gland | VNC tracheal arborization | Alary muscles | |
| RS2 | Salivary Glands | Salivary Glands | |
| Posterior Spiracles | |||
| CNS | CNS | Body wall muscles (subset) | |
| Body wall muscles (subset) | · Neurons | Heart | |
| Healing Epithelia | · Glia | Alary muscles | |
| Midgut | Anal pad periphery | ||
| RS3 | Salivary Glands | Salivary Glands | |
| Posterior Spiracles | Posterior Spiracles | ||
| Epidermis | Epidermis | ||
| Tracheal System | Tracheal System | ||
| · Dorsal trunks | |||
| · Distal arborization | |||
| RS4 | Salivary Glands | Salivary Glands | |
| Posterior Spiracles | Posterior Spiracles | ||
| Tracheal System | |||
| Anal pad | |||
| Maxillary primordia | |||
| PG4 | Salivary Glands | Salivary Glands | |
| Posterior Spiracles | Posterior Spiracles | ||
| Epidermis (patchy) | Brain lobes/VNC Interneurons | Epidermis (patchy) | |
| Ring gland | Nerves axonal projections | Ring gland | |
| Astrocytes | Posterior midgut | ||
| Ensheathing glia | Tracheal junctions | ||
| PG5 | Salivary Glands | Salivary Glands | |
| Posterior Spiracles | Posterior Spiracles | ||
| Epidermis (scattered) | Epidermis (scattered) | ||
| Maxillary primordia? |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karkali, K.; Martin-Blanco, E. Dissection of the Regulatory Elements of the Complex Expression Pattern of Puckered, a Dual-Specificity JNK Phosphatase. Int. J. Mol. Sci. 2021, 22, 12205. https://doi.org/10.3390/ijms222212205
Karkali K, Martin-Blanco E. Dissection of the Regulatory Elements of the Complex Expression Pattern of Puckered, a Dual-Specificity JNK Phosphatase. International Journal of Molecular Sciences. 2021; 22(22):12205. https://doi.org/10.3390/ijms222212205
Chicago/Turabian StyleKarkali, Katerina, and Enrique Martin-Blanco. 2021. "Dissection of the Regulatory Elements of the Complex Expression Pattern of Puckered, a Dual-Specificity JNK Phosphatase" International Journal of Molecular Sciences 22, no. 22: 12205. https://doi.org/10.3390/ijms222212205






