Antibacterial Activity and Amphidinol Profiling of the Marine Dinoflagellate Amphidinium carterae (Subclade III)
Abstract
:1. Introduction
2. Results
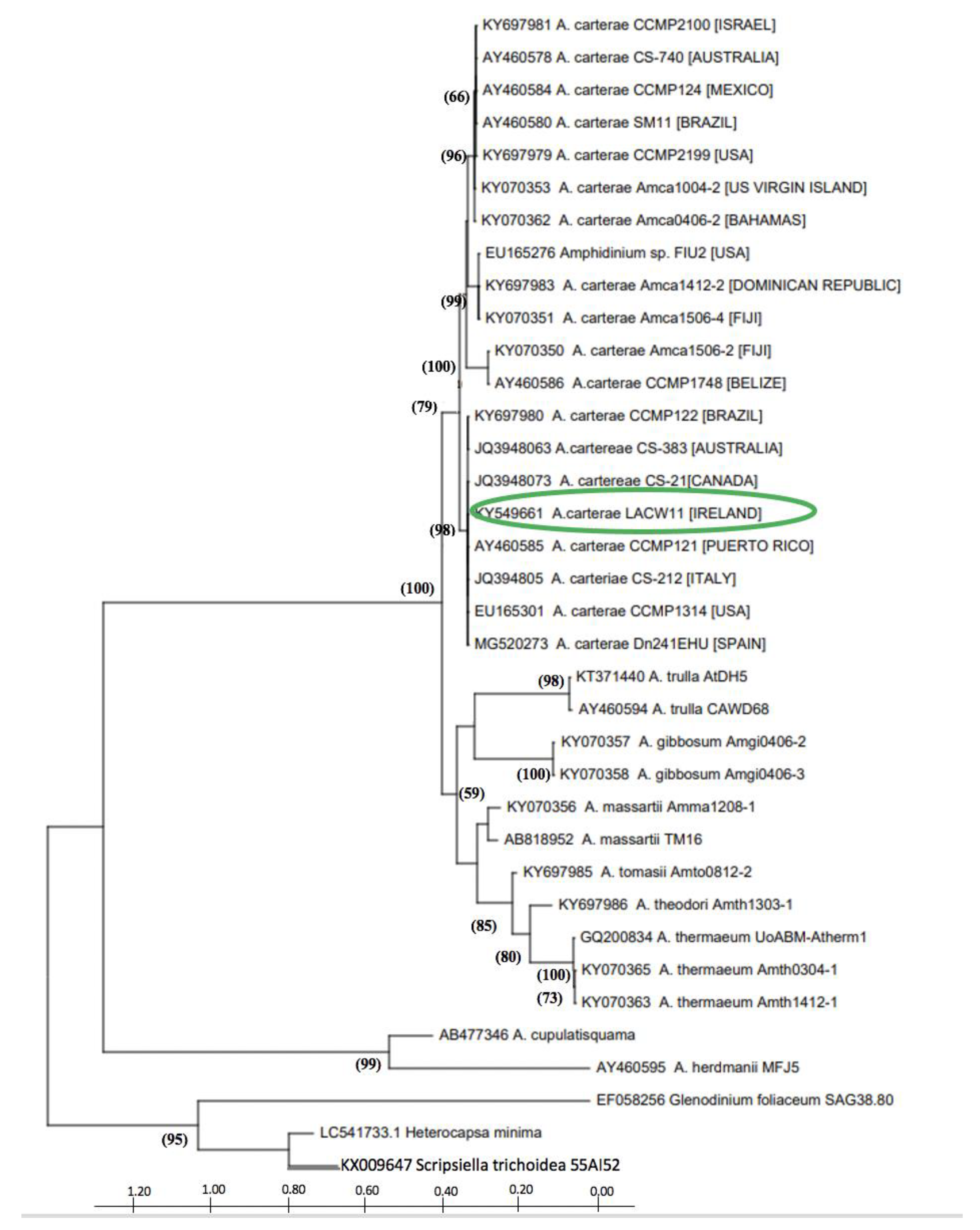
2.1. Phylogenetic Characterisation of Strain A. carterae LACW11
2.2. Amphidinium carterae Culture
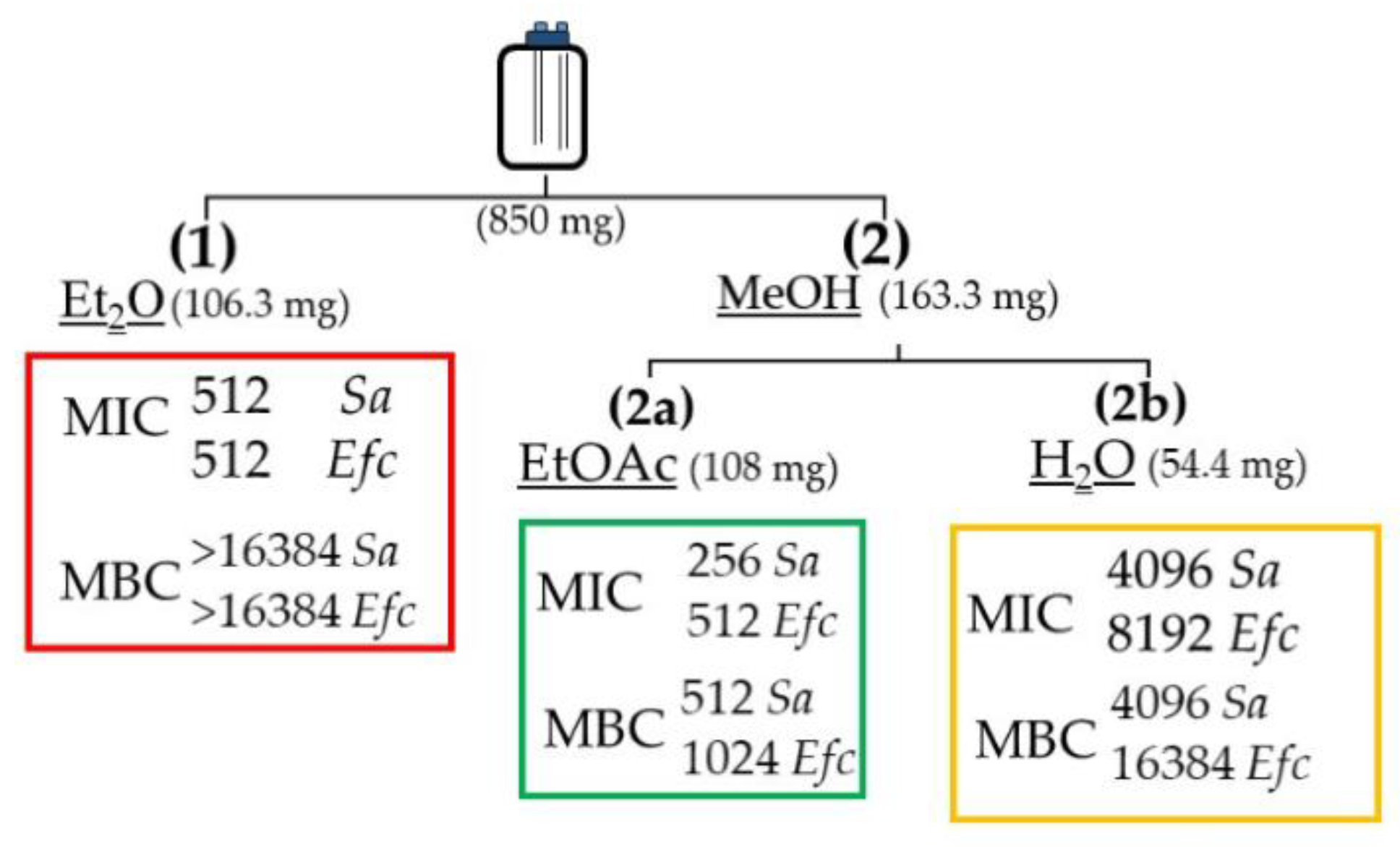
2.3. Stage-1: Bioactivity Assays on the Extracts
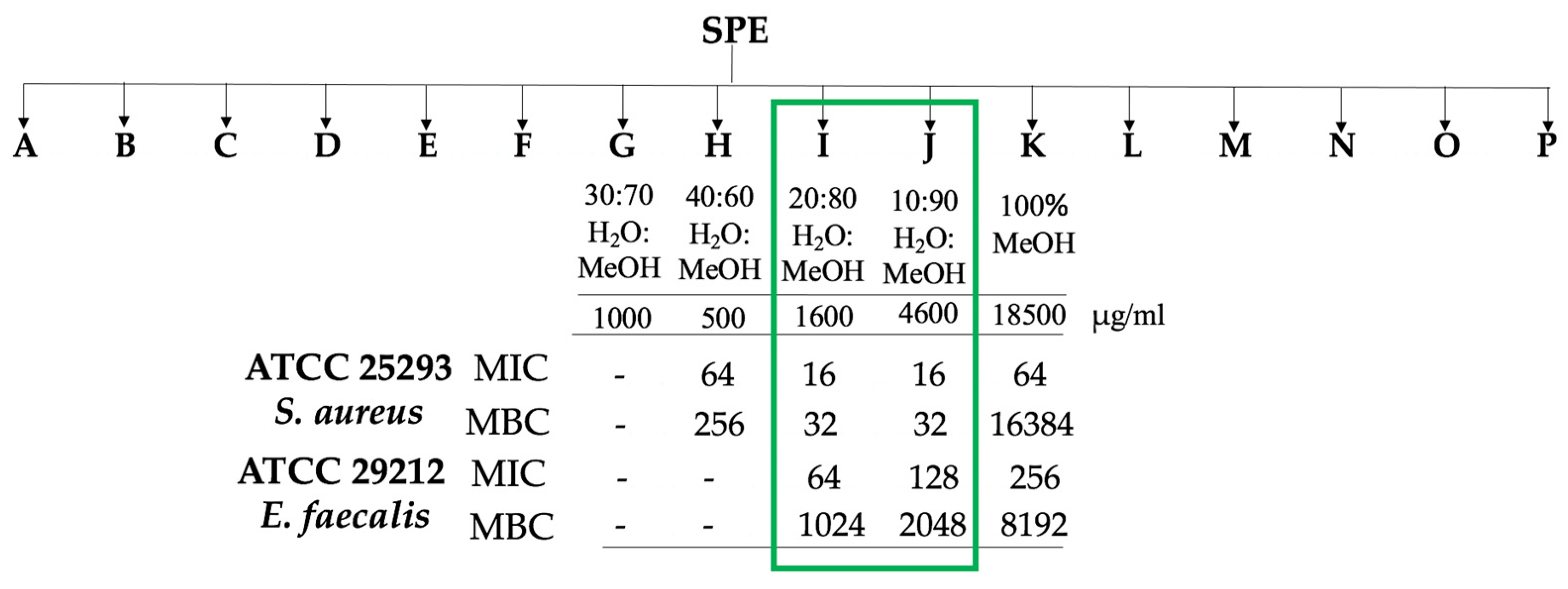
2.4. Stage-2: Bioactivity Assays on the Fractions
2.4.1. Microbial Assays
2.4.2. Chemical Profiling of the Bioactive Fractions
3. Discussion
4. Materials and Methods
4.1. Amphidinium carterae Cultivation
4.2. DNA Extraction, Partial 28SrDNA Gene PCR and Sequencing
4.3. Phylogenetic Inference
4.4. Extraction and Fractionation of the Biomass
4.5. Chemical Profiling of the Fractions by LC-MS
4.6. Microbiological Assays
4.6.1. Bacterial Strains
4.6.2. Determination of Minimum Inhibitory Concentrations (MIC)
4.6.3. Determination of Minimum Bactericidal Concentration (MBC)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization (WHO). Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report 2021; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Beceiro, A.; Tomás, M.; Bou, G. Antimicrobial Resistance and Virulence: A Successful or Deleterious Association in the Bacterial World? Clin. Microbiol. Rev. 2013, 26, 185–230. [Google Scholar] [CrossRef] [Green Version]
- Landecker, H. Antibiotic Resistance and Biology of History. Body Soc. 2015, 22, 1–34. [Google Scholar] [CrossRef] [Green Version]
- European Centre for Disease Prevention and Control (ECDC). Antimicrobial Resistance in the EU/EEA (EARS-Net) Annual Epidemiological Report 2019; ECDC: Solna kommun, Sweden, 2019. [Google Scholar]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2015, 32, 116–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cepas, V.; López, Y.; Gabasa, Y.; Martins, C.B.; Ferreira, J.D.; Correia, M.J.; Santos, L.M.A.; Oliveira, F.; Ramos, V.; Reis, M.; et al. Inhibition of Bacterial and Fungal Biofilm Formation by 675 Extracts from Microalgae and Cyanobacteria. Antibiotics 2019, 8, 77. [Google Scholar] [CrossRef] [Green Version]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine Natural Products. Nat. Prod. Rep. 2021, 38, 362–413. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, F.; Pinto, E.; Kijjoa, A.; Pinto, M.; Sousa, E. Targeting antimicrobial drug resistance with marine natural products. Int. J. Antimicrob. Agents 2020, 56, 106005. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.; Shin, J.H.; Kim, J.D. The promising future of microalgae: Current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb. Cell Factories 2018, 17, 36. [Google Scholar] [CrossRef] [PubMed]
- Brennan, L.; Owende, P. Biofuels from microalgae—A review of technologies for production, processing, and extractions of biofuels and co-products. Renew. Suistain. Energy Rev. 2010, 14, 557–577. [Google Scholar] [CrossRef]
- Shannon, E.; Abu-Ghannam, N. Antibacterial Derivatives of Marine Algae: An Overview of Pharmacological Mechanisms and Applications. Mar. Drugs 2016, 14, 81. [Google Scholar] [CrossRef]
- Sanmukh, S.; Bruno, B.; Ramakrishnan, U.; Khairnar, K.; Swaminathan, S.; Paunikar, W. Bioactive Compounds Derived from Microalgae Showing Antimicrobial Activities. J. Aquac. Res. Dev. 2014, 5, 3. [Google Scholar] [CrossRef] [Green Version]
- Karpiński, T.M. Marine Macrolides with Antibacterial and/or Antifungal Activity (Review). Mar. Drugs 2019, 17, 241. [Google Scholar] [CrossRef] [Green Version]
- Karpiński, T.M.; Adamczak, A. Fucoxanthin—An Antibacterial Carotenoids. Antioxidants 2019, 8, 239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dewi, C.; Falaise, C.; Hellio, C.; Bouigougnon, N.; Mouget, J. Chapter 12—Anticancer, Antiviral, Antibacterial, and Antifungal Properties in Microalgae. Microalgae Health Dis. Prev. 2018, 12, 235–261. [Google Scholar]
- Falaise, C.; François, C.; Travers, M.; Morga, B.; Haure, J.; Tremblay, R.; Turcotte, F.; Pasetto, P.; Gastineau, R.; Hardivillier, Y.; et al. Antimicrobial Compounds from Eukaryotic Microalgae against Human Pathogens and Diseases in Aquaculture. Mar. Drugs 2016, 14, 159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desbois, A.P.; Mearns-Spragg, A.; Smith, V.J. A fatty acid from the diatom Phaeodactylum tricornutum is antibacterial against diverse bacteria including multi-resistant Staphylococcus aureus (MRSA). Mar. Biotechnol. 2009, 11, 45–52. [Google Scholar] [CrossRef]
- Berstein, P.S.; Li, B.; Vachali, P.P.; Gorusupudi, A.; Shyam, R.; Henriksen, B.S.; Nolan, J.M. Lutein, Zeaxanthin, and meso-Zeaxanthin: The basic and clinical science underlying carotenoid-base nutritional interventions against ocular disease. Prog. Retin. Eye Res. 2016, 50, 34–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venugopal, V.C.; Thakur, A.; Chennabasappa, L.K.; Mishra, G.; Singh, K.; Rathee, P.; Ranjan, A. Phycocyanin Extracted from Oscillatoria minima Shows Antimicrobial, Algicidal, and Antiradical Activities: In silico and in vitro Analysis. Antiinflamm. Antiallergy Agents Med. Chem. 2020, 19, 240–253. [Google Scholar] [CrossRef]
- Garcia Camacho, F.; Gallardo Rodrígez, J.; Sánchez Mirón, A.; Cerón García, M.C.; Belarbi, E.H.; Chisti, Y.; Molina Grima, E. Biotechnological significance of toxic marine dinoflagellates. Biotechnol. Adv. 2007, 25, 176–194. [Google Scholar] [CrossRef]
- Kobayashi, J.; Kubota, T. Bioactive macrolides and polyketides from marine dinoflagellates of the genus Amphidinium. J. Nat. Prod. 2007, 70, 451–460. [Google Scholar] [CrossRef]
- Kellmann, R.; Stüken, A.; Orr, R.J.S.; Svendesen, H.M.; Jakobsen, K.S. Biosynthesis and Molecular Genetics of Polyketides in Marine Dinoflagellates. Mar. Drugs 2010, 8, 1011–1048. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, J.; Shimbo, K.; Kubota, T.; Tsuda, M. Bioactive macrolides and polyketides from marine dinoflagellates. Pure Appl. Chem. 2003, 75, 337–342. [Google Scholar] [CrossRef]
- Kubota, T.; Takahashi, A.; Tsuda, M.; Kobayashi, J. Luteophanol D, New Polyhydroxyl Metabolite from Marine Dinoflagellate Amphidinium sp. Mar. Drugs 2005, 3, 113–118. [Google Scholar] [CrossRef] [Green Version]
- Martinez, K.A.; Lauritano, C.; Druka, D.; Romano, G.; Grohmann, T.; Jaspars, M.; Martín, J.; Díaz, C.; Cautain, B.; De la Cruz, M.; et al. Amphidinol 22, a New Cytotoxic and Antifungal Amphidinol from the Dinoflagellate Amphidinium carterae. Mar. Drugs 2019, 17, 385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satake, M.; Murata, M.; Yasumoto, T.; Naoki, H. Amphidinol, a polyhydroxy-polyene antifungal agent with an unprecedented structure, from a marine dinoflagellate, Amphidinium klebsi. J. Am. Chem. Soc. 1991, 112, 9859–9861. [Google Scholar] [CrossRef]
- Houdai, T.; Matsuoka, S.; Matsumori, N.; Murata, M. Membrane-permeabilizing activities of amphidinol 3, polyene-polyhydroxy antifungal from a marine dinoflagellate. Biochim. Biophys. Acta 2004, 1667, 91–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cutignano, A.; Nuzzo, G.; Sardo, A.; Fontana, A. The Missing Piece in Biosynthesis of Amphidinols: First Evidence of Glycolate as a Starter Unit in New Polyketides from Amphidinium carterae. Mar. Drugs 2017, 15, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wellkamp, M.; Garcis-Camacho, F.; Duran-Riveroll, L.M.; Tebben, J.; Tillmann, U.; Krock, B. LC-MS/MS Method Development for the Discovery and Identification of Amphidinols Produced by Amphidinium. Mar. Drugs 2020, 18, 497. [Google Scholar] [CrossRef]
- Bradley, J.; Glasser, C.; Patino, H.; Arnold, S.R.; Arrieta, A.; Congeni, B.; Daum, R.S.; Kojaoghlanian, T.; Yoon, M.; Anastasiou, D.; et al. Daptomycin for Complicated Skin Infections: A Randomized Trial. Pediatrics 2017, 139, e20162477. [Google Scholar] [CrossRef] [Green Version]
- Campanile, F.; Bongiorno, D.; Borbone, S.; Venditti, M.; Giannella, M.; Franchi, C.; Stefani, S. Characterization of a Variant of the SCCmec Element in a Bloodstream Isolate od Staphylococcus intermedius. Microb. Drug Resist. 2007, 13, 7–10. [Google Scholar] [CrossRef]
- Murray, S.; Jørgensen, M.F.; Daugbjerg, N.; Rhodes, L. Amphidinium revisited. ii. resolving species boundaries in the Amphidinium operculatum species complex (dinophyceae), including the descriptions of Amphidinium trulla sp. nov. and Amphidinium gibbosum. comb. Nov. 1. J. Phycol. 2004, 40, 366–382. [Google Scholar] [CrossRef] [Green Version]
- Desbois, A.P.; Smith, V.J. Antibacterial free fatty acids: Activities, mechanisms of action and biotechnological potential. Appl. Microbiol. Biotechnol. 2010, 85, 1629–1642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molina-Miras, A.; López-Rosales, L.; Sánchez-Mirón, A.; Cerón-García, M.C.; Seoane-Parra, S.; García-Camacho, F.; Molina-Grima, E. Long-term culture of the marine dinoflagellate microalga Amphidinium carterae in an indoor LED-lighted raceway photobioreactor: Production of carotenoids and fatty acids. Bioresour. Technol. 2018, 265, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Grünewald, C.; Bayliss, C.; Fonlut, F.; Chapuli, E. Long-term dinoflagellate culture performance in a commercial photobioreactor: Amphidinium carterae case. Bioresour. Technol. 2016, 218, 533–540. [Google Scholar] [CrossRef] [Green Version]
- Lim, A.S.; Jeong, H.J.; You, J.H.; Park, S.A. Semi-continuous cultivation of the mixotrophic dinoflagellate Gymnodinium smaydae, a new promising microalga for omega-3 production. Algae 2020, 35, 277–292. [Google Scholar] [CrossRef]
- Takahashi, Y.; Kubota, T.; Kobayashi, J. Amphidinolactone B, a New 26-Membered Macrolide from Dinoflagellate Amphidinium sp. J. Antibiot. 2007, 60, 376–379. [Google Scholar] [CrossRef] [Green Version]
- Song, H.Y.; Joo, J.M.; Kang, J.W.; Kim, D.; Jung, C.; Kwak, H.S.; Park, J.H.; Lee, E.; Hong, C.Y.; Jeong, S.; et al. Lasonolide A: Structural Revision and Total Synthesis. J. Org. Chem. 2003, 68, 8080–8087. [Google Scholar] [CrossRef]
- Sakamoto, K.; Hakamata, A.; Tsuda, M.; Fuwa, H. Total Synthesis and Stereochemical Revision of Irimoteolide-2a. Angew. Chem. Int. Ed. 2018, 57, 3801–3805. [Google Scholar] [CrossRef]
- Akakabe, M.; Kumagai, K.; Tsuda, M.; Konica, Y.; Tominaga, A.; Kaneno, D.; Fukushi, E.; Kawabata, J.; Masuda, A.; Tsuda, M. Iriomoteolides-10a and 12a, Cytotoxic Macrolides from Marine Dinoflagellate Amphidinium Species. Chem. Pharm. Bull. 2016, 64, 1019–1023. [Google Scholar] [CrossRef]
- Washida, K.; Koyama, T.; Yamada, K.; Kita, M.; Uemure, D. Karatungiols A and B, two novel antimicrobial polyol compounds, from the symbiotic marine dinoflagellate Amphidinium sp. Tetrahedron Lett. 2006, 47, 2521–2525. [Google Scholar] [CrossRef]
- Kohli, G.S.; John, U.; Van Dolah, F.M.; Murray, S.A. Evolutionary distinctiveness of fatty acid and polyketide synthesis in eukaryotes. ISME J. 2016, 10, 1877–1890. [Google Scholar] [CrossRef] [Green Version]
- Karafas, S.; Teng, S.T.; Leaw, C.P.; Alves-de-Souza, C. An evaluation of the genus Amphidinium (Dinophyceae) combining evidence from morphology, phylogenetics, and toxin production, with the introduction of six novel species. Harmful Algae 2017, 68, 128–151. [Google Scholar] [CrossRef] [PubMed]
- Lilly, E.L.; Halanych, K.M.; Anderson, D.M. Species boundaries and global biogeography of Alexandrium tamarense complex (Dinophyceae). J. Phycol. 2007, 43, 1329–1338. [Google Scholar] [CrossRef]
- Touzet, N.; Franco, J.M.; Raine, R. Characterization of Nontoxic and Toxin-Producin Strains of Alexandrium minutum (Dinophyceae) in Irish Coastal Waters. Appl. Environ. Microbiol. 2007, 73, 3333–3342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tillmann, U.; Wietkamp, S.; Gu, H.; Krock, B.; Salas, R.; Clarke, D. Multiple New Strains of Amphidomataceae (Dinophyceae) from the North Atlantic Revealed a High Toxin Profile Variability of Azadinium spinosum and a New Non-Toxigenic Az. Cf. spinosum. Microorganisms 2021, 9, 134. [Google Scholar] [CrossRef] [PubMed]
- Morales-Amador, A.; Molina-Miras, A.; López-Rosales, L.; Sánchez-Mirón, A.; García-Camacho, F.; Souto, M.L.; Fernández, J.J. Isolation and Structural Elucidation of New Amphidinol Analogues from Amphidinium carterae Cultivated in a Pilot-Scale Photobioreactor. Mar. Drugs 2021, 19, 432. [Google Scholar] [CrossRef]
- Tsuda, M.; Endo, T.; Kobayashi, J. Amphidinolide T, Novel 19-Membered Macrolide from Marine Dinoflagellate Amphidinium sp. J. Org. Chem. 2000, 65, 1349–1352. [Google Scholar] [CrossRef] [PubMed]
- Kubota, T.; Iwai, T.; Sakai, K.; Gonoi, T.; Kobayashi, J. Amphidinins C-F, amphidinolide Q analogues from marine dinoflagellate Amphidinium sp. Org. Lett. 2014, 7, 5624–5627. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, K.; Minamida, M.; Akakabe, M.; Tsuda, M.; Konishi, Y.; Tominaga, A.; Tsuda, M.; Fukushi, E.; Kawabata, J. Amphirionin-2, a novel linear polyketide with potent cytotoxic activity from a marine dinoflagellate Amphidinium species. Bioorg. Med. Chem. Lett. 2015, 25, 635–638. [Google Scholar] [CrossRef]
- Espiritu, R.A.; Matsumori, N.; Tsuda, M.; Murata, M. Direct and Stereospecific Interaction of Amphidinol 3 with Sterol in Lipid Bilayers. Biochemistry 2014, 53, 3287–3293. [Google Scholar] [CrossRef]
- Morsy, N.; Houdai, T.; Konoki, K.; Matsumori, N.; Osihi, T.; Murata, M. Effects of lipid constituents on membrane permeabilizing activities of amphidinols. Bioorg. Med. Chem. 2008, 16, 3084–3090. [Google Scholar] [CrossRef]
- Patel, P.H.; Hashmi, M.F. Macrolides. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK551495/ (accessed on 4 November 2021).
- Vázquez-Laslop, N.; Mankin, A.S. How macrolide antibiotic work. Trends Biochem. Sci. 2018, 43, 668–684. [Google Scholar] [CrossRef] [PubMed]
- Guillard, R.L.; Ryther, J.H. Studies of Marine Planktonic Diatoms: I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can. J. Microbiol. 1962, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Guillard, R.L. Culture of Marine Invertebrate Animals: Culture of Phytoplankton for Feeding Marine Invertebrates; Smith, W.L., Chanley, M.H., Eds.; Springer: Boston, MA, USA, 1975; pp. 29–60. [Google Scholar]
- Lenaers, G.; Maroteaux, L.; Michot, B.; Herzog, M. Dinoflagellates in evolution. A molecular phylogenetic analysis of large subunit ribosomal RNA. J. Mol. Evol. 1989, 1, 40–51. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [PubMed]
- Clinical Laboratory Standard Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; CLSI guideline M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Urban, T.; Jarstrand, C. Rapid determination of the susceptibility of bacteria to antibiotics with “Sensititre” plates and nitroblue tetrazolium. J. Antimicrob. Chemother. 1981, 8, 363–369. [Google Scholar] [CrossRef]


| Compound | Fraction | m/z Observed | Elemental Composition | RT (min) | ±ppm |
|---|---|---|---|---|---|
| AM-B | G, H | 1463.7927 | C69H125Na2O27S | 4.125 | 0.192 |
| AM-22 | I | 1667.9270 | C84H140NaO31 | 3.473 | 0.705 |
| AM-A | I, J | 1361.8547 | C69H126NaO24 | 3.649 | 1.395 |
| Dehydro-AM-A | J | 1343.8447 | C71H122O23 | 3.766 | 1.015 |
| Stage-2 Fractions | |||||
|---|---|---|---|---|---|
| G | H | I | J | K | |
| MIC S. aureus | - | 64 | 16 | 16 | 64 |
| MIC E. faecalis | - | - | 64 | 128 | 256 |
| AM-B | 100 | 100 | 0 | 0 | 0 |
| AM-22 | 0 | 0 | 8 | 0 | 0 |
| AM-A | 0 | 0 | 92 | 8 | 0 |
| DehydroAM-A | 0 | 0 | 0 | 92 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barone, M.E.; Murphy, E.; Parkes, R.; Fleming, G.T.A.; Campanile, F.; Thomas, O.P.; Touzet, N. Antibacterial Activity and Amphidinol Profiling of the Marine Dinoflagellate Amphidinium carterae (Subclade III). Int. J. Mol. Sci. 2021, 22, 12196. https://doi.org/10.3390/ijms222212196
Barone ME, Murphy E, Parkes R, Fleming GTA, Campanile F, Thomas OP, Touzet N. Antibacterial Activity and Amphidinol Profiling of the Marine Dinoflagellate Amphidinium carterae (Subclade III). International Journal of Molecular Sciences. 2021; 22(22):12196. https://doi.org/10.3390/ijms222212196
Chicago/Turabian StyleBarone, Maria Elena, Elliot Murphy, Rachel Parkes, Gerard T. A. Fleming, Floriana Campanile, Olivier P. Thomas, and Nicolas Touzet. 2021. "Antibacterial Activity and Amphidinol Profiling of the Marine Dinoflagellate Amphidinium carterae (Subclade III)" International Journal of Molecular Sciences 22, no. 22: 12196. https://doi.org/10.3390/ijms222212196
APA StyleBarone, M. E., Murphy, E., Parkes, R., Fleming, G. T. A., Campanile, F., Thomas, O. P., & Touzet, N. (2021). Antibacterial Activity and Amphidinol Profiling of the Marine Dinoflagellate Amphidinium carterae (Subclade III). International Journal of Molecular Sciences, 22(22), 12196. https://doi.org/10.3390/ijms222212196








