The Potential Role of Human NME1 in Neuronal Differentiation of Porcine Mesenchymal Stem Cells: Application of NB-hNME1 as a Human NME1 Suppressor
Abstract
1. Introduction
2. Results
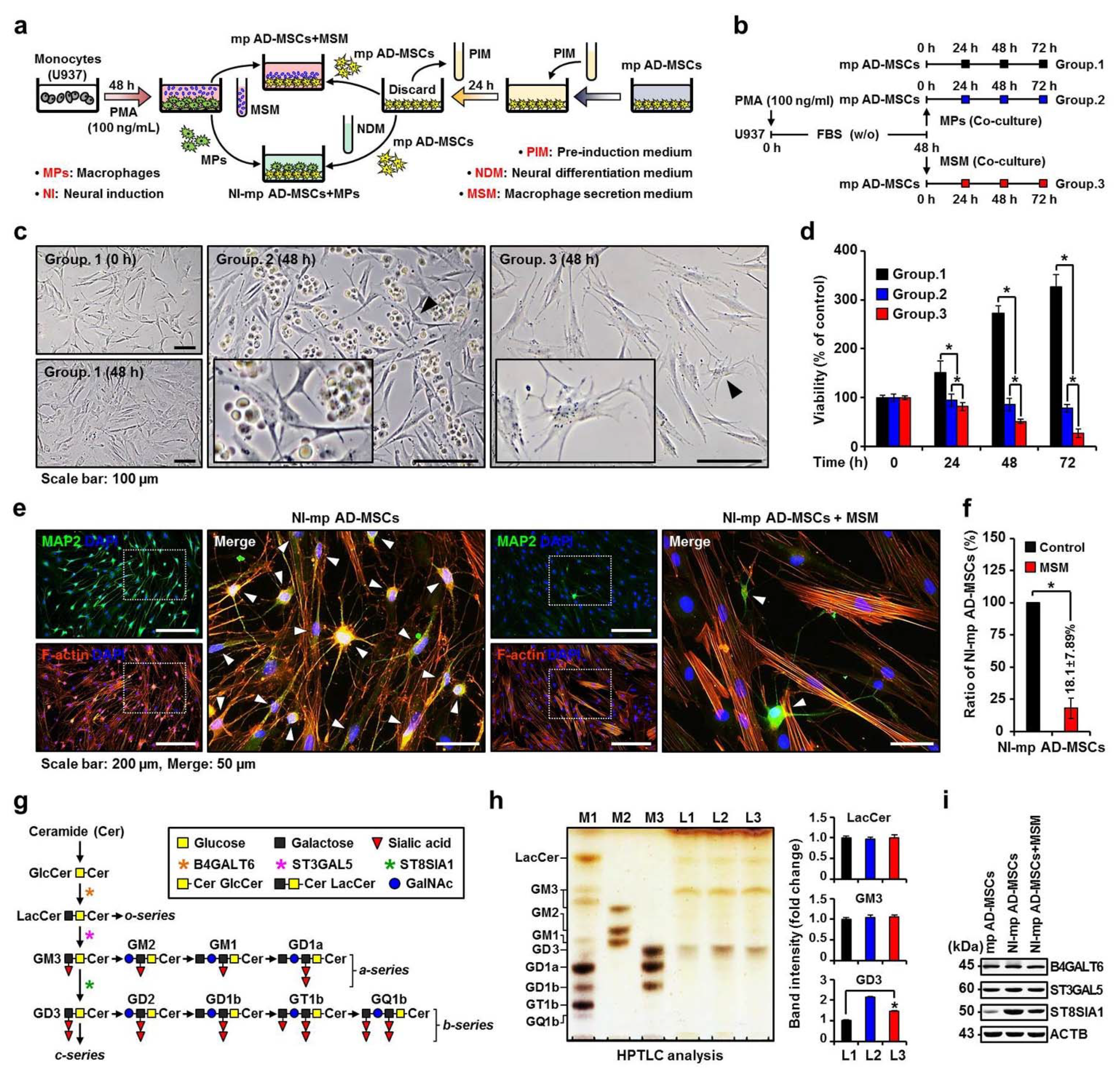
2.1. Effect of the MP Secretome on Changes in Ganglioside Expression and Neuronal Differentiation of Mp AD-MSCs
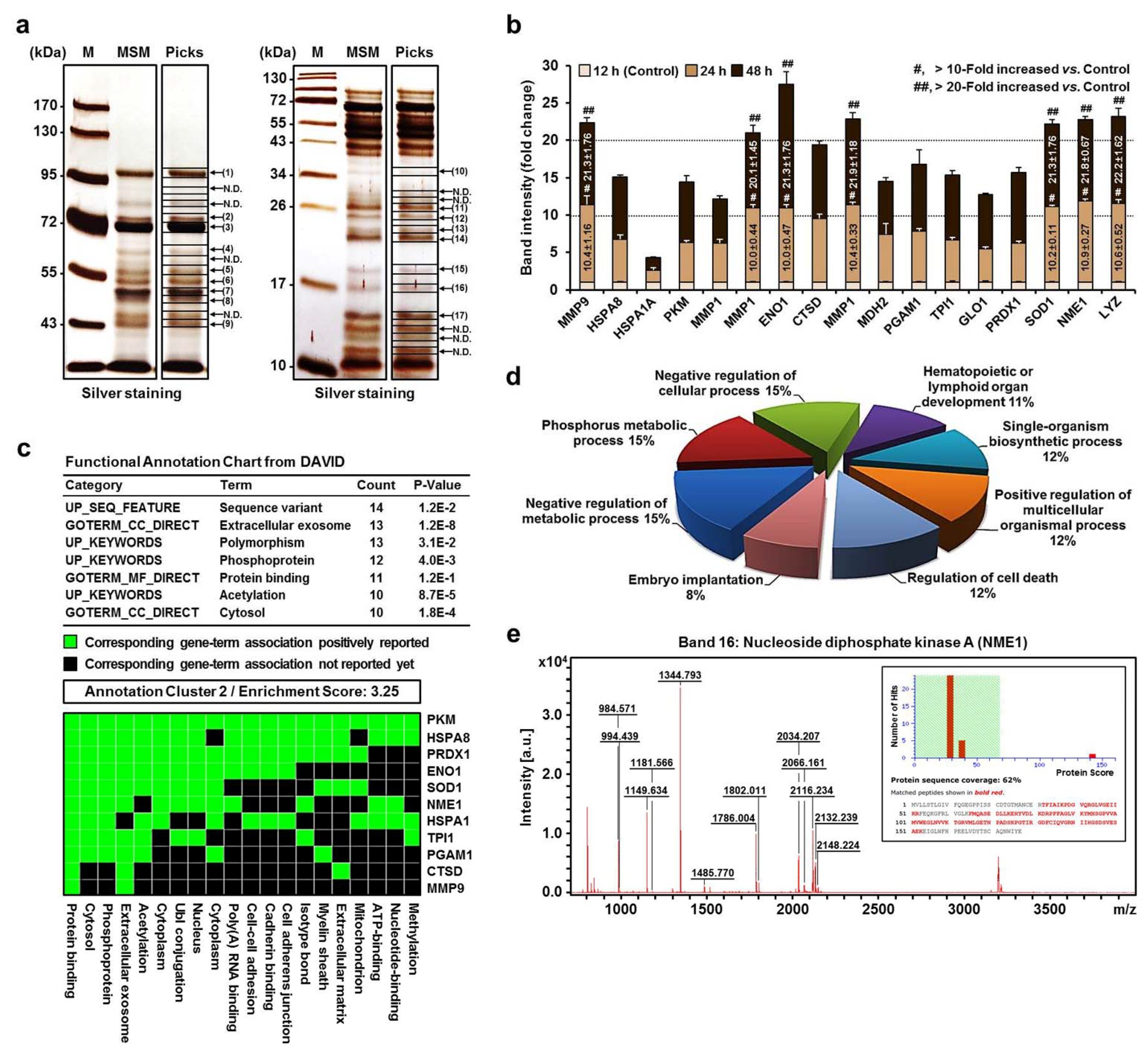
2.2. MALDI-TOF MS and Proteomic Analysis of the Secretome of MPs in MSM
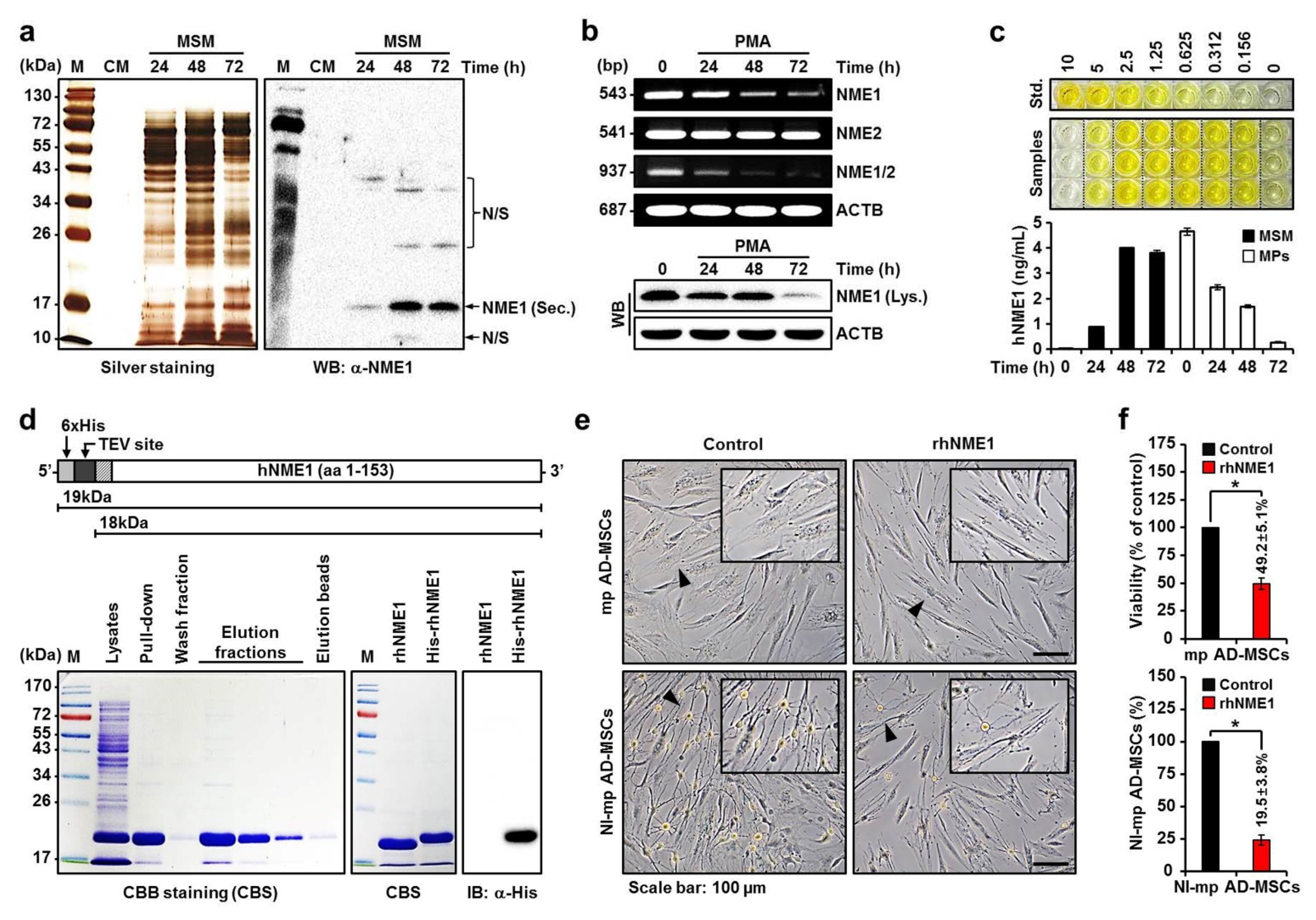
2.3. Production of Recombinant hNME1 to Verity the Effect of hNME1 on the Neuronal Differentiation of mp AD-MSCs
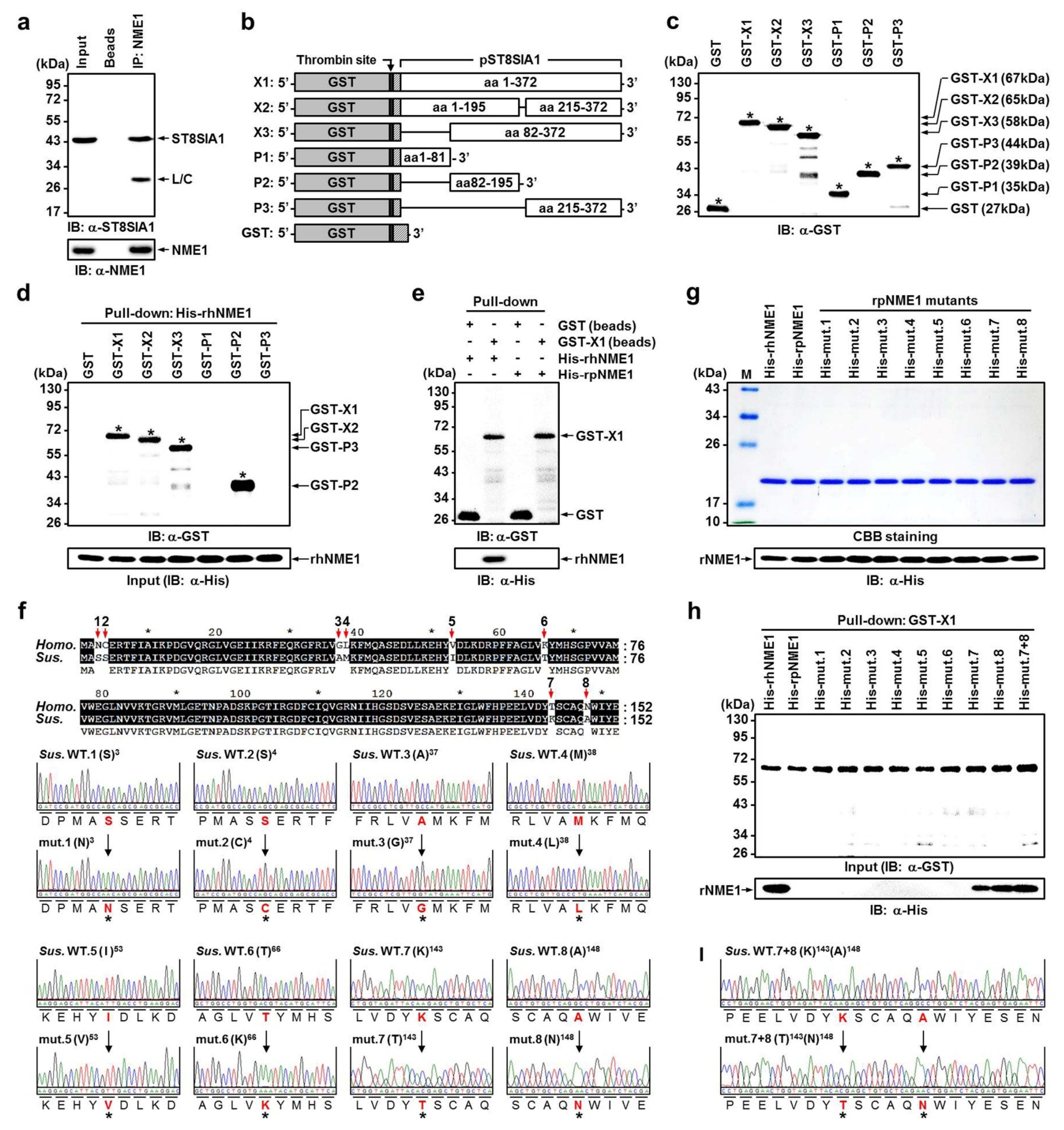
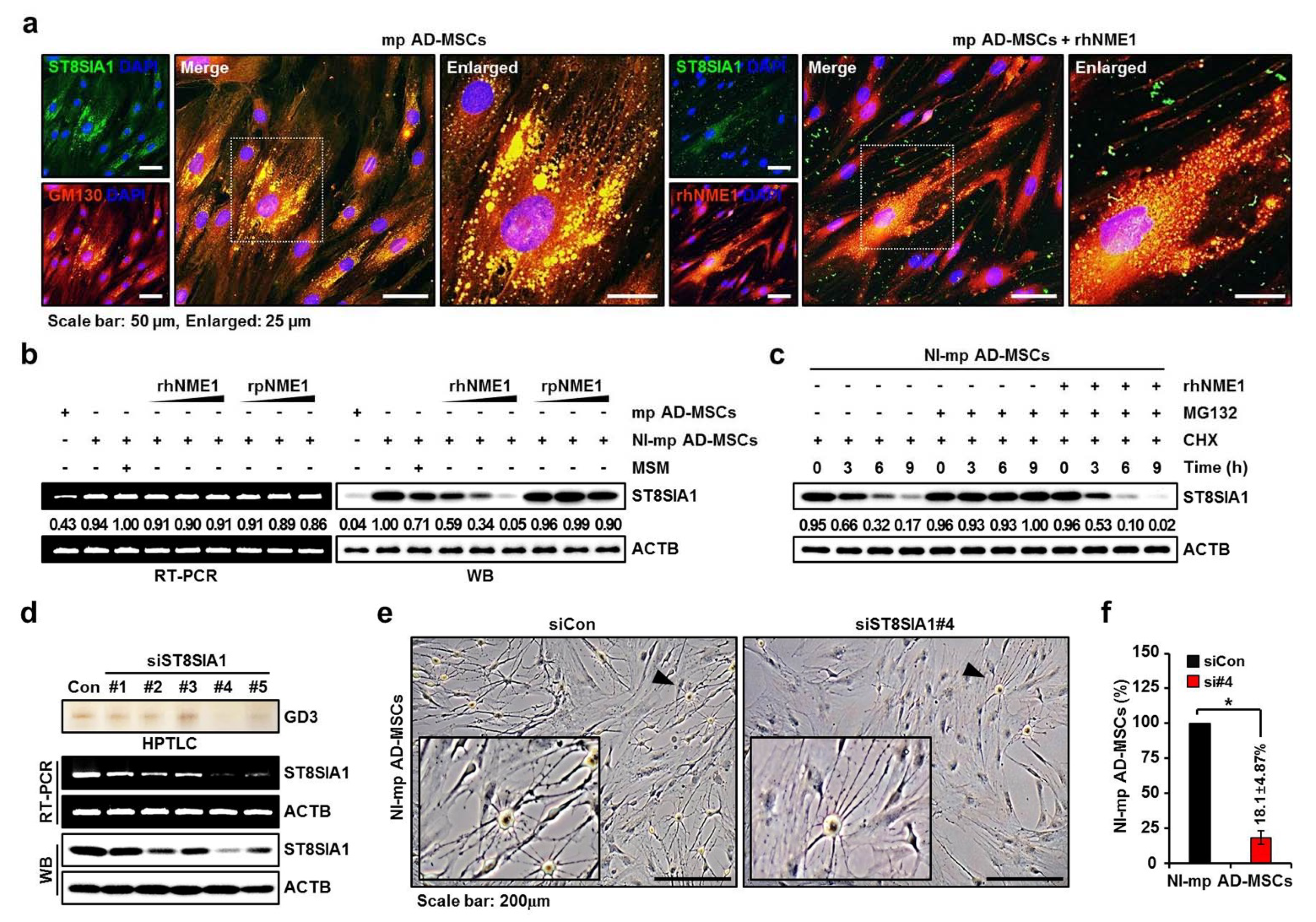
2.4. Verification of Newly Identified Interactions between hNME1 and pST8SIA1
2.5. Correlation Analysis of the Neuronal Differentiation of mp AD-MSCs with Degradation of pST8SIA1 by hNME1
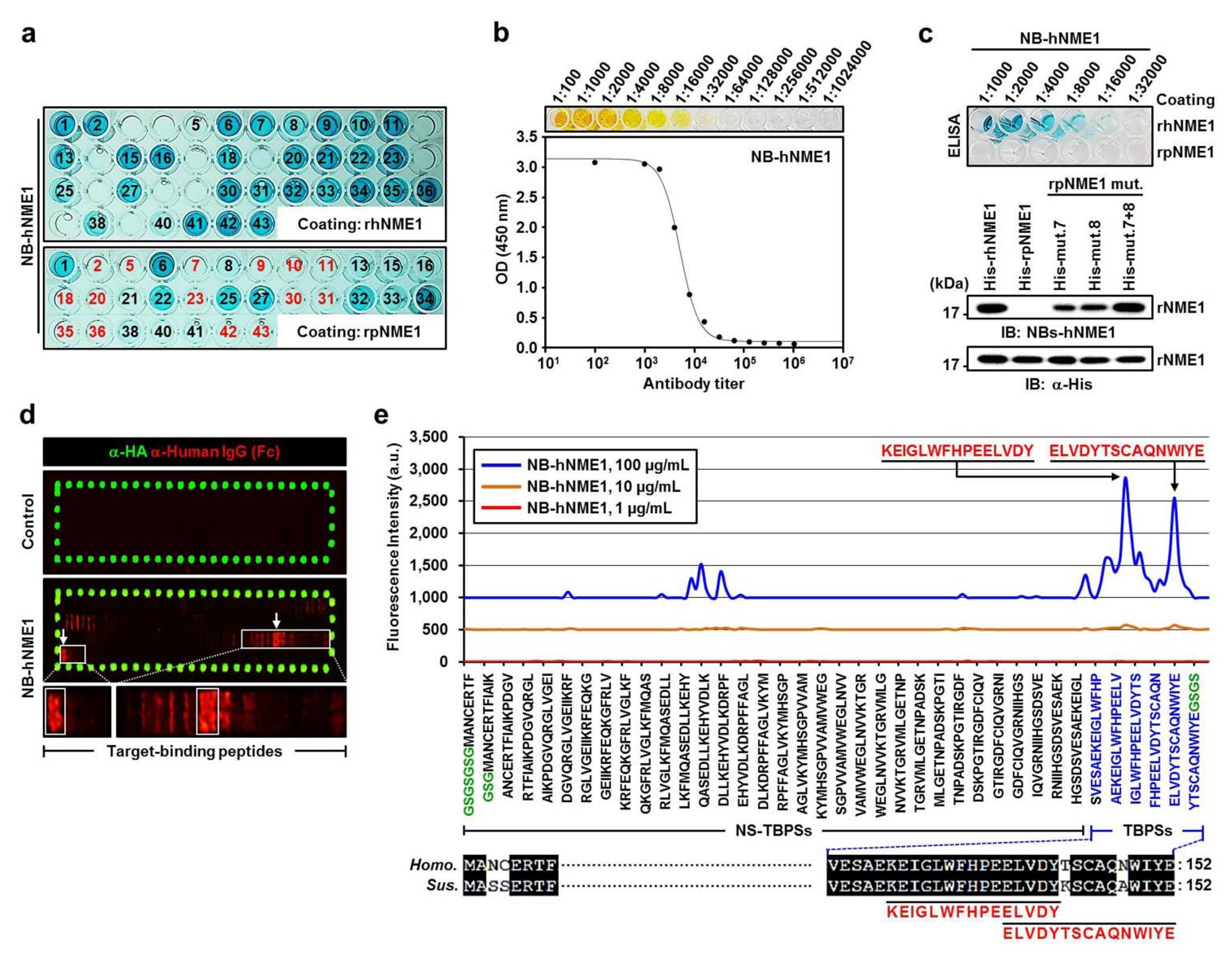
2.6. Production of NB-hNME1 for Inhibiting the Binding Capacity of hNME1 with pST8SIA1
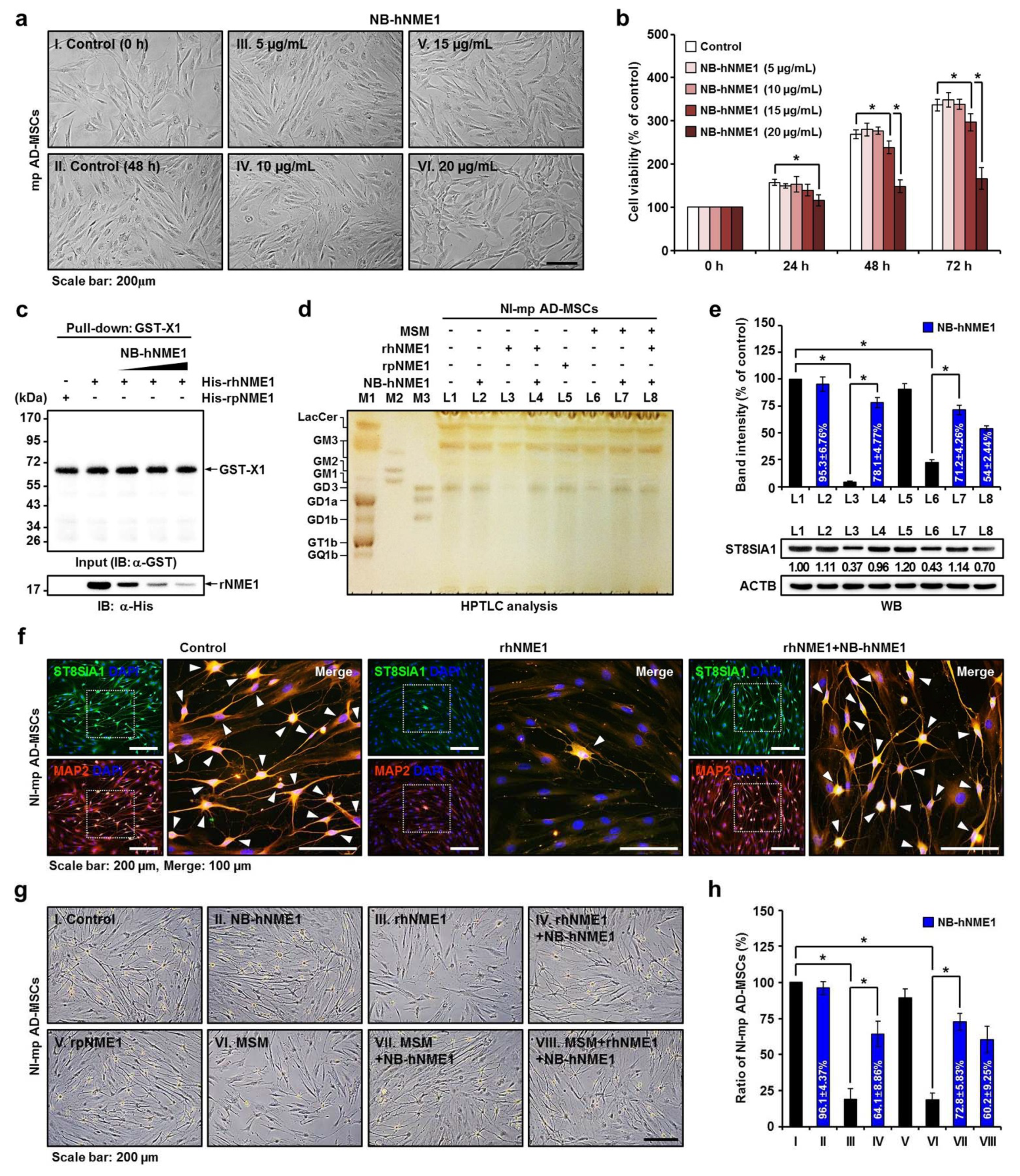
2.7. Verification of the Recovery of Neuronal Differentiation of mp AD-MSCs by Treatment with NB-hNME1 as an hNME1 Suppressor
3. Discussion
4. Materials and Methods
4.1. Cultivation and Characterization of mp AD-MSCs
4.2. Neuronal Differentiation of mp AD-MSCs
4.3. Morphological Evaluation and PMA-Induced Differentiation of U937
4.4. RT-PCR
4.5. IB Analyses
4.6. Immunocytochemistry
4.7. Ganglioside Extraction and Purification
4.8. HPTLC
4.9. Protein Identification by Mass Spectrometry
4.10. ELISA
4.11. Expression Constructs
4.12. Transfection with siRNA
4.13. Screening Peptide Microarray
4.14. Peptide Microarray Spot Quantification
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lu, T.; Yang, B.; Wang, R.; Qin, C. Xenotransplantation: Current Status in Preclinical Research. Front. Immunol. 2019, 10, 3060. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.K.; Satyananda, V.; Ekser, B.; van der Windt, D.J.; Hara, H.; Ezzelarab, M.B.; Schuurman, H.J. Progress in pig-to-non-human primate transplantation models (1998–2013): A comprehensive review of the literature. Xenotransplantation 2014, 21, 397–419. [Google Scholar] [CrossRef] [PubMed]
- Satyananda, V.; Hara, H.; Ezzelarab, M.B.; Phelps, C.; Ayares, D.; Cooper, D.K. New concepts of immune modulation in xenotransplantation. Transplantation 2013, 96, 937–945. [Google Scholar] [CrossRef]
- Cooper, D.K.C.; Pierson, R.N., 3rd; Hering, B.J.; Mohiuddin, M.M.; Fishman, J.A.; Denner, J.; Ahn, C.; Azimzadeh, A.M.; Buhler, L.H.; Cowan, P.J.; et al. Regulation of Clinical Xenotransplantation-Time for a Reappraisal. Transplantation 2017, 101, 1766–1769. [Google Scholar] [CrossRef]
- Stehlik, J.; Mehra, M.R.; Sweet, S.C.; Kirklin, J.K.; Cypel, M.; Kirk, R.; Dipchand, A.I.; Van Raemdonck, D.; Edwards, L.B.; Mario Bertolotti, A.; et al. The International Society for Heart and Lung Transplantation Registries in the Era of Big Data with Global Reach. J. Heart Lung Transplant. 2015, 34, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Martin-Gandul, C.; Mueller, N.J.; Pascual, M.; Manuel, O. The Impact of Infection on Chronic Allograft Dysfunction and Allograft Survival after Solid Organ Transplantation. Am. J. Transplant. 2015, 15, 3024–3040. [Google Scholar] [CrossRef] [PubMed]
- Nankivell, B.J.; Kuypers, D.R. Diagnosis and prevention of chronic kidney allograft loss. Lancet 2011, 378, 1428–1437. [Google Scholar] [CrossRef]
- Murphy, S.P.; Porrett, P.M.; Turka, L.A. Innate immunity in transplant tolerance and rejection. Immunol. Rev. 2011, 241, 39–48. [Google Scholar] [CrossRef]
- Lakkis, F.G.; Li, X.C. Innate allorecognition by monocytic cells and its role in graft rejection. Am. J. Transplant. 2018, 18, 289–292. [Google Scholar] [CrossRef]
- Oberbarnscheidt, M.H.; Lakkis, F.G. Innate allorecognition. Immunol. Rev. 2014, 258, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Kloc, M.; Li, X.C. Macrophages as Effectors of Acute and Chronic Allograft Injury. Curr. Transplant. Rep. 2016, 3, 303–312. [Google Scholar] [CrossRef]
- Morelli, A.E.; Thomson, A.W. Tolerogenic dendritic cells and the quest for transplant tolerance. Nat. Rev. Immunol. 2007, 7, 610–621. [Google Scholar] [CrossRef]
- van den Bosch, T.P.; Kannegieter, N.M.; Hesselink, D.A.; Baan, C.C.; Rowshani, A.T. Targeting the Monocyte-Macrophage Lineage in Solid Organ Transplantation. Front. Immunol. 2017, 8, 153. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Xu, X.; Vu, M.D.; Kilpatrick, E.D.; Li, X.C. NK cells promote transplant tolerance by killing donor antigen-presenting cells. J. Exp. Med. 2006, 203, 1851–1858. [Google Scholar] [CrossRef]
- Lu, L.F.; Lind, E.F.; Gondek, D.C.; Bennett, K.A.; Gleeson, M.W.; Pino-Lagos, K.; Scott, Z.A.; Coyle, A.J.; Reed, J.L.; Van Snick, J.; et al. Mast cells are essential intermediaries in regulatory T-cell tolerance. Nature 2006, 442, 997–1002. [Google Scholar] [CrossRef]
- Matheson, P.J.; Dittmer, I.D.; Beaumont, B.W.; Merrilees, M.J.; Pilmore, H.L. The macrophage is the predominant inflammatory cell in renal allograft intimal arteritis. Transplantation 2005, 79, 1658–1662. [Google Scholar] [CrossRef]
- Xu, L.; Collins, J.; Drachenberg, C.; Kukuruga, D.; Burke, A. Increased macrophage density of cardiac allograft biopsies is associated with antibody-mediated rejection and alloantibodies to HLA antigens. Clin. Transplant. 2014, 28, 554–560. [Google Scholar] [CrossRef]
- Bergler, T.; Jung, B.; Bourier, F.; Kuhne, L.; Banas, M.C.; Rummele, P.; Wurm, S.; Banas, B. Infiltration of Macrophages Correlates with Severity of Allograft Rejection and Outcome in Human Kidney Transplantation. PLoS ONE 2016, 11, e0156900. [Google Scholar] [CrossRef] [PubMed]
- Brasen, J.H.; Khalifa, A.; Schmitz, J.; Dai, W.; Einecke, G.; Schwarz, A.; Hallensleben, M.; Schmidt, B.M.W.; Kreipe, H.H.; Haller, H.; et al. Macrophage density in early surveillance biopsies predicts future renal transplant function. Kidney Int. 2017, 92, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, C.; Zhuang, Q.; Peng, B.; Zhu, Y.; Ye, Q.; Ming, Y. The Evolving Roles of Macrophages in Organ Transplantation. J. Immunol. Res. 2019, 2019, 5763430. [Google Scholar] [CrossRef]
- Dai, H.; Friday, A.J.; Abou-Daya, K.I.; Williams, A.L.; Mortin-Toth, S.; Nicotra, M.L.; Rothstein, D.M.; Shlomchik, W.D.; Matozaki, T.; Isenberg, J.S.; et al. Donor SIRPalpha polymorphism modulates the innate immune response to allogeneic grafts. Sci. Immunol. 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xiao, X.; Demirci, G.; Madsen, J.; Li, X.C. Innate NK cells and macrophages recognize and reject allogeneic nonself in vivo via different mechanisms. J. Immunol. 2012, 188, 2703–2711. [Google Scholar] [CrossRef]
- Candinas, D.; Belliveau, S.; Koyamada, N.; Miyatake, T.; Hechenleitner, P.; Mark, W.; Bach, F.H.; Hancock, W.W. T cell independence of macrophage and natural killer cell infiltration, cytokine production, and endothelial activation during delayed xenograft rejection. Transplantation 1996, 62, 1920–1927. [Google Scholar] [CrossRef]
- Cadili, A.; Kneteman, N. The role of macrophages in xenograft rejection. Transplant. Proc. 2008, 40, 3289–3293. [Google Scholar] [CrossRef]
- Murray, P.J.; Wynn, T.A. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 2011, 11, 723–737. [Google Scholar] [CrossRef]
- Shi, C.; Pamer, E.G. Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 2011, 11, 762–774. [Google Scholar] [CrossRef] [PubMed]
- Unanue, E.R. Antigen-presenting function of the macrophage. Annu. Rev. Immunol. 1984, 2, 395–428. [Google Scholar] [CrossRef] [PubMed]
- Davies, L.C.; Jenkins, S.J.; Allen, J.E.; Taylor, P.R. Tissue-resident macrophages. Nat. Immunol. 2013, 14, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Kaewarpai, T.; Thongboonkerd, V. High-glucose-induced changes in macrophage secretome: Regulation of immune response. Mol. Cell Biochem. 2019, 452, 51–62. [Google Scholar] [CrossRef]
- Dupont, A.; Tokarski, C.; Dekeyzer, O.; Guihot, A.L.; Amouyel, P.; Rolando, C.; Pinet, F. Two-dimensional maps and databases of the human macrophage proteome and secretome. Proteomics 2004, 4, 1761–1778. [Google Scholar] [CrossRef]
- Ohman, T.; Teirila, L.; Lahesmaa-Korpinen, A.M.; Cypryk, W.; Veckman, V.; Saijo, S.; Wolff, H.; Hautaniemi, S.; Nyman, T.A.; Matikainen, S. Dectin-1 pathway activates robust autophagy-dependent unconventional protein secretion in human macrophages. J. Immunol. 2014, 192, 5952–5962. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, J.J.; Matikainen, S.; Nyman, T.A. Global secretome characterization of herpes simplex virus 1-infected human primary macrophages. J. Virol. 2012, 86, 12770–12778. [Google Scholar] [CrossRef] [PubMed]
- Singhto, N.; Kanlaya, R.; Nilnumkhum, A.; Thongboonkerd, V. Roles of Macrophage Exosomes in Immune Response to Calcium Oxalate Monohydrate Crystals. Front. Immunol. 2018, 9, 316. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.K.; Tsai, Y.T.; Ariga, T.; Yanagisawa, M. Structures, biosynthesis, and functions of gangliosides—An overview. J. Oleo Sci. 2011, 60, 537–544. [Google Scholar] [CrossRef]
- Moussavou, G.; Kwak, D.H.; Lim, M.U.; Kim, J.S.; Kim, S.U.; Chang, K.T.; Choo, Y.K. Role of gangliosides in the differentiation of human mesenchymal-derived stem cells into osteoblasts and neuronal cells. BMB Rep. 2013, 46, 527–532. [Google Scholar] [CrossRef]
- Yu, R.K.; Nakatani, Y.; Yanagisawa, M. The role of glycosphingolipid metabolism in the developing brain. J. Lipid Res. 2009, 50, S440–S445. [Google Scholar] [CrossRef]
- Allende, M.L.; Proia, R.L. Lubricating cell signaling pathways with gangliosides. Curr. Opin. Struct. Biol. 2002, 12, 587–592. [Google Scholar] [CrossRef]
- Spiegel, S.; Fishman, P.H. Gangliosides as bimodal regulators of cell growth. Proc. Natl. Acad. Sci. USA 1987, 84, 141–145. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Bell, R.M. Functions of sphingolipids and sphingolipid breakdown products in cellular regulation. Science 1989, 243, 500–507. [Google Scholar] [CrossRef]
- Kondo, C. Studies on the spatial velocity electrocardiogram in ventricular septal defect (authors’s transl). Jpn. Circ. J. 1977, 41, 29–30. [Google Scholar]
- Oliver, S.P.; Lewis, M.J.; Gillespie, B.E.; Dowlen, H.H. Influence of prepartum antibiotic therapy on intramammary infections in primigravid heifers during early lactation. J. Dairy Sci. 1992, 75, 406–414. [Google Scholar] [CrossRef]
- Nan, C.; Shi, Y.; Zhao, Z.; Ma, S.; Liu, J.; Yan, D.; Song, G.; Liu, H. Monosialoteterahexosyl ganglioside induces the differentiation of human umbilical cord-derived mesenchymal stem cells into neuron-like cells. Int. J. Mol. Med. 2015, 36, 1057–1062. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kwak, D.H.; Yu, K.; Kim, S.M.; Lee, D.H.; Kim, S.M.; Jung, J.U.; Seo, J.W.; Kim, N.; Lee, S.; Jung, K.Y.; et al. Dynamic changes of gangliosides expression during the differentiation of embryonic and mesenchymal stem cells into neural cells. Exp. Mol. Med. 2006, 38, 668–676. [Google Scholar] [CrossRef] [PubMed]
- Nakatani, Y.; Yanagisawa, M.; Suzuki, Y.; Yu, R.K. Characterization of GD3 ganglioside as a novel biomarker of mouse neural stem cells. Glycobiology 2010, 20, 78–86. [Google Scholar] [CrossRef]
- Wang, J.; Cheng, A.; Wakade, C.; Yu, R.K. Ganglioside GD3 Is Required for Neurogenesis and Long-Term Maintenance of Neural Stem Cells in the Postnatal Mouse Brain. J. Neurosci. 2014, 34, 13790–13800. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.L.; Wang, J.; Itokazu, Y.; Yu, R.K. Ganglioside GD3 regulates dendritic growth in newborn neurons in adult mouse hippocampus via modulation of mitochondrial dynamics. J. Neurochem. 2021, 156, 819–833. [Google Scholar] [CrossRef] [PubMed]
- Ramos, T.L.; Sanchez-Abarca, L.I.; Muntion, S.; Preciado, S.; Puig, N.; Lopez-Ruano, G.; Hernandez-Hernandez, A.; Redondo, A.; Ortega, R.; Rodriguez, C.; et al. MSC surface markers (CD44, CD73, and CD90) can identify human MSC-derived extracellular vesicles by conventional flow cytometry. Cell Commun. Signal. 2016, 14, 2. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Zhou, N.; Li, J.; Shi, D.; Cao, H.; Li, J.; Li, L. Porcine Adipose-Derived Mesenchymal Stem Cells Retain Their Stem Cell Characteristics and Cell Activities While Enhancing the Expression of Liver-Specific Genes after Acute Liver Failure. Int. J. Mol. Sci. 2016, 17, 62. [Google Scholar] [CrossRef]
- Arrizabalaga, J.H.; Nollert, M.U. Properties of porcine adipose-derived stem cells and their applications in preclinical models. Adipocyte 2017, 6, 217–223. [Google Scholar] [CrossRef]
- Lv, F.J.; Tuan, R.S.; Cheung, K.M.; Leung, V.Y. Concise review: The surface markers and identity of human mesenchymal stem cells. Stem Cells 2014, 32, 1408–1419. [Google Scholar] [CrossRef]
- Gurel Pekozer, G.; Ramazanoglu, M.; Schlegel, K.A.; Kok, F.N.; Torun Kose, G. Role of STRO-1 sorting of porcine dental germ stem cells in dental stem cell-mediated bone tissue engineering. Artif. Cells Nanomed. Biotechnol. 2018, 46, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Johnston, P.V.; Hwang, C.W.; Bogdan, V.; Mills, K.J.; Eggan, E.R.; Leszczynska, A.; Wu, K.C.; Herzka, D.A.; Brinker, J.A.; Schulman, S.P.; et al. Intravascular Stem Cell Bioreactor for Prevention of Adverse Remodeling After Myocardial Infarction. J. Am. Heart Assoc. 2019, 8, e012351. [Google Scholar] [CrossRef] [PubMed]
- Baek, Y.S.; Haas, S.; Hackstein, H.; Bein, G.; Hernandez-Santana, M.; Lehrach, H.; Sauer, S.; Seitz, H. Identification of novel transcriptional regulators involved in macrophage differentiation and activation in U937 cells. BMC Immunol. 2009, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Kim, G.; Kabir, M.H.; Park, S.J.; Lee, S.T.; Lee, C. Use of composite protein database including search result sequences for mass spectrometric analysis of cell secretome. PLoS ONE 2015, 10, e0121692. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Rhim, J.; Kwon, Y.; Choi, S.Y.; Shin, S.; Ha, C.W.; Lee, C. Comparative analysis of differentially secreted proteins in serum-free and serum-containing media by using BONCAT and pulsed SILAC. Sci. Rep. 2019, 9, 3096. [Google Scholar] [CrossRef]
- Pirkmajer, S.; Chibalin, A.V. Serum starvation: Caveat emptor. Am. J. Physiol. Cell Physiol. 2011, 301, C272–C279. [Google Scholar] [CrossRef]
- Maccioni, H.J. Glycosylation of glycolipids in the Golgi complex. J. Neurochem. 2007, 103 (Suppl. S1), 81–90. [Google Scholar] [CrossRef]
- Ngounou Wetie, A.G.; Sokolowska, I.; Woods, A.G.; Roy, U.; Loo, J.A.; Darie, C.C. Investigation of stable and transient protein-protein interactions: Past, present, and future. Proteomics 2013, 13, 538–557. [Google Scholar] [CrossRef]
- Chamberlain, P.P.; Hamann, L.G. Development of targeted protein degradation therapeutics. Nat. Chem. Biol. 2019, 15, 937–944. [Google Scholar] [CrossRef]
- Pamidimukkala, N.V.; Leonard, M.K.; Snyder, D.; McCorkle, J.R.; Kaetzel, D.M. Metastasis Suppressor NME1 Directly Activates Transcription of the ALDOC Gene in Melanoma Cells. Anticancer Res. 2018, 38, 6059–6068. [Google Scholar] [CrossRef]
- Khan, I.; Gril, B.; Steeg, P.S. Metastasis Suppressors NME1 and NME2 Promote Dynamin 2 Oligomerization and Regulate Tumor Cell Endocytosis, Motility, and Metastasis. Cancer Res. 2019, 79, 4689–4702. [Google Scholar] [CrossRef]
- Iqbal, B.; Masood, A.; Lone, M.M.; Lone, A.R.; Dar, N.A. Polymorphism of Metastasis Suppressor Genes MKK4 and NME1 in Kashmiri Patients with Breast Cancer. Breast J. 2016, 22, 673–677. [Google Scholar] [CrossRef]
- Adam, K.; Lesperance, J.; Hunter, T.; Zage, P.E. The Potential Functional Roles of NME1 Histidine Kinase Activity in Neuroblastoma Pathogenesis. Int. J. Mol. Sci. 2020, 21, 3319. [Google Scholar] [CrossRef]
- Khan, I.; Steeg, P.S. The relationship of NM23 (NME) metastasis suppressor histidine phosphorylation to its nucleoside diphosphate kinase, histidine protein kinase and motility suppression activities. Oncotarget 2018, 9, 10185–10202. [Google Scholar] [CrossRef]
- Yugandhar, K.; Gromiha, M.M. Protein-protein binding affinity prediction from amino acid sequence. Bioinformatics 2014, 30, 3583–3589. [Google Scholar] [CrossRef] [PubMed]
- Moal, I.H.; Fernandez-Recio, J. Comment on ‘protein-protein binding affinity prediction from amino acid sequence’. Bioinformatics 2015, 31, 614–615. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yugandhar, K.; Gromiha, M.M. Response to the comment on ‘protein-protein binding affinity prediction from amino acid sequence’. Bioinformatics 2015, 31, 978. [Google Scholar] [CrossRef]
- Eren, E.; Berber, M.; Ozoren, N. NLRC3 protein inhibits inflammation by disrupting NALP3 inflammasome assembly via competition with the adaptor protein ASC for pro-caspase-1 binding. J. Biol. Chem. 2017, 292, 12691–12701. [Google Scholar] [CrossRef]
- Satake, T.; Otsuki, K.; Banba, Y.; Suenaga, J.; Hirano, H.; Yamanaka, Y.; Ohno, S.; Hirai, S. The interaction of Kinesin-1 with its adaptor protein JIP1 can be regulated via proteins binding to the JIP1-PTB domain. BMC Cell Biol. 2013, 14, 12. [Google Scholar] [CrossRef] [PubMed]
- Elliott, E.; Tsvetkov, P.; Ginzburg, I. BAG-1 associates with Hsc70.Tau complex and regulates the proteasomal degradation of Tau protein. J. Biol. Chem. 2007, 282, 37276–37284. [Google Scholar] [CrossRef] [PubMed]
- Gunawardana, D. PDZ Binding Domains, Structural Disorder and Phosphorylation: A Menage-a-trois Tailing Dcp2 mRNA Decapping Enzymes. Protein Pept. Lett. 2016, 23, 612–618. [Google Scholar] [CrossRef] [PubMed]
- Brender, J.R.; Zhang, Y. Predicting the Effect of Mutations on Protein-Protein Binding Interactions through Structure-Based Interface Profiles. PLoS Comput. Biol. 2015, 11, e1004494. [Google Scholar] [CrossRef]
- Deng, C.; Capecchi, M.R. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol. Cell Biol. 1992, 12, 3365–3371. [Google Scholar]
- Green, L.L.; Hardy, M.C.; Maynard-Currie, C.E.; Tsuda, H.; Louie, D.M.; Mendez, M.J.; Abderrahim, H.; Noguchi, M.; Smith, D.H.; Zeng, Y.; et al. Antigen-specific human monoclonal antibodies from mice engineered with human Ig heavy and light chain YACs. Nat. Genet. 1994, 7, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Lonberg, N. Fully human antibodies from transgenic mouse and phage display platforms. Curr. Opin. Immunol. 2008, 20, 450–459. [Google Scholar] [CrossRef]
- Klee, G.G. Human anti-mouse antibodies. Arch. Pathol. Lab. Med. 2000, 124, 921–923. [Google Scholar] [CrossRef] [PubMed]
- Birch, J.R.; Racher, A.J. Antibody production. Adv. Drug Deliv. Rev. 2006, 58, 671–685. [Google Scholar] [CrossRef]
- Frenzel, A.; Schirrmann, T.; Hust, M. Phage display-derived human antibodies in clinical development and therapy. MAbs 2016, 8, 1177–1194. [Google Scholar] [CrossRef]
- Chan, C.E.; Lim, A.P.; MacAry, P.A.; Hanson, B.J. The role of phage display in therapeutic antibody discovery. Int. Immunol. 2014, 26, 649–657. [Google Scholar] [CrossRef]
- Pucca, M.B.; Cerni, F.A.; Janke, R.; Bermudez-Mendez, E.; Ledsgaard, L.; Barbosa, J.E.; Laustsen, A.H. History of Envenoming Therapy and Current Perspectives. Front. Immunol. 2019, 10, 1598. [Google Scholar] [CrossRef]
- Khodabakhsh, F.; Behdani, M.; Rami, A.; Kazemi-Lomedasht, F. Single-Domain Antibodies or Nanobodies: A Class of Next-Generation Antibodies. Int. Rev. Immunol. 2018, 37, 316–322. [Google Scholar] [CrossRef]
- Henry, K.A.; MacKenzie, C.R. Editorial: Single-Domain Antibodies-Biology, Engineering and Emerging Applications. Front. Immunol. 2018, 9, 41. [Google Scholar] [CrossRef]
- Yu, J.M.; Bunnell, B.A.; Kang, S.K. Neural differentiation of human adipose tissue-derived stem cells. Methods Mol. Biol. 2011, 702, 219–231. [Google Scholar]
- Liu, Y.; Liu, L.; Ma, X.; Yin, Y.; Tang, B.; Li, Z. Characteristics and neural-like differentiation of mesenchymal stem cells derived from foetal porcine bone marrow. Biosci. Rep. 2013, 33, e00032. [Google Scholar] [CrossRef] [PubMed]
- Elgamal, A.; Althani, A.A.; Abd-Elmaksoud, A.; Kassab, M.; Farag, A.; Lashen, S.; Gabr, M.M.; Zakaria, M.M.; Alissawi, M.M.; Ismail, H.E.A.; et al. Xeno-free trans-differentiation of adipose tissue-derived mesenchymal stem cells into glial and neuronal cells. Am. J. Stem Cells 2019, 8, 38–51. [Google Scholar] [PubMed]
- Mohammad, M.H.; Al-Shammari, A.M.; Al-Juboory, A.A.; Yaseen, N.Y. Characterization of neural stemness status through the neurogenesis process for bone marrow mesenchymal stem cells. Stem Cells Cloning 2016, 9, 1–15. [Google Scholar]
- Yuan, S.H.; Martin, J.; Elia, J.; Flippin, J.; Paramban, R.I.; Hefferan, M.P.; Vidal, J.G.; Mu, Y.; Killian, R.L.; Israel, M.A.; et al. Cell-surface marker signatures for the isolation of neural stem cells, glia and neurons derived from human pluripotent stem cells. PLoS ONE 2011, 6, e17540. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Xie, G.; Ding, F.; Zhou, X. LPS-induced MMP-9 expression is mediated through the MAPKs-AP-1 dependent mechanism in BEAS-2B and U937 cells. Exp. Lung Res. 2018, 44, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Grkovich, A.; Johnson, C.A.; Buczynski, M.W.; Dennis, E.A. Lipopolysaccharide-induced cyclooxygenase-2 expression in human U937 macrophages is phosphatidic acid phosphohydrolase-1-dependent. J. Biol. Chem. 2006, 281, 32978–32987. [Google Scholar] [CrossRef] [PubMed]
- Song, M.G.; Ryoo, I.G.; Choi, H.Y.; Choi, B.H.; Kim, S.T.; Heo, T.H.; Lee, J.Y.; Park, P.H.; Kwak, M.K. NRF2 Signaling Negatively Regulates Phorbol-12-Myristate-13-Acetate (PMA)-Induced Differentiation of Human Monocytic U937 Cells into Pro-Inflammatory Macrophages. PLoS ONE 2015, 10, e0134235. [Google Scholar] [CrossRef]
- Camilli, G.; Cassotta, A.; Battella, S.; Palmieri, G.; Santoni, A.; Paladini, F.; Fiorillo, M.T.; Sorrentino, R. Regulation and trafficking of the HLA-E molecules during monocyte-macrophage differentiation. J. Leukoc. Biol. 2016, 99, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Kwak, D.H.; Ryu, J.S.; Kim, C.H.; Ko, K.; Ma, J.Y.; Hwang, K.A.; Choo, Y.K. Relationship between ganglioside expression and anti-cancer effects of the monoclonal antibody against epithelial cell adhesion molecule in colon cancer. Exp. Mol. Med. 2011, 43, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, J.; Gharahdaghi, F.; Mische, S.M. Routine identification of proteins from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels or polyvinyl difluoride membranes using matrix assisted laser desorption/ionization-time of flight-mass spectrometry (MALDI-TOF-MS). Electrophoresis 1998, 19, 1036–1045. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, J.H.; Ju, W.S.; Seo, S.Y.; Kim, B.H.; Kim, J.-S.; Kim, J.-G.; Park, S.J.; Choo, Y.-K. The Potential Role of Human NME1 in Neuronal Differentiation of Porcine Mesenchymal Stem Cells: Application of NB-hNME1 as a Human NME1 Suppressor. Int. J. Mol. Sci. 2021, 22, 12194. https://doi.org/10.3390/ijms222212194
Cho JH, Ju WS, Seo SY, Kim BH, Kim J-S, Kim J-G, Park SJ, Choo Y-K. The Potential Role of Human NME1 in Neuronal Differentiation of Porcine Mesenchymal Stem Cells: Application of NB-hNME1 as a Human NME1 Suppressor. International Journal of Molecular Sciences. 2021; 22(22):12194. https://doi.org/10.3390/ijms222212194
Chicago/Turabian StyleCho, Jin Hyoung, Won Seok Ju, Sang Young Seo, Bo Hyun Kim, Ji-Su Kim, Jong-Geol Kim, Soon Ju Park, and Young-Kug Choo. 2021. "The Potential Role of Human NME1 in Neuronal Differentiation of Porcine Mesenchymal Stem Cells: Application of NB-hNME1 as a Human NME1 Suppressor" International Journal of Molecular Sciences 22, no. 22: 12194. https://doi.org/10.3390/ijms222212194
APA StyleCho, J. H., Ju, W. S., Seo, S. Y., Kim, B. H., Kim, J.-S., Kim, J.-G., Park, S. J., & Choo, Y.-K. (2021). The Potential Role of Human NME1 in Neuronal Differentiation of Porcine Mesenchymal Stem Cells: Application of NB-hNME1 as a Human NME1 Suppressor. International Journal of Molecular Sciences, 22(22), 12194. https://doi.org/10.3390/ijms222212194





