Chromosome Folding Promotes Intrachromosomal Aberrations under Radiation- and Nuclease-Induced DNA Breakage
Abstract
1. Introduction
2. Results
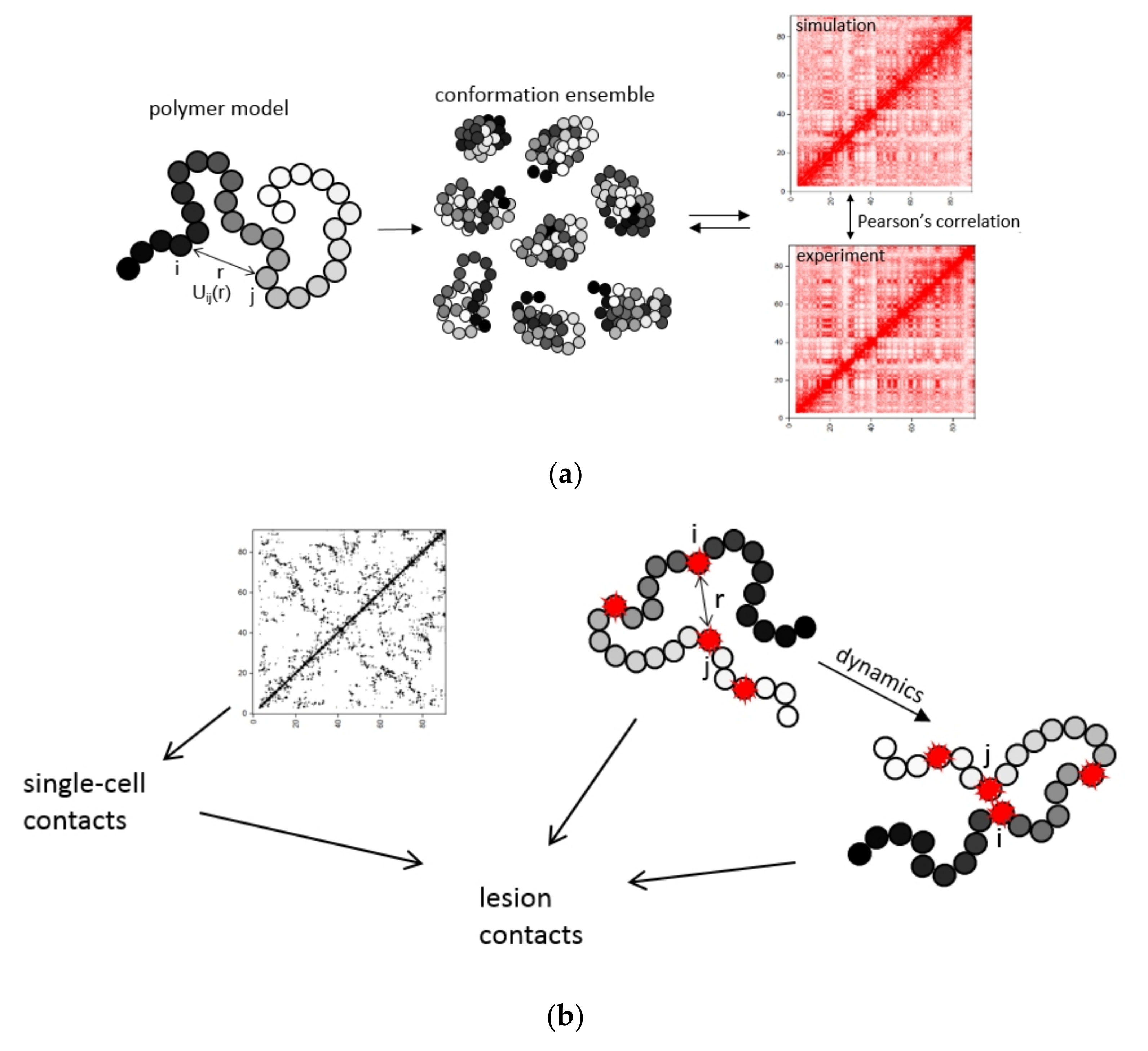
2.1. Modeling of Chromosome Aberrations Based on Polymer Physics of Chromosomes
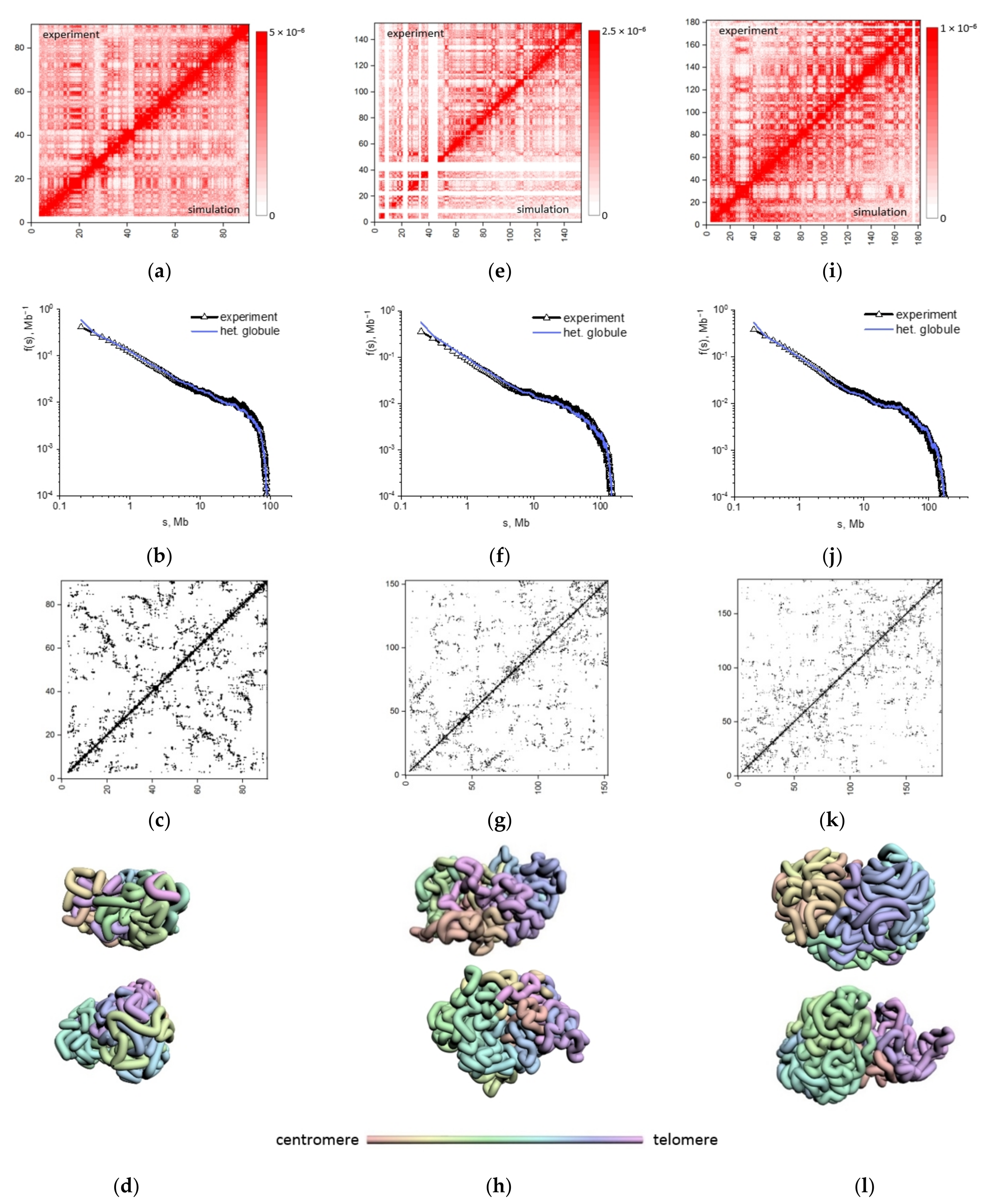
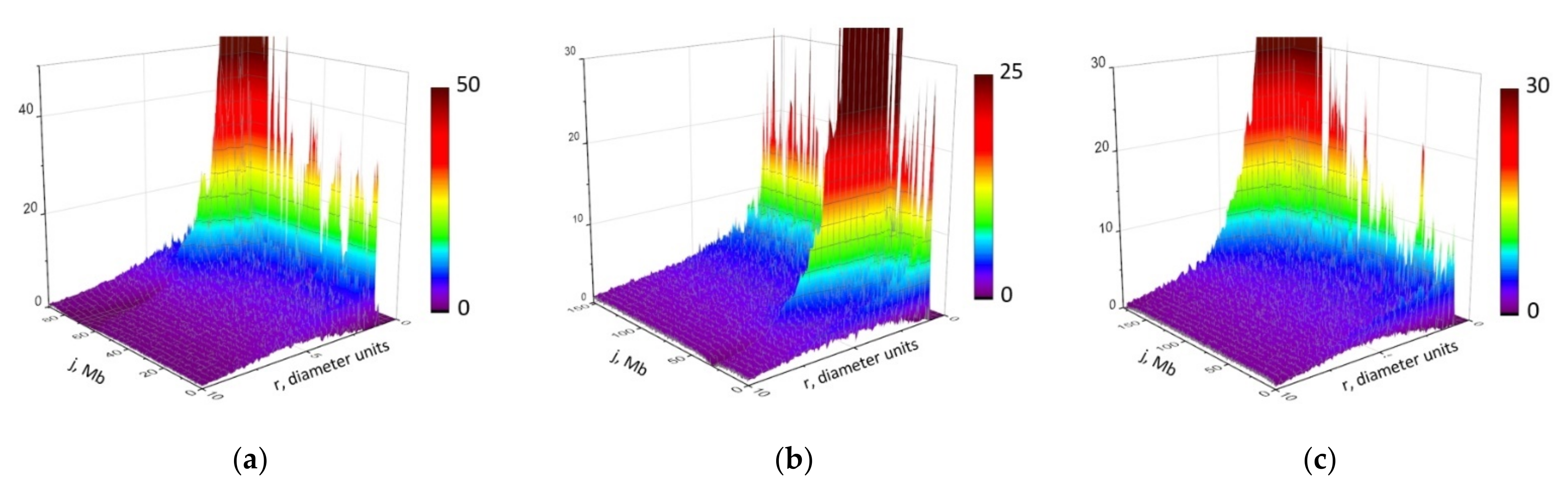
2.2. Physical Modeling of Hi-C Data Reveals Heteropolymer Globule Organization of Mouse Chromosomes
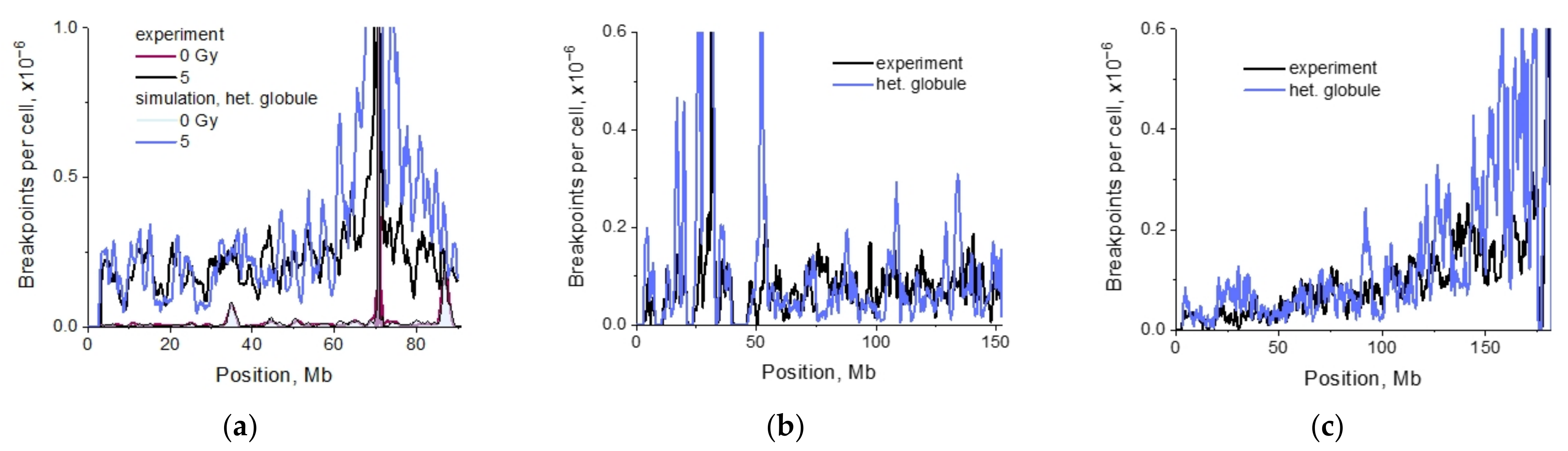
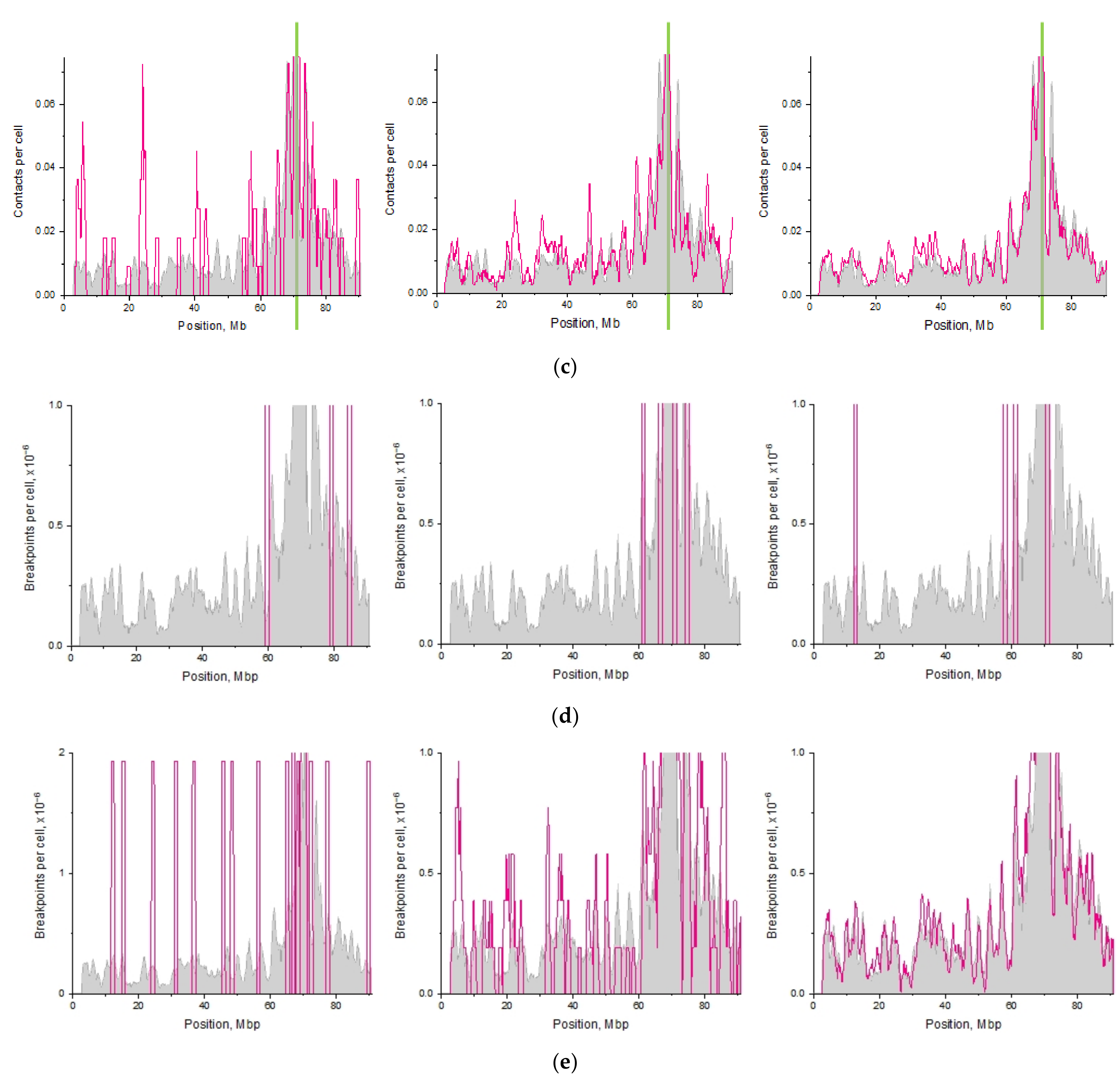
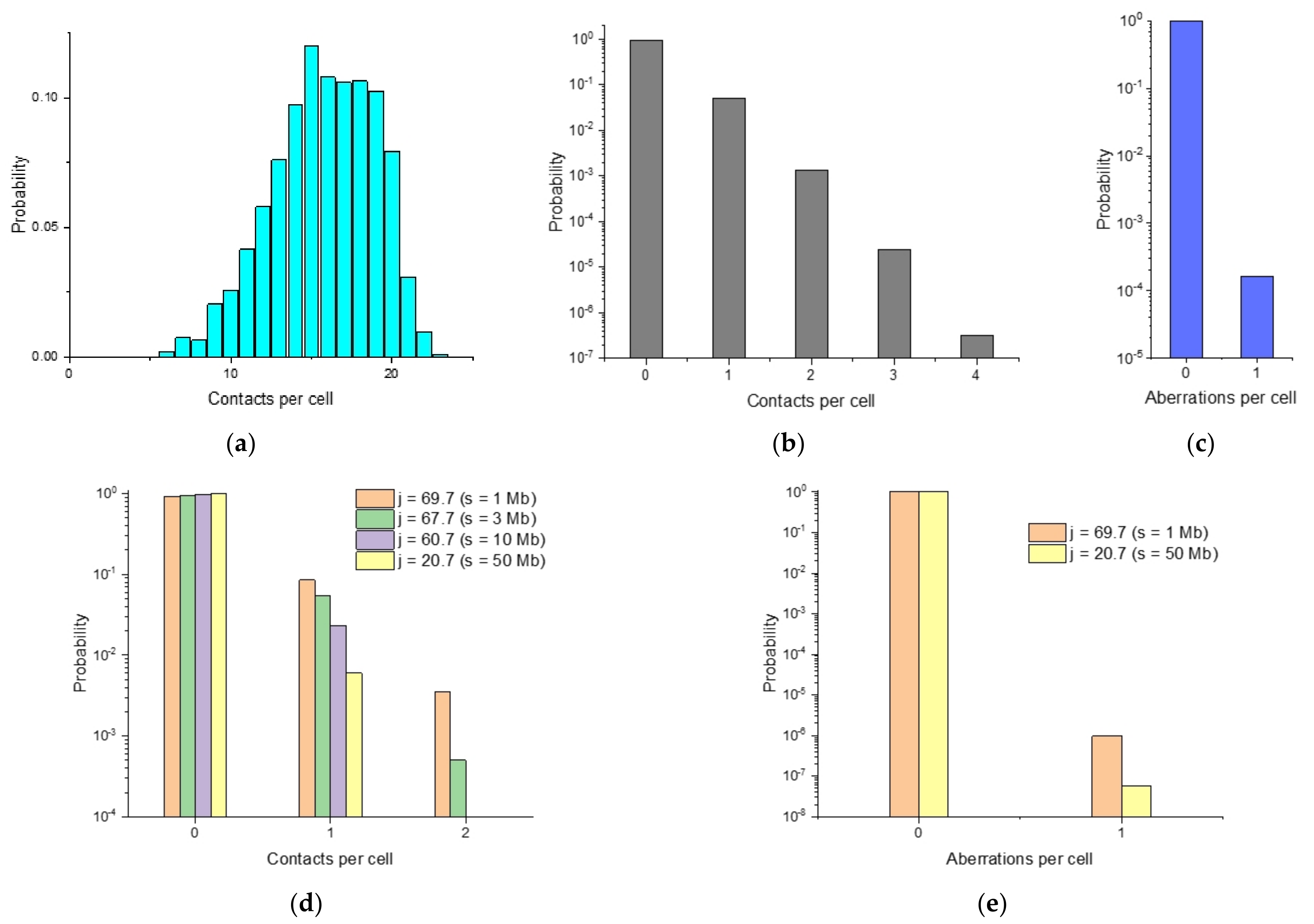
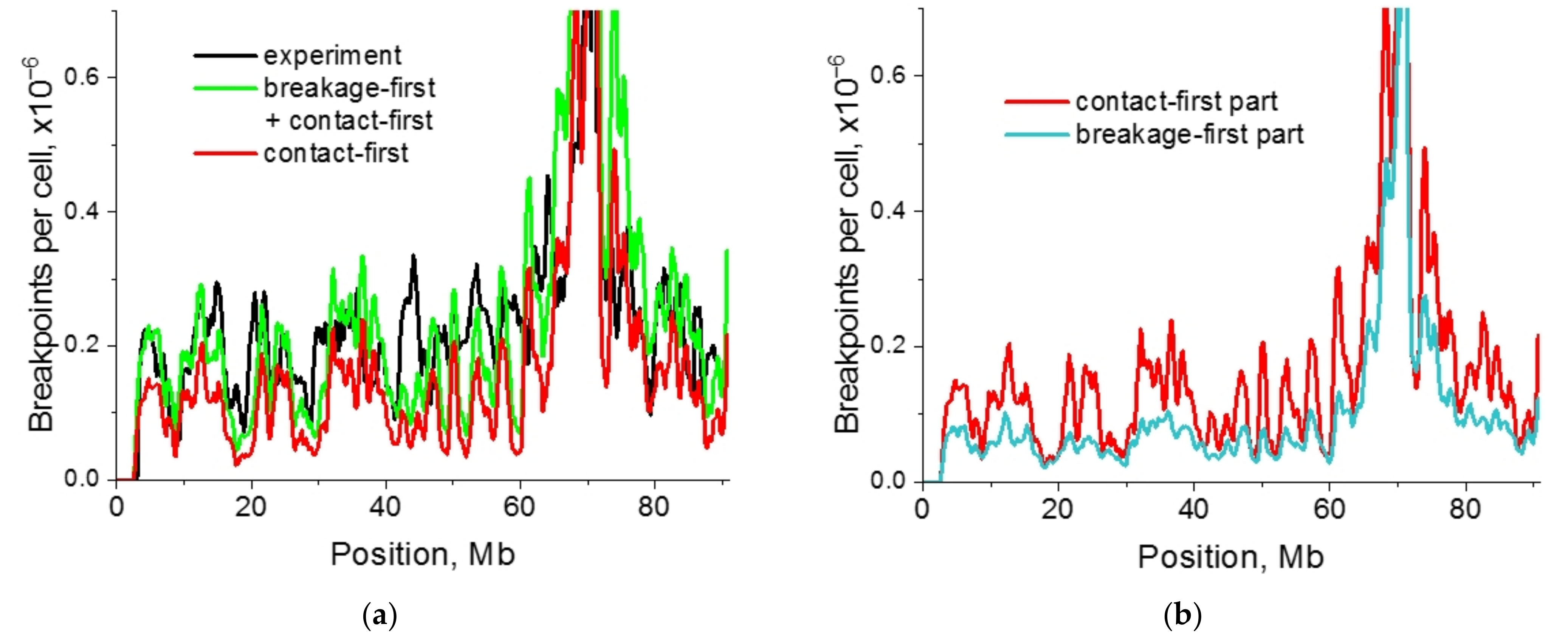
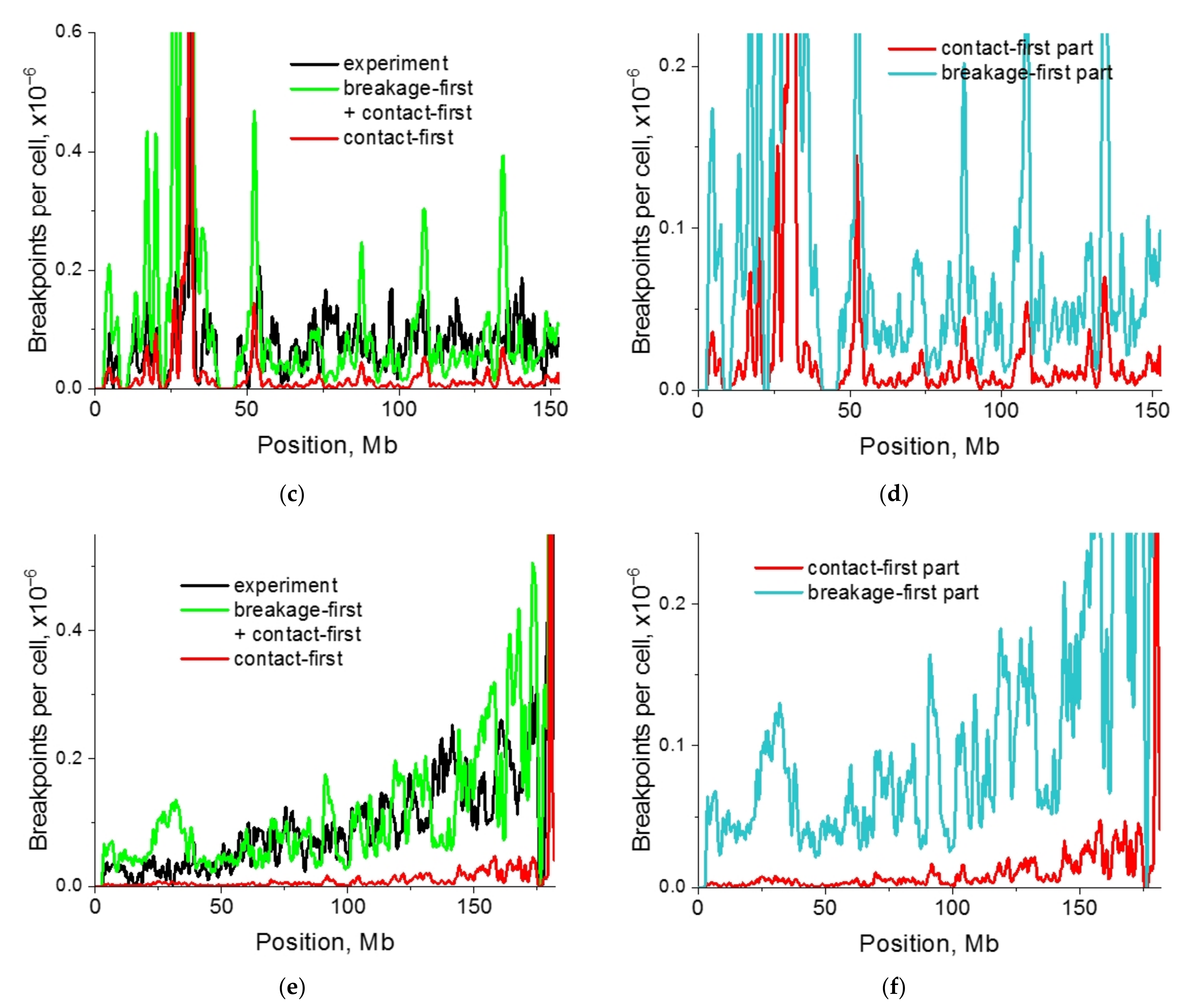
2.3. Structural Organization of Mouse Chromosomes Promotes IR-Recurrent Aberrations
2.4. Impact of Targeted vs. Nontargeted DSBs on Translocation Breakpoint Distribution
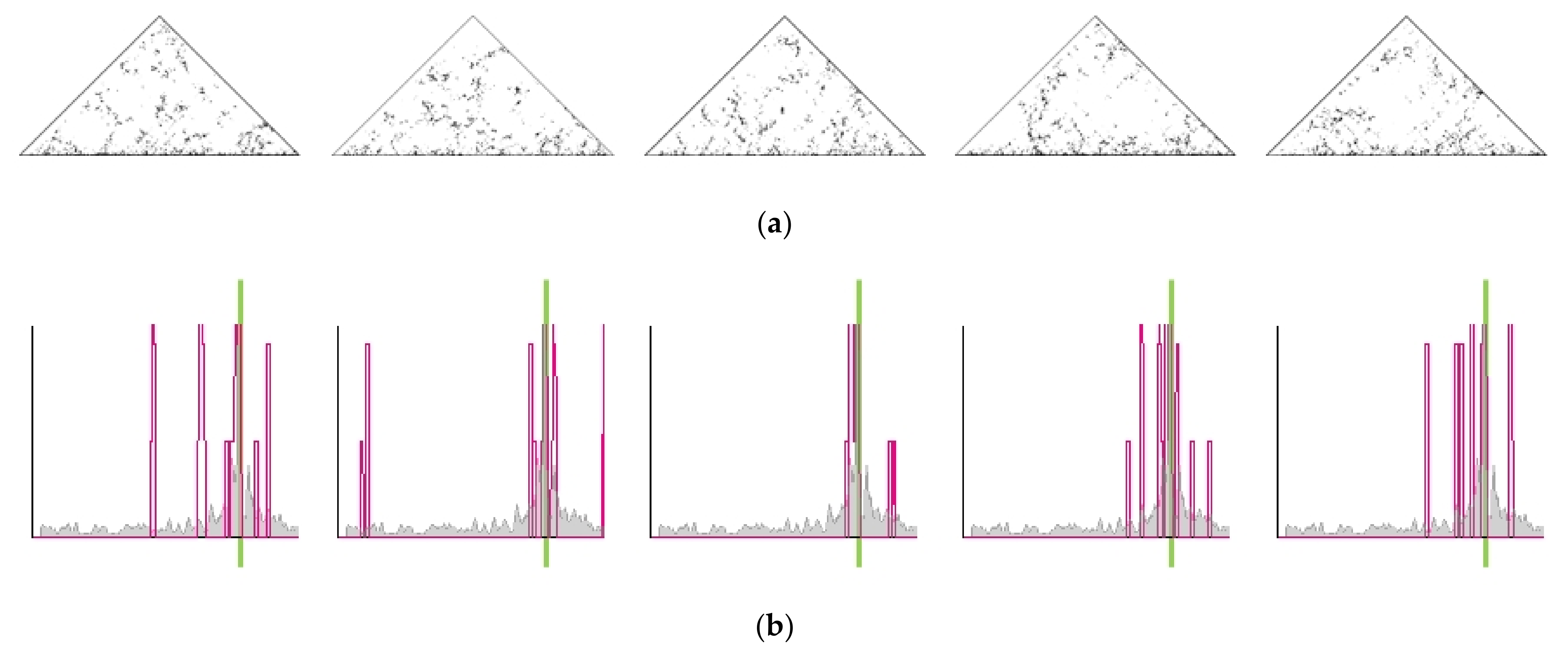
2.5. Single-Cell Variability of Cis-Translocation Breakpoints
2.6. Heterogeneity in Spatial Chromosome Organization among Individual Cells and Breakpoint Distributions
2.7. Translocation Breakpoint Distribution Depends on Movement of Damaged Loci
3. Discussion
4. Materials and Methods
4.1. Polymer Modeling of Chromosome Structure
- The initial coefficients are equal for all element pairs, U0 = 1.2 kT.
- The ensemble of up to 4000 conformations is simulated.
- The contact map with 100 kb resolution is obtained. Pearson’s correlation between the simulated map and the experimental one is calculated.
- If Pearson’s correlation between the contact maps ceases to improve, the simulation is stopped. Otherwise, for each element pair (i,j), the coefficient in the attraction potential increases if the simulated contact frequency is lower than the experimental one and decreases if it is higher.
- Return to step 2.
4.2. Simulation of Trial Structures
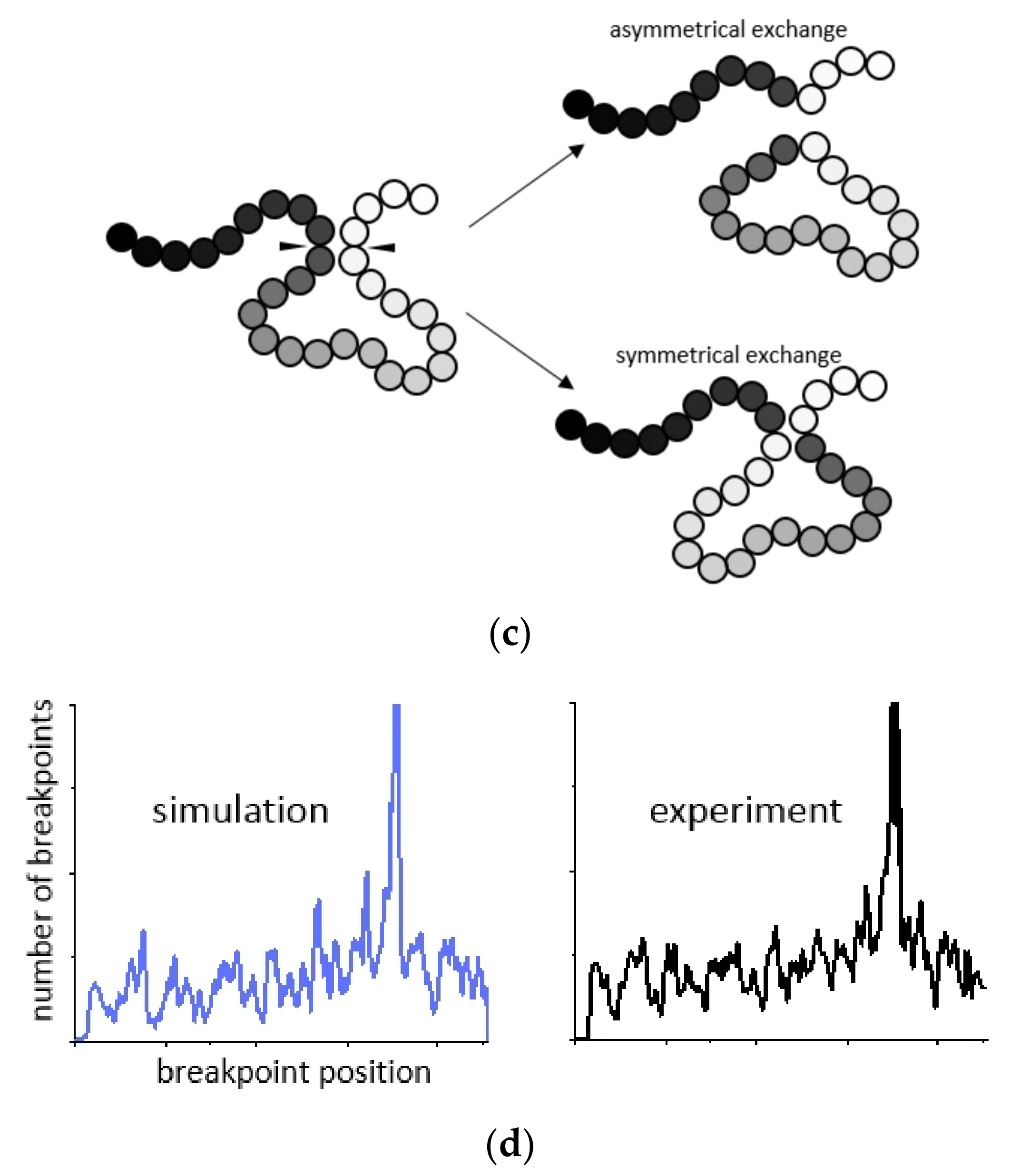
4.3. Modeling of Intrachromosomal Aberrations by the Contact-First Mechanism
4.4. Aberration Modeling in the Absence of Irradiation
4.5. CA Modeling by the Breakage-First Mechanism
4.6. CA Modeling Incorporating Conformational Transitions
4.7. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nagano, T.; Lubling, Y.; Stevens, T.J.; Schoenfelder, S.; Yaffe, E.; Dean, W.; Laue, E.D.; Tanay, A.; Fraser, P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 2013, 502, 59–64. [Google Scholar] [CrossRef]
- Nagano, T.; Lubling, Y.; Várnai, C.; Dudley, C.; Leung, W.; Baran, Y.; Cohen, N.M.; Wingett, S.; Fraser, P.; Tanay, A. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 2017, 547, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Ulianov, S.V.; Zakharova, V.V.; Aleksandra, A.; Galitsyna, A.A.; Polovnikov, K.E.; Flyamer, I.M.; Mikhaleva, E.A.; Khrameeva, E.E.; Germini, D.; Logacheva, M.D.; et al. Order and stochasticity in the folding of individual Drosophila genomes. Nat. Commun. 2021, 12, 41. [Google Scholar] [CrossRef] [PubMed]
- Hakim, O.; Resch, W.; Yamane, A.; Klein, I.; Kieffer-Kwon, K.-R.; Jankovic, M.; Oliveira, T.; Bothmer, A.; Voss, T.C.; Ansarah-Sobrinho, C.; et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature 2012, 484, 69–74. [Google Scholar] [CrossRef]
- Alt, F.W.; Zhang, Y.; Meng, F.-L.; Guo, C.; Schwer, B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell 2013, 152, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Chiarle, R. Translocations in normal B cells and cancers: Insights from new technical approaches. Adv. Immunol. 2013, 117, 39–71. [Google Scholar]
- McCord, R.P.; Balajee, A. 3D genome organization influences the chromosome translocation pattern. Adv. Exp. Med. Biol. 2018, 1044, 113–133. [Google Scholar]
- Savage, J.R.K. A brief survey of aberration origin theories. Mutat. Res. 1998, 404, 139–147. [Google Scholar] [CrossRef]
- Zhang, Y.; Gostissa, M.; Hildebrand, D.G.; Becker, M.S.; Boboila, C.; Chiarle, R.; Lewis, S.; Alt, F.W. The role of mechanistic factors in promoting chromosomal translocations found in lymphoid and other cancers. Adv. Immunol. 2010, 106, 93–133. [Google Scholar]
- Roukos, V.; Voss, T.C.; Schmidt, C.K.; Lee, S.; Wangsa, D.; Misteli, T. Spatial dynamics of chromosome translocations in living cells. Science 2013, 341, 660–664. [Google Scholar] [CrossRef]
- Iarovaia, O.V.; Rubtsov, M.; Ioudinkova, E.; Tsfasman, T.; Razin, S.V.; Vassetzky, Y.S. Dynamics of double strand breaks and chromosomal translocations. Mol. Cancer 2014, 13, 249. [Google Scholar] [CrossRef]
- Cornforth, M.N.; Durante, M. Radiation quality and intra-chromosomal aberrations: Size matters. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2018, 836 Pt A, 28–35. [Google Scholar] [CrossRef]
- Misteli, T.; Soutoglou, E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat. Rev. Mol. Cell Biol. 2009, 10, 243–254. [Google Scholar] [CrossRef]
- Hlatky, L.; Sachs, R.K.; Vazquez, M.; Cornforth, M.N. Radiation-induced chromosome aberrations: Insights gained from biophysical modeling. BioEssays 2002, 24, 714–723. [Google Scholar] [CrossRef] [PubMed]
- Balajee, A.S.; Sanders, J.T.; Golloshi, R.; Shuryak, I.; McCord, R.P.; Dainiak, N. Investigation of spatial organization of chromosome territories in chromosome exchange aberrations after ionizing radiation exposure. Health Phys. 2018, 115, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Nikiforova, M.N.; Stringer, J.R.; Blough, R.; Medvedovic, M.; Fagin, J.A.; Nikiforov, Y.E. Proximity of chromosomal loci that participate in radiation-induced rearrangements in human cells. Science 2000, 290, 138–141. [Google Scholar] [CrossRef] [PubMed]
- Savage, J.R. Cancer. Proximity matters. Science 2000, 290, 62–63. [Google Scholar] [CrossRef]
- Horstmann, M.; Durante, M.; Obe, G. Distribution of breakpoints and fragment sizes in human chromosome 5 after heavy-ion bombardment. Int. J. Radiat. Biol. 2004, 80, 437–443. [Google Scholar] [CrossRef]
- Hada, M.; Zhang, Y.; Feiveson, A.; Cucinotta, F.A.; Wu, H. Association of inter- and intrachromosomal exchanges with the distribution of low- and high-LET radiation-induced breaks in chromosomes. Rad. Res. 2011, 176, 25–37. [Google Scholar] [CrossRef]
- Chiarle, R.; Zhang, Y.; Frock, R.L.; Lewis, S.M.; Molinie, B.; Ho, Y.-J.; Myers, D.R.; Choi, V.W.; Compagno, M.; Malkin, D.J.; et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 2011, 147, 107–119. [Google Scholar] [CrossRef]
- Zhang, Y.; McCord, R.P.; Ho, Y.J.; Lajoie, B.R.; Hildebrand, D.G.; Simon, A.C.; Becker, M.S.; Alt, F.W.; Dekker, J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 2012, 148, 908–921. [Google Scholar] [CrossRef]
- Finn, E.H.; Pegoraro, G.; Brandão, H.B.; Valton, A.-L.; Oomen, M.E.; Dekker, J.; Mirny, L.; Misteli, T. Extensive heterogeneity and intrinsic variation in spatial genome organization. Cell 2019, 176, 1502–1515.e10. [Google Scholar] [CrossRef]
- Giorgetti, L.; Galupa, R.; Nora, E.P.; Piolot, T.; Lam, F.; Dekker, J.; Tiana, G.; Heard, E. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell 2014, 157, 950–963. [Google Scholar] [CrossRef] [PubMed]
- Ballarini, F.; Merzagora, M.; Monforit, F.; Durante, M.; Gialanella, G.; Grossi, G.F.; Pugliese, M.; Ottolenghi, A. Chromosome aberrations induced by light ions: Monte Carlo simulations based on a mechanistic model. Int. J. Radiat. Biol. 1999, 75, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Ballarini, F.; Biaggi, M.; Ottolenghi, A. Nuclear architecture and radiation induced chromosome aberrations: Models and simulations. Radiat. Prot. Dosim. 2002, 99, 175–182. [Google Scholar] [CrossRef]
- Grosberg, A.Y.; Khokhlov, A.R. Statistical Physics of Macromolecules, 1st ed.; AIP-Press: Woodbury, NY, USA, 1994; pp. 14–41. [Google Scholar]
- Ponomarev, A.L.; Sachs, R.K. Radiation breakage of DNA: A model based on random-walk chromatin structure. J. Math. Biol. 2001, 43, 356–376. [Google Scholar] [CrossRef]
- Holley, W.R.; Mian, I.S.; Park, S.J.; Rydberg, B.; Chatterjee, A. A model for interphase chromosomes and evaluation of radiation-induced aberrations. Radiat. Res. 2002, 158, 568–580. [Google Scholar] [CrossRef][Green Version]
- Kreth, G.; Pazhanisamy, S.K.; Hausmann, M.; Cremer, C. Cell type-specific quantitative predictions of radiation-induced chromosome aberrations: A computer model approach. Radiat. Res. 2007, 167, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Friedland, W.; Paretzke, H.G.; Ballarini, F.; Ottolenghi, A.; Kreth, G.; Cremer, C. First steps towards systems radiation biology studies concerned with DNA and chromosome structure within living cells. Radiat. Environ. Biophys. 2008, 47, 49–61. [Google Scholar] [CrossRef]
- Friedland, W.; Kundrát, P. Track structure based modelling of chromosome aberrations after photon and alpha-particle irradiation. Mutat. Res. 2013, 756, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Kreth, G.; Finsterle, J.; von Hase, J.; Cremer, M.; Cremer, C. Radial arrangement of chromosome territories in human cell nuclei: A computer model approach based on gene density indicates a probabilistic global positioning code. Biophys. J. 2004, 86, 2803–2812. [Google Scholar] [CrossRef]
- Andreev, S.G.; Eidelman, Y. Intrachromosomal exchange aberrations predicted on the basis of the globular interphase chromosome model. Radiat. Prot. Dosim. 2002, 99, 193–196. [Google Scholar] [CrossRef]
- Eidelman, Y.A.; Salnikov, I.V.; Slanina, S.V.; Andreev, S.G. Highly heterogeneous long-range interactions explain chromosome compartmentalization. bioRxiv 2019. [Google Scholar] [CrossRef]
- Revell, S.H. The breakage-and-reunion theory and the exchange theory for chromosomal aberrations induced by ionizing radiations: A short history. Adv. Radiat. Biol. 1974, 4, 367–416. [Google Scholar]
- Soutoglou, E.; Dorn, J.F.; Sengupta, K.; Jasin, M.; Nussenzweig, A.; Ried, T.; Danuser, G.; Misteli, T. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 2007, 9, 675–682. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef]
- Rao, S.S.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef] [PubMed]
- Di Pierro, M.; Zhang, B.; Aiden, E.L.; Wolynes, P.G.; Onuchic, J.N. Transferable model for chromosome architecture. Proc. Natl Acad. Sci. USA 2016, 113, 12168–12173. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.J.; Meng, L.; Liu, S.; Zhang, L.; Cai, X.; Gao, Y.Q. Structural modeling of chromatin integrates genome features and reveals chromosome folding principle. Sci. Rep. 2017, 7, 2818. [Google Scholar] [CrossRef] [PubMed]
- Imakaev, M.V.; Fudenberg, G.; Mirny, L. Modeling chromosomes: Beyond pretty pictures. FEBS Lett. 2015, 589, 3031–3036. [Google Scholar] [CrossRef]
- Krawczyk, P.M.; Stap, J.; Hoebe, R.A.; van Oven, C.H.; Kanaar, R.; Aten, J.A. Analysis of the mobility of DNA double-strand break-containing chromosome domains in living mammalian cells. In The Nucleus; Methods in Molecular Biology; Hancock, R., Ed.; Humana Press: Totowa, NJ, USA, 2008; Volume 463, pp. 309–320. [Google Scholar]
- Miné-Hattab, J.; Chiolo, I. Complex chromatin motions for DNA repair. Front. Genet. 2020, 11, 800. [Google Scholar] [CrossRef]
- Sanders, J.; Freeman, T.; Xu, Y.; Golloshi, R.; Stallard, M.A.; Hill, A.M.; San Martin, R.; Balajee, A.S.; McCord, R.P. Radiation-induced DNA damage and repair effects on 3D genome organization. Nat. Commun. 2020, 11, 6178. [Google Scholar] [CrossRef]
- Eidelman, Y.; Slanina, S.; Aleshchenko, A.; Sen’Ko, O.; Kononkova, A.D.; Andreev, S.G. Nuclear 3D organization and radiosensitivity. J. Phys. Conf. Series 2017, 784, 012009. [Google Scholar] [CrossRef]
- Khvostunov, I.K.; Andreev, S.G. Microdosimetric distributions for target volumes of complex topology. In Microdosimetry. An Interdisciplinary Approach; Goodhead, D.T., O’Neill, P., Menzel, H.G., Eds.; Royal Society of Chemistry: London, UK, 1997; pp. 47–50. [Google Scholar]
- Durante, M.; Pignalosa, D.; Jansen, J.A.; Walboomers, X.F.; Ritter, S. Influence of nuclear geometry on the formation of genetic rearrangements in human cells. Radiat. Res. 2010, 174, 20–26. [Google Scholar] [CrossRef]
- Andreev, S.G.; Eidelman, Y.A.; Salnikov, I.V.; Khvostunov, I.K. Mechanistic modelling of genetic and epigenetic events in radiation carcinogenesis. Radiat. Prot. Dosim. 2006, 122, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Eidelman, Y.A.; Slanina, S.V.; Pyatenko, V.S.; Andreev, S.G. DNA damage induced chromosomal instability. Computational modeling view. bioRxiv 2017. [Google Scholar] [CrossRef]
- Andreev, S.G.; Eidelman, Y.A.; Salnikov, I.V.; Slanina, S.V. Modeling study of dose-response relationships for radiation-induced chromosomal instability. Dokl. Biochem. Biophys. 2013, 451, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Xu, J.; Dileep, V.; Zhan, Y.; Song, F.; Le, V.T.; Yardımcı, G.G.; Chakraborty, A.; Bann, D.V.; Wang, J.; et al. Integrative detection and analysis of structural variation in cancer genomes. Nat. Genet. 2018, 50, 1388–1398. [Google Scholar] [CrossRef] [PubMed]
- Akdemir, K.C.; Le, V.T.; Chandran, S.; Li, Y.; Verhaak, R.G.; Beroukhim, R.; Campbell, P.J.; Chin, L.; Dixon, J.R.; Futreal, P.A.; et al. Disruption of chromatin folding domains by somatic genomic rearrangements in human cancer. Nat. Genet. 2020, 52, 294–305. [Google Scholar] [CrossRef]
- Akdemir, K.C.; Le, V.T.; Kim, J.M.; Killcoyne, S.; King, D.A.; Lin, Y.-P.; Tian, Y.; Inoue, A.; Amin, S.B.; Robinson, F.S.; et al. Somatic mutation distributions in cancer genomes vary with three-dimensional chromatin structure. Nat. Genet. 2020, 52, 1178–1188. [Google Scholar] [CrossRef] [PubMed]
- Swenson, K.M.; Blanchette, M. Large-scale mammalian genome rearrangements coincide with chromatin interactions. Bioinformatics 2019, 35, i117–i126. [Google Scholar] [CrossRef] [PubMed]
- Eastman, P.; Swails, J.; Chodera, J.D.; McGibbon, R.T.; Zhao, Y.; Beauchamp, K.A.; Wang, L.-P.; Simmonett, A.C.; Harrigan, M.P.; Stern, C.D.; et al. OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLoS Comp. Biol. 2017, 13, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Hoglund, E.; Blomquist, E.; Carlsson, J.; Stenerlow, B. DNA damage induced by radiation of different linear energy transfer: Initial fragmentation. Int. J. Rad. Biol. 2000, 76, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Eidelman, Y.A.; Salnikov, I.V.; Slanina, S.V.; Andreev, S.G. Spatial organization of the mouse chromosomes influences the landscape of intrachromosomal exchange aberration breakpoints. bioRxiv 2019. [Google Scholar] [CrossRef]
- Nuebler, J.; Fudenberg, G.; Imakaev, M.; Abdennur, N.; Mirny, L.A. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc. Natl Acad. Sci. USA 2018, 115, E6697–E6706. [Google Scholar] [CrossRef] [PubMed]
- Mirnylab: Openmm-Polymer-Legacy. Available online: https://github.com/mirnylab/openmm-polymer-legacy (accessed on 26 September 2019).
- Kremer, K.; Grest, G.S. Dynamics of entangled linear polymer melts: A molecular-dynamics simulation. J. Chem. Phys. 1990, 92, 5057–5086. [Google Scholar] [CrossRef]




| Characteristics | Relative Fluctuations, SD/Mean |
|---|---|
| Number of total contacts in chromosome | 0.014 |
| Number of contacts with I-SceI site | 0.33 |
| Number of lesion contacts with I-SceI site (5 Gy) | 4.32 |
| Number of breakpoints (5 Gy) | 110.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eidelman, Y.; Salnikov, I.; Slanina, S.; Andreev, S. Chromosome Folding Promotes Intrachromosomal Aberrations under Radiation- and Nuclease-Induced DNA Breakage. Int. J. Mol. Sci. 2021, 22, 12186. https://doi.org/10.3390/ijms222212186
Eidelman Y, Salnikov I, Slanina S, Andreev S. Chromosome Folding Promotes Intrachromosomal Aberrations under Radiation- and Nuclease-Induced DNA Breakage. International Journal of Molecular Sciences. 2021; 22(22):12186. https://doi.org/10.3390/ijms222212186
Chicago/Turabian StyleEidelman, Yuri, Ilya Salnikov, Svetlana Slanina, and Sergey Andreev. 2021. "Chromosome Folding Promotes Intrachromosomal Aberrations under Radiation- and Nuclease-Induced DNA Breakage" International Journal of Molecular Sciences 22, no. 22: 12186. https://doi.org/10.3390/ijms222212186
APA StyleEidelman, Y., Salnikov, I., Slanina, S., & Andreev, S. (2021). Chromosome Folding Promotes Intrachromosomal Aberrations under Radiation- and Nuclease-Induced DNA Breakage. International Journal of Molecular Sciences, 22(22), 12186. https://doi.org/10.3390/ijms222212186





