There Is Strength in Numbers: Quantitation of Fc Gamma Receptors on Murine Tissue-Resident Macrophages
Abstract
:1. Introduction
2. Results
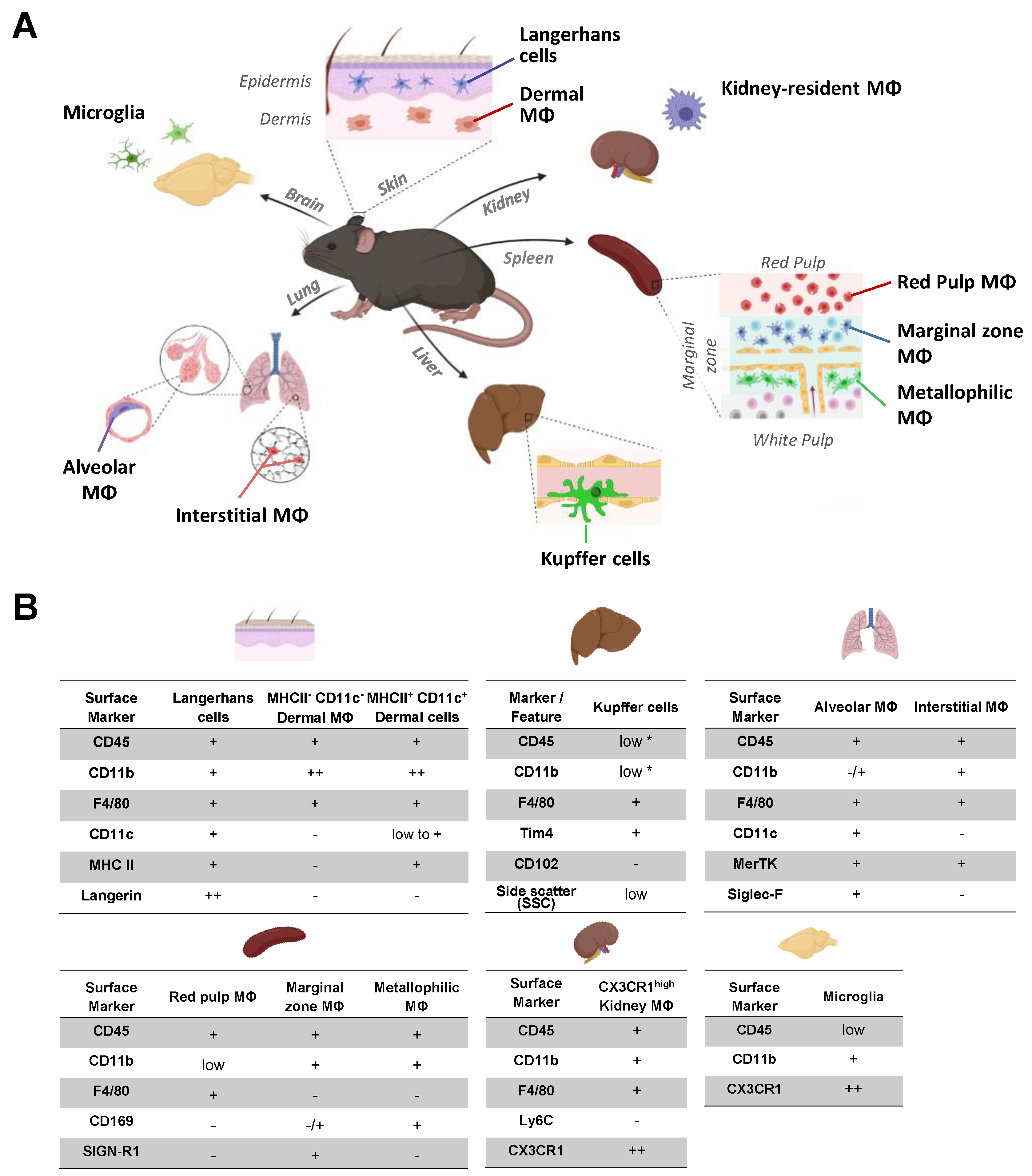
2.1. Identification of Organ Resident Macrophage Subsets
2.2. Impact of Organ Digestion on FcγR Expression
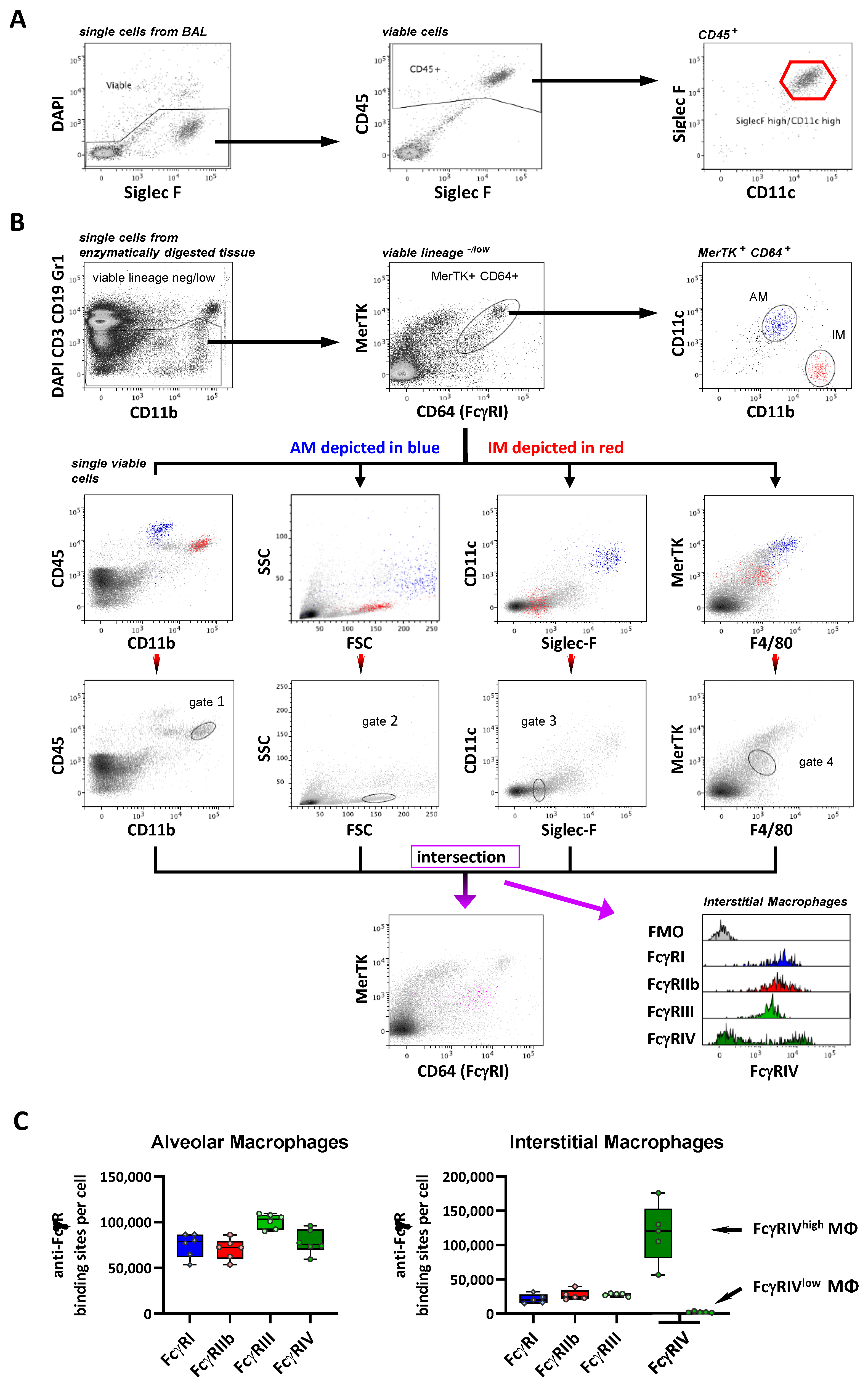
2.3. FcγR Quantification on Lung Macrophage Subpopulations
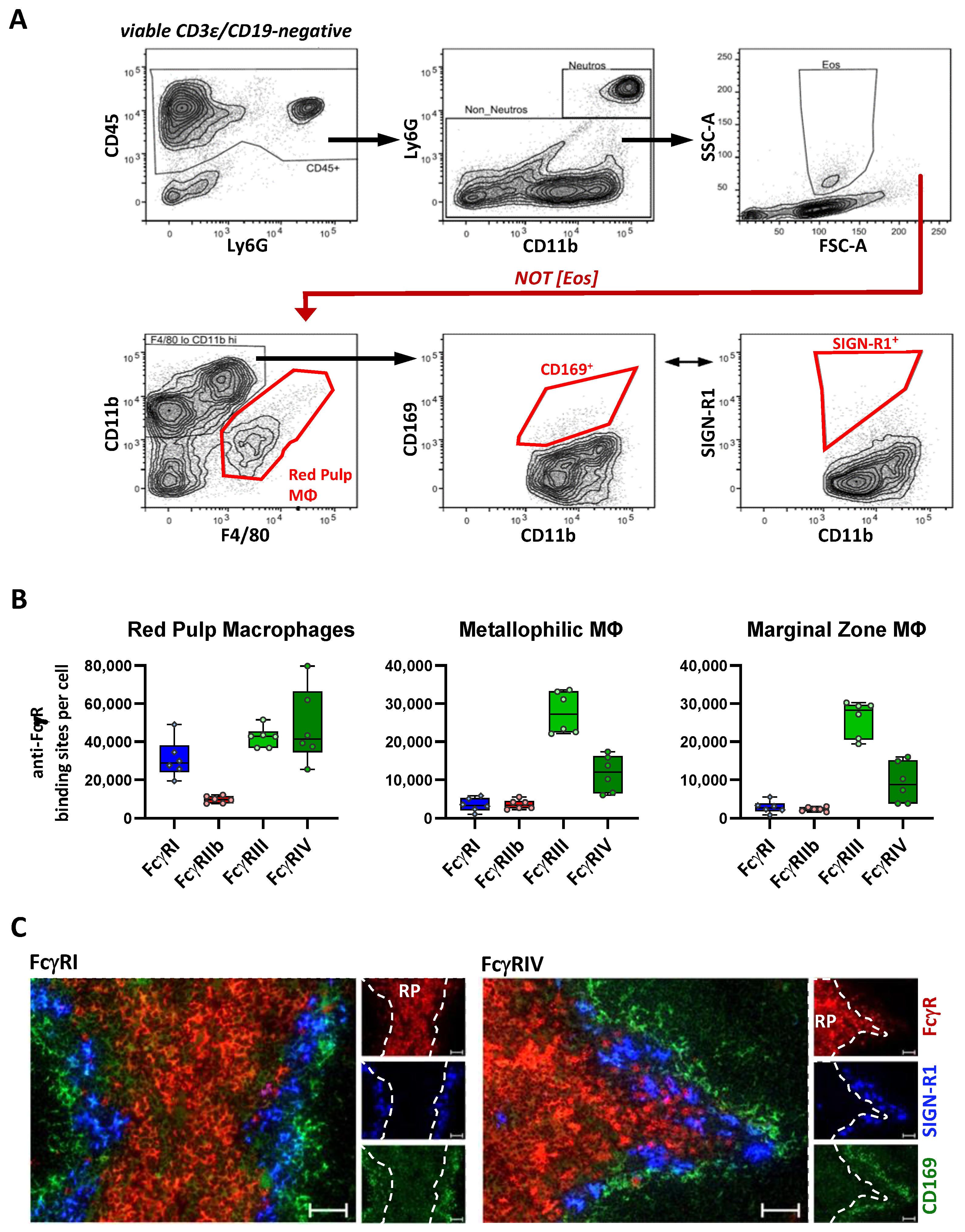
2.4. FcγR Quantification on Splenic Macrophage Subsets
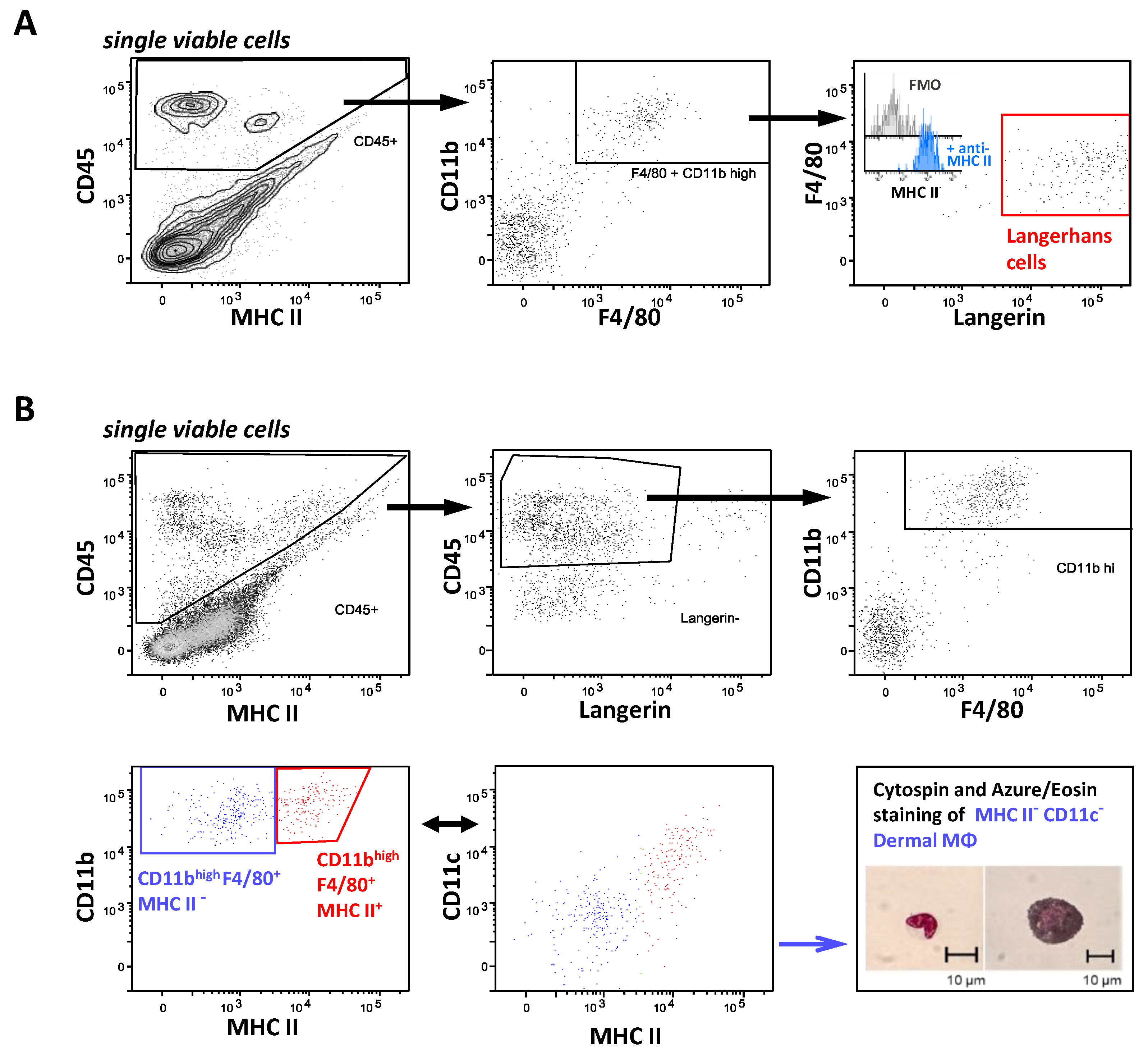
2.5. FcγR Quantification on Skin Resident Macrophage/DC Subsets
2.6. FcγR Quantification on Liver Resident Macrophages
2.7. FcγR Quantification on Kidney Macrophages
2.8. FcγR Quantification on Brain Macrophages
3. Discussion
3.1. Importance of FcγR Quantification on Tissue-Resident Macrophage Subsets
3.2. Comparison to FcγR Expression on Human Tissue Macrophage Subsets
4. Material and Methods
4.1. Mice
4.2. Preparation of Murine Tissue Macrophages
4.3. Flow Cytometric Characterization of Murine Tissue Macrophages
4.4. Bright Field Microscopy
4.5. Fluorescence Microscopy
4.6. Quantification of Fcγ Receptors
4.7. Software
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hirayama, D.; Iida, T.; Nakase, H. The phagocytic function of macrophage-enforcing innate immunity and tissue homeostasis. Int. J. Mol. Sci. 2018, 19, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, S.; Alexander, M.; Misharin, A.V.; Budinger, G.R.S. The role of macrophages in the resolution of inflammation. J. Clin. Investig. 2019, 129, 2619–2628. [Google Scholar] [CrossRef] [Green Version]
- Hine, A.M.; Loke, P. Intestinal Macrophages in Resolving Inflammation. J. Immunol. 2019, 203, 593–599. [Google Scholar] [CrossRef]
- Anfray, C.; Ummarino, A.; Andón, F.T.; Allavena, P. Current Strategies to Target Tumor-Associated-Macrophages to Improve Anti-Tumor Immune Responses. Cells 2019, 9, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaiswal, S.; Chao, M.P.; Majeti, R.; Weissman, I.L. Macrophages as mediators of tumor immunosurveillance. Trends Immunol. 2010, 31, 212–219. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, B.; Biburger, M.; Brückner, C.; Ipsen-Escobedo, A.; Gordan, S.; Lehmann, C.; Voehringer, D.; Winkler, T.; Schaft, N.; Dudziak, D.; et al. Tumor location determines tissue-specific recruitment of tumor-associated macrophages and antibody-dependent immunotherapy response. Sci. Immunol. 2017, 2, eaah6413. [Google Scholar] [CrossRef]
- Montalvao, F.; Garcia, Z.; Celli, S.; Breart, B.; Deguine, J.; Van Rooijen, N.; Bousso, P. The mechanism of anti-CD20–mediated B cell depletion revealed by intravital imaging. J. Clin. Investig. 2013, 123, 5098–5103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krenkel, O.; Tacke, F. Liver macrophages in tissue homeostasis and disease. Nat. Rev. Immunol. 2017, 17, 306–321. [Google Scholar] [CrossRef]
- Allard, B.; Panariti, A.; Martin, J.G. Alveolar Macrophages in the Resolution of Inflammation, Tissue Repair, and Tolerance to Infection. Front. Immunol. 2018, 9, 1777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosser, D.M.; Hamidzadeh, K.; Goncalves, R. Macrophages and the maintenance of homeostasis. Cell. Mol. Immunol. 2021, 18, 579–587. [Google Scholar] [CrossRef]
- Granata, F.; Frattini, A.; Loffredo, S.; Staiano, R.I.; Petraroli, A.; Ribatti, D.; Oslund, R.; Gelb, M.H.; Lambeau, G.; Marone, G.; et al. Production of Vascular Endothelial Growth Factors from Human Lung Macrophages Induced by Group IIA and Group X Secreted Phospholipases A2. J. Immunol. 2010, 184, 5232–5241. [Google Scholar] [CrossRef] [Green Version]
- Staiano, R.I.; Loffredo, S.; Borriello, F.; Iannotti, F.A.; Piscitelli, F.; Orlando, P.; Secondo, A.; Granata, F.; Lepore, M.T.; Fiorelli, A.; et al. Human lung-resident macrophages express CB 1 and CB 2 receptors whose activation inhibits the release of angiogenic and lymphangiogenic factors. J. Leukoc. Biol. 2016, 99, 531–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swirski, F.K.; Nahrendorf, M. Cardioimmunology: The immune system in cardiac homeostasis and disease. Nat. Rev. Immunol. 2018, 18, 733–744. [Google Scholar] [CrossRef]
- Ardain, A.; Marakalala, M.J.; Leslie, A. Tissue-resident innate immunity in the lung. Immunology 2020, 159, 245–256. [Google Scholar] [CrossRef] [Green Version]
- Tesch, G.H. Role of macrophages in complications of Type 2 diabetes. Clin. Exp. Pharmacol. Physiol. 2007, 34, 1016–1019. [Google Scholar] [CrossRef]
- Cassetta, L.; Pollard, J.W. Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug Discov. 2018, 17, 887–904. [Google Scholar] [CrossRef] [PubMed]
- Bogunovic, M.; Ginhoux, F.; Helft, J.; Shang, L.; Greter, M.; Liu, K.; Jakubzick, C.; Ingersoll, M.A.; Leboeuf, M.; Stanley, E.R.; et al. Origin of the Lamina Propria Dendritic Cell Networkc Access. Immunity 2009, 31, 513–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamoutounour, S.; Henri, S.; Lelouard, H.; de Bovis, B.; de Haar, C.; van der Woude, C.J.; Woltman, A.M.; Reyal, Y.; Bonnet, D.; Sichien, D.; et al. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur. J. Immunol. 2012, 42, 3150–3166. [Google Scholar] [CrossRef]
- Ginhoux, F.; Greter, M.; Leboeuf, M.; Nandi, S.; See, P.; Gokhan, S.; Mehler, M.F.; Conway, S.J.; Ng, L.G.; Stanley, E.R.; et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 2010, 330, 841–845. [Google Scholar] [CrossRef] [Green Version]
- Schulz, C.; Gomez Perdiguero, E.; Chorro, L.; Szabo-Rogers, H.; Cagnard, N.; Kierdorf, K.; Prinz, M.; Wu, B.; Jacobsen, S.E.; Pollard, J.W.; et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 2012, 336, 86–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoeffel, G.; Wang, Y.; Greter, M.; See, P.; Teo, P.; Malleret, B.; Leboeuf, M.; Low, D.; Oller, G.; Almeida, F.; et al. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J. Exp. Med. 2012, 209, 1167–1181. [Google Scholar] [CrossRef] [Green Version]
- Guilliams, M.; De Kleer, I.; Henri, S.; Post, S.; Vanhoutte, L.; De Prijck, S.; Deswarte, K.; Malissen, B.; Hammad, H.; Lambrecht, B.N. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J. Exp. Med. 2013, 210, 1977–1992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, D.; Chow, A.; Noizat, C.; Teo, P.; Beasley, M.B.; Leboeuf, M.; Becker, C.D.; See, P.; Price, J.; Lucas, D.; et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 2013, 38, 792–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yona, S.; Kim, K.-W.; Wolf, Y.; Mildner, A.; Varol, D.; Breker, M.; Strauss-Ayali, D.; Viukov, S.; Guilliams, M.; Misharin, A.; et al. Fate Mapping Reveals Origins and Dynamics of Monocytes and Tissue Macrophages under Homeostasis. Immunity 2013, 38, 79–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Zou, W.; Du, J.; Zhao, Y. The origins and homeostasis of monocytes and tissue-resident macrophages in physiological situation. J. Cell. Physiol. 2018, 233, 6425–6439. [Google Scholar] [CrossRef] [PubMed]
- Brandsma, A.M.; Hogarth, P.M.; Nimmerjahn, F.; Leusen, J.H.W. Clarifying the Confusion between Cytokine and Fc Receptor “Common Gamma Chain”. Immunity 2016, 45, 225–226. [Google Scholar] [CrossRef] [Green Version]
- Nimmerjahn, F.; Ravetch, J.V. Fcgamma receptors: Old friends and new family members. Immunity 2006, 24, 19–28. [Google Scholar] [CrossRef] [Green Version]
- Hargreaves, C.E.; Rose-Zerilli, M.J.J.; Machado, L.R.; Iriyama, C.; Hollox, E.J.; Cragg, M.S.; Strefford, J.C. Fcγ receptors: Genetic variation, function, and disease. Immunol. Rev. 2015, 268, 6–24. [Google Scholar] [CrossRef]
- Guilliams, M.; Bruhns, P.; Saeys, Y.; Hammad, H.; Lambrecht, B.N. The function of Fcγ receptors in dendritic cells and macrophages. Nat. Rev. Immunol. 2014, 14, 94–108. [Google Scholar] [CrossRef]
- Joller, N.; Weber, S.S.; Muller, A.J.; Sporri, R.; Selchow, P.; Sander, P.; Hilbi, H.; Oxenius, A. Antibodies protect against intracellular bacteria by Fc receptor-mediated lysosomal targeting. Proc. Natl. Acad. Sci. USA 2010, 107, 20441–20446. [Google Scholar] [CrossRef] [Green Version]
- Nimmerjahn, F.; Gordan, S.; Lux, A. FcγR dependent mechanisms of cytotoxic, agonistic, and neutralizing antibody activities. Trends Immunol. 2015, 36, 325–336. [Google Scholar] [CrossRef] [Green Version]
- Roghanian, A.; Hu, G.; Fraser, C.; Singh, M.; Foxall, R.B.; Meyer, M.J.; Lees, E.; Huet, H.; Glennie, M.J.; Beers, S.A.; et al. Cyclophosphamide Enhances Cancer Antibody Immunotherapy in the Resistant Bone Marrow Niche by Modulating Macrophage FcγR Expression. Cancer Immunol. Res. 2019, 7, 1876–1890. [Google Scholar] [CrossRef]
- Gordan, S.; Albert, H.; Danzer, H.; Lux, A.; Biburger, M.; Nimmerjahn, F. The Immunological Organ Environment Dictates the Molecular and Cellular Pathways of Cytotoxic Antibody Activity. Cell Rep. 2019, 29, 3033–3046. [Google Scholar] [CrossRef] [Green Version]
- Gerber, J.S.; Mosser, D.M. Reversing Lipopolysaccharide Toxicity by Ligating the Macrophage Fcγ Receptors. J. Immunol. 2001, 166, 6861–6868. [Google Scholar] [CrossRef] [PubMed]
- Sutterwala, F.S.; Noel, G.J.; Clynes, R.; Mosser, D.M. Selective Suppression of Interleukin-12 Induction after Macrophage Receptor Ligation. J. Exp. Med. 1997, 185, 1977–1985. [Google Scholar] [CrossRef] [Green Version]
- Guzman, M.G.; Vazquez, S. The Complexity of Antibody-Dependent Enhancement of Dengue Virus Infection. Viruses 2010, 2, 2649–2662. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.T.; Sewatanon, J.; Memoli, M.J.; Wrammert, J.; Bournazos, S.; Bhaumik, S.K.; Pinsky, B.A.; Chokephaibulkit, K.; Onlamoon, N.; Pattanapanyasat, K.; et al. IgG antibodies to dengue enhanced for FcγRIIIA binding determine disease severity. Science 2017, 355, 395–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bournazos, S.; Vo, H.T.M.; Duong, V.; Auerswald, H.; Ly, S.; Sakuntabhai, A.; Dussart, P.; Cantaert, T.; Ravetch, J.V. Antibody fucosylation predicts disease severity in secondary dengue infection. Science 2021, 372, 1102–1105. [Google Scholar] [CrossRef] [PubMed]
- Lux, A.; Yu, X.; Scanlan, C.N.; Nimmerjahn, F. Impact of immune complex size and glycosylation on IgG binding to human FcγRs. J. Immunol. 2013, 190, 4315–4323. [Google Scholar] [CrossRef] [Green Version]
- Nimmerjahn, F.; Ravetch, J.V. Divergent Immunoglobulin G Subclass Activity Through Selective Fc Receptor Binding. Science 2005, 310, 1510–1512. [Google Scholar] [CrossRef] [Green Version]
- Robinett, R.A.; Guan, N.; Lux, A.; Biburger, M.; Nimmerjahn, F.; Meyer, A.S. Dissecting FcγR Regulation through a Multivalent Binding Model. Cell Syst. 2018, 7, 41–48. [Google Scholar] [CrossRef] [Green Version]
- Kerntke, C.; Nimmerjahn, F.; Biburger, M. There Is (Scientific) Strength in Numbers: A Comprehensive Quantitation of Fc Gamma Receptor Numbers on Human and Murine Peripheral Blood Leukocytes. Front. Immunol. 2020, 11, 118. [Google Scholar] [CrossRef]
- Hobday, P.M.; Auger, J.L.; Schuneman, G.R.; Haasken, S.; Verbeek, J.S.; Binstadt, B.A. Fcγ Receptor III and Fcγ Receptor IV on Macrophages Drive Autoimmune Valvular Carditis in Mice. Arthritis Rheumatol. 2014, 66, 852–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mancardi, D.A.; Iannascoli, B.; Hoos, S.; England, P.; Daëron, M.; Bruhns, P. FcγRIV is a mouse IgE receptor that resembles macrophage FcεRI in humans and promotes IgE-induced lung inflammation. J. Clin. Investig. 2008, 118, 3738–3750. [Google Scholar] [CrossRef]
- Shushakova, N.; Skokowa, J.; Schulman, J.; Baumann, U.; Zwirner, J.; Schmidt, R.E.; Gessner, J.E. C5a anaphylatoxin is a major regulator of activating versus inhibitory FcγRs in immune complex–induced lung disease. J. Clin. Investig. 2002, 110, 1823–1830. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Ali, S.R.; Konrad, S.; Zwirner, J.; Verbeek, J.S.; Schmidt, R.E.; Gessner, J.E. Cell-derived anaphylatoxins as key mediators of antibody-dependent type II autoimmunity in mice. J. Clin. Investig. 2006, 116, 512–520. [Google Scholar] [CrossRef] [Green Version]
- Syed, S.N.; Konrad, S.; Wiege, K.; Nieswandt, B.; Nimmerjahn, F.; Schmidt, R.E.; Gessner, J.E. Both FcγRIV and FcγRIII are essential receptors mediating type II and type III autoimmune responses via FcRγ-LAT-dependent generation of C5a. Eur. J. Immunol. 2009, 39, 3343–3356. [Google Scholar] [CrossRef] [PubMed]
- Lunnon, K.; Teeling, J.L.; Tutt, A.L.; Cragg, M.S.; Glennie, M.J.; Perry, V.H. Systemic Inflammation Modulates Fc Receptor Expression on Microglia during Chronic Neurodegeneration. J. Immunol. 2011, 186, 7215–7224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lever, J.M.; Hull, T.D.; Boddu, R.; Pepin, M.E.; Black, L.M.; Adedoyin, O.O.; Yang, Z.; Traylor, A.M.; Jiang, Y.; Li, Z.; et al. Resident macrophages reprogram toward a developmental state after acute kidney injury. JCI Insight 2019, 4, e125503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suwanichkul, A.; Wenderfer, S.E. Differential expression of functional Fc-receptors and additional immune complex receptors on mouse kidney cells. Mol. Immunol. 2013, 56, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Schyns, J.; Bureau, F.; Marichal, T. Lung Interstitial Macrophages: Past, Present, and Future. J. Immunol. Res. 2018, 2018, 5160794. [Google Scholar] [CrossRef]
- Gibbings, S.L.; Thomas, S.M.; Atif, S.M.; McCubbrey, A.L.; Desch, A.N.; Danhorn, T.; Leach, S.M.; Bratton, D.L.; Henson, P.M.; Janssen, W.J.; et al. Three Unique Interstitial Macrophages in the Murine Lung at Steady State. Am. J. Respir. Cell Mol. Biol. 2017, 57, 66–76. [Google Scholar] [CrossRef]
- Merad, M.; Ginhoux, F.; Collin, M. Origin, homeostasis and function of Langerhans cells and other langerin-expressing dendritic cells. Nat. Rev. Immunol. 2008, 8, 935–947. [Google Scholar] [CrossRef] [PubMed]
- Tamoutounour, S.; Guilliams, M.; MontananaSanchis, F.; Liu, H.; Terhorst, D.; Malosse, C.; Pollet, E.; Ardouin, L.; Luche, H.; Sanchez, C.; et al. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity 2013, 39, 925–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganesan, L.P.; Kim, J.; Wu, Y.; Mohanty, S.; Phillips, G.S.; Birmingham, D.J.; Robinson, J.M.; Anderson, C.L. FcγRIIb on Liver Sinusoidal Endothelium Clears Small Immune Complexes. J. Immunol. 2012, 189, 4981–4988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, R.W.; Hawley, C.A.; Pellicoro, A.; Bain, C.C.; Iredale, J.P.; Jenkins, S.J. An efficient method to isolate Kupffer cells eliminating endothelial cell contamination and selective bias. J. Leukoc. Biol. 2018, 104, 579–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otten, M.A.; van der Bij, G.J.; Verbeek, S.J.; Nimmerjahn, F.; Ravetch, J.V.; Beelen, R.H.J.; van de Winkel, J.G.J.; van Egmond, M. Experimental antibody therapy of liver metastases reveals functional redundancy between Fc gammaRI and Fc gammaRIV. J. Immunol. 2008, 181, 6829–6836. [Google Scholar] [CrossRef] [Green Version]
- Jenne, C.N.; Kubes, P. Immune surveillance by the liver. Nat. Immunol. 2013, 14, 996–1006. [Google Scholar] [CrossRef]
- Fossati-Jimack, L.; Ioan-Facsinay, A.; Reininger, L.; Chicheportiche, Y.; Watanabe, N.; Saito, T.; Hofhuis, F.M.A.; Gessner, J.E.; Schiller, C.; Schmidt, R.E.; et al. Markedly Different Pathogenicity of Four Immunoglobulin G Isotype-Switch Variants of an Antierythrocyte Autoantibody Is Based on Their Capacity to Interact in Vivo with the Low-Affinity Fcγ Receptor III. J. Exp. Med. 2000, 191, 1293–1302. [Google Scholar] [CrossRef] [Green Version]
- Bevaart, L.; Jansen, M.J.H.; van Vugt, M.J.; Verbeek, J.S.; van de Winkel, J.G.J.; Leusen, J.H.W. The High-Affinity IgG Receptor, FcγRI, Plays a Central Role in Antibody Therapy of Experimental Melanoma. Cancer Res. 2006, 66, 1261–1264. [Google Scholar] [CrossRef] [Green Version]
- DiLillo, D.J.; Palese, P.; Wilson, P.C.; Ravetch, J.V. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Investig. 2016, 126, 605–610. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Chen, C.-J.; Mullarkey, C.E.; Hamilton, J.R.; Wong, C.K.; Leon, P.E.; Uccellini, M.B.; Chromikova, V.; Henry, C.; Hoffman, K.W.; et al. Alveolar macrophages are critical for broadly-reactive antibody-mediated protection against influenza A virus in mice. Nat. Commun. 2017, 8, 846. [Google Scholar] [CrossRef]
- El Bakkouri, K.; Descamps, F.; De Filette, M.; Smet, A.; Festjens, E.; Birkett, A.; Van Rooijen, N.; Verbeek, S.; Fiers, W.; Saelens, X. Universal Vaccine Based on Ectodomain of Matrix Protein 2 of Influenza A: Fc Receptors and Alveolar Macrophages Mediate Protection. J. Immunol. 2011, 186, 1022–1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasui, F.; Kohara, M.; Kitabatake, M.; Nishiwaki, T.; Fujii, H.; Tateno, C.; Yoneda, M.; Morita, K.; Matsushima, K.; Koyasu, S.; et al. Phagocytic cells contribute to the antibody-mediated elimination of pulmonary-infected SARS coronavirus. Virology 2014, 454–455, 157–168. [Google Scholar] [CrossRef] [Green Version]
- Koga, H.; Prost-Squarcioni, C.; Iwata, H.; Jonkman, M.F.; Ludwig, R.J.; Bieber, K. Epidermolysis Bullosa Acquisita: The 2019 Update. Front. Med. 2019, 5, 362. [Google Scholar] [CrossRef]
- Hirose, M.; Kasprick, A.; Beltsiou, F.; Dieckhoff, K.S.; Schulze, F.S.; Samavedam, U.K.J.S.R.L.; Hundt, J.E.; Pas, H.H.; Jonkman, M.F.; Schmidt, E.; et al. Reduced Skin Blistering in Experimental Epidermolysis Bullosa Acquisita After Anti-TNF Treatment. Mol. Med. 2016, 22, 918–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovacs, B.; Tillmann, J.; Freund, L.-C.; Nimmerjahn, F.; Sadik, C.D.; Bieber, K.; Ludwig, R.J.; Karsten, C.M.; Köhl, J. Fcγ Receptor IIB Controls Skin Inflammation in an Active Model of Epidermolysis Bullosa Acquisita. Front. Immunol. 2020, 10, 3012. [Google Scholar] [CrossRef]
- Stamatiades, E.G.; Tremblay, M.E.; Bohm, M.; Crozet, L.; Bisht, K.; Kao, D.; Coelho, C.; Fan, X.; Yewdell, W.T.; Davidson, A.; et al. Immune Monitoring of Trans-endothelial Transport by Kidney-Resident Macrophages. Cell 2016, 166, 991–1003. [Google Scholar] [CrossRef] [Green Version]
- Kaneko, Y.; Nimmerjahn, F.; Madaio, M.P.; Ravetch, J.V. Pathology and protection in nephrotoxic nephritis is determined by selective engagement of specific Fc receptors. J. Exp. Med. 2006, 203, 789–797. [Google Scholar] [CrossRef] [Green Version]
- Chauhan, P.; Hu, S.; Sheng, W.S.; Prasad, S.; Lokensgard, J.R. Modulation of Microglial Cell Fcγ Receptor Expression Following Viral Brain Infection. Sci. Rep. 2017, 7, 41889. [Google Scholar] [CrossRef] [Green Version]
- Bruggeman, C.W.; Houtzager, J.; Dierdorp, B.; Kers, J.; Pals, S.T.; Lutter, R.; van Gulik, T.; den Haan, J.M.M.; van den Berg, T.K.; van Bruggen, R.; et al. Tissue-specific expression of IgG receptors by human macrophages ex vivo. PLoS ONE 2019, 14, e0223264. [Google Scholar] [CrossRef]
- Nagelkerke, S.Q.; Bruggeman, C.W.; den Haan, J.M.M.; Mul, E.P.J.; van den Berg, T.K.; van Bruggen, R.; Kuijpers, T.W. Red pulp macrophages in the human spleen are a distinct cell population with a unique expression of Fc-γ receptors. Blood Adv. 2018, 2, 941–953. [Google Scholar] [CrossRef] [Green Version]
- Unkeless, J.C. Characterization of a monoclonal antibody directed against mouse macrophage and lymphocyte Fc receptors. J. Exp. Med. 1979, 150, 580–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biburger, M.; Trenkwald, I.; Nimmerjahn, F. Three blocks are not enough-Blocking of the murine IgG receptor FcγRIV is crucial for proper characterization of cells by FACS analysis. Eur. J. Immunol. 2015, 45, 2694–2697. [Google Scholar] [CrossRef] [PubMed]
- Deka, C.; Lehnert, B.E.; Lehnert, N.M.; Jones, G.M.; Sklar, L.A.; Steinkamp, J.A. Analysis of fluorescence lifetime and quenching of FITC-conjugated antibodies on cells by phase-sensitive flow cytometry. Cytometry 1996, 25, 271–279. [Google Scholar] [CrossRef]


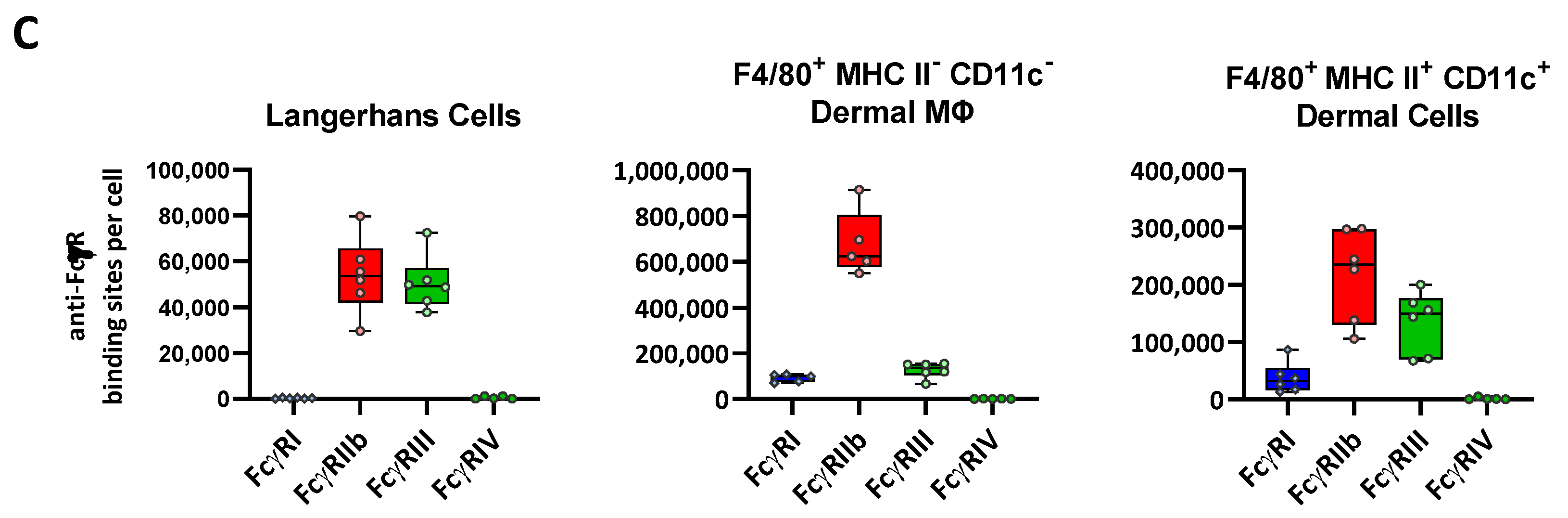
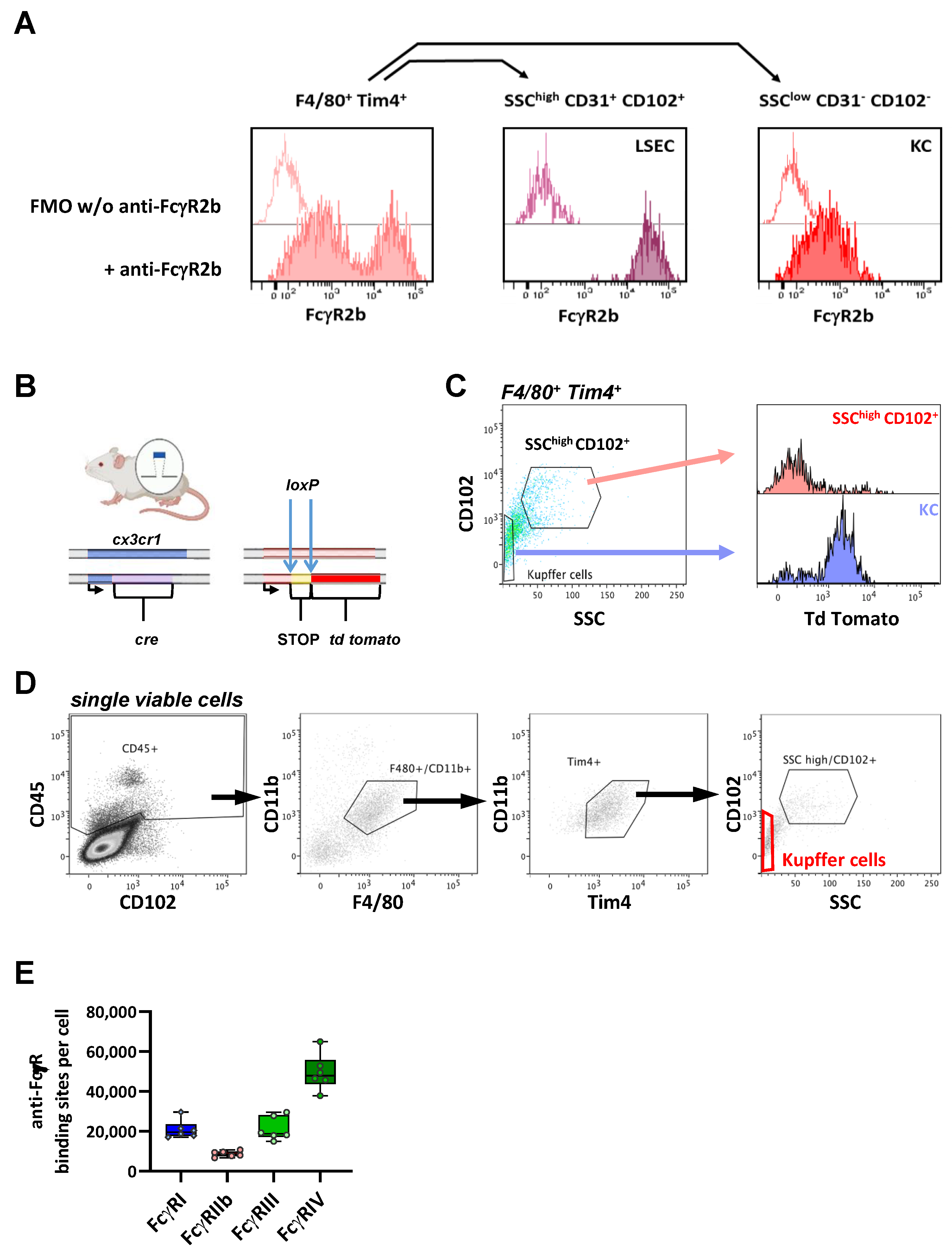
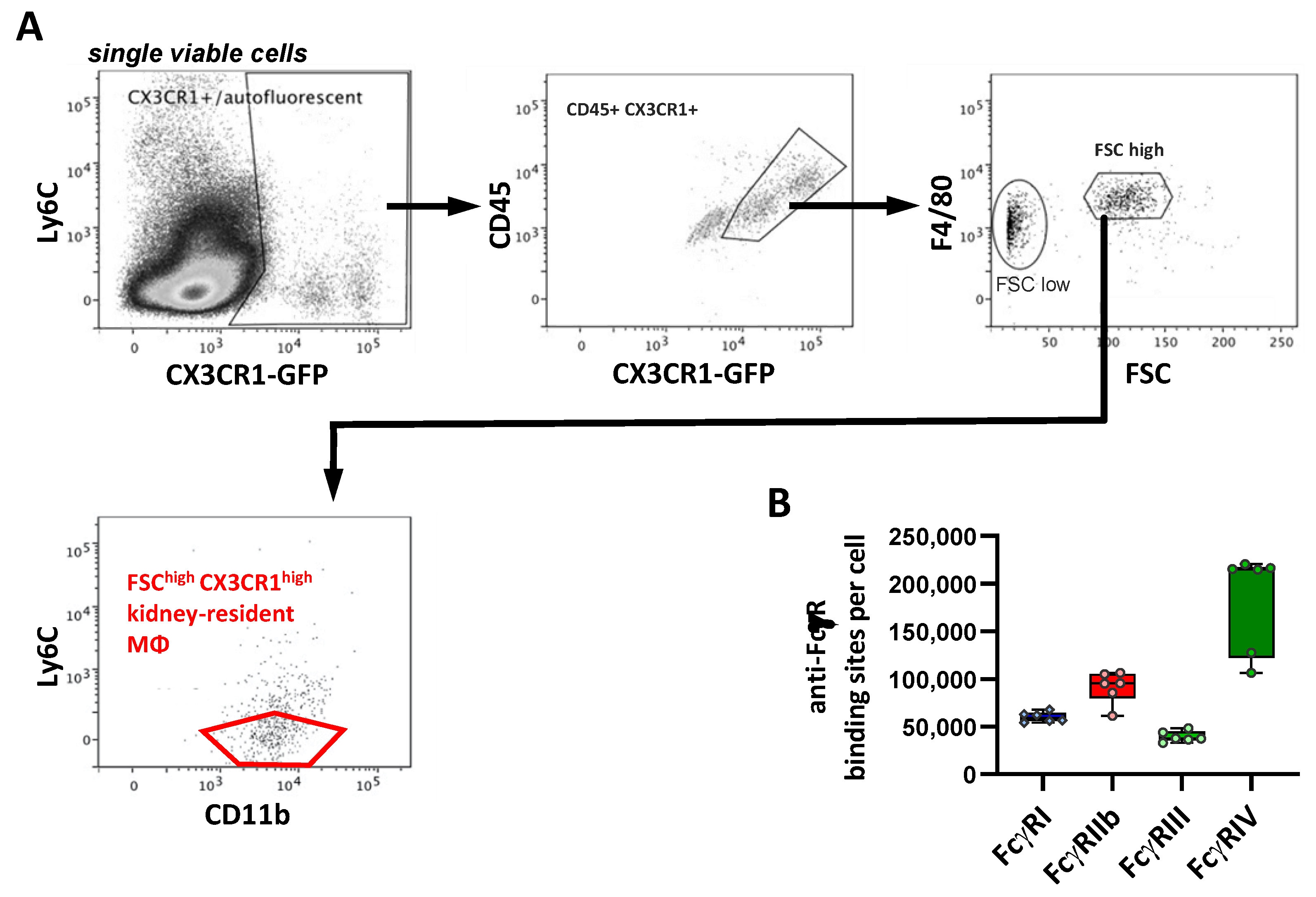

| Median ABC (×10³ Binding Sites/Cell) | FcγRI | FcγRIIb | FcγRIII | FcγRIV | |
|---|---|---|---|---|---|
 | Kupffer Cells | 20 | 9 | 19 | 48 |
 | Alveolar Macrophages | 79 | 73 | 103 | 76 |
| Interstitial Macrophages | 20 | 24 | 29 | 121/2 * | |
 | Red Pulp Macrophages | 29 | 10 | 43 | 41 |
| Metallophilic Macrophages | 3 | 3 | 27 | 12 | |
| Marginal Zone Macrophages | 3 | 2 | 28 | 9 | |
 | Langerhans Cells | <1 | 54 | 49 | <1 |
| MHC II− CD11c− Dermal Macrophages | 99 | 624 | 135 | <1 | |
| MHC II+ CD11clow to + Dermal Cells | 32 | 236 | 150 | <1 | |
 | Kidney-resident Macrophages | 59 | 95 | 38 | 215 |
 | Microglia | 46 | 55 | 28 | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vorsatz, C.; Friedrich, N.; Nimmerjahn, F.; Biburger, M. There Is Strength in Numbers: Quantitation of Fc Gamma Receptors on Murine Tissue-Resident Macrophages. Int. J. Mol. Sci. 2021, 22, 12172. https://doi.org/10.3390/ijms222212172
Vorsatz C, Friedrich N, Nimmerjahn F, Biburger M. There Is Strength in Numbers: Quantitation of Fc Gamma Receptors on Murine Tissue-Resident Macrophages. International Journal of Molecular Sciences. 2021; 22(22):12172. https://doi.org/10.3390/ijms222212172
Chicago/Turabian StyleVorsatz, Christof, Niklas Friedrich, Falk Nimmerjahn, and Markus Biburger. 2021. "There Is Strength in Numbers: Quantitation of Fc Gamma Receptors on Murine Tissue-Resident Macrophages" International Journal of Molecular Sciences 22, no. 22: 12172. https://doi.org/10.3390/ijms222212172
APA StyleVorsatz, C., Friedrich, N., Nimmerjahn, F., & Biburger, M. (2021). There Is Strength in Numbers: Quantitation of Fc Gamma Receptors on Murine Tissue-Resident Macrophages. International Journal of Molecular Sciences, 22(22), 12172. https://doi.org/10.3390/ijms222212172







