Transcriptome and Metabolome Analyses Provide Insights into the Stomium Degeneration Mechanism in Lily
Abstract
:1. Introduction
2. Results
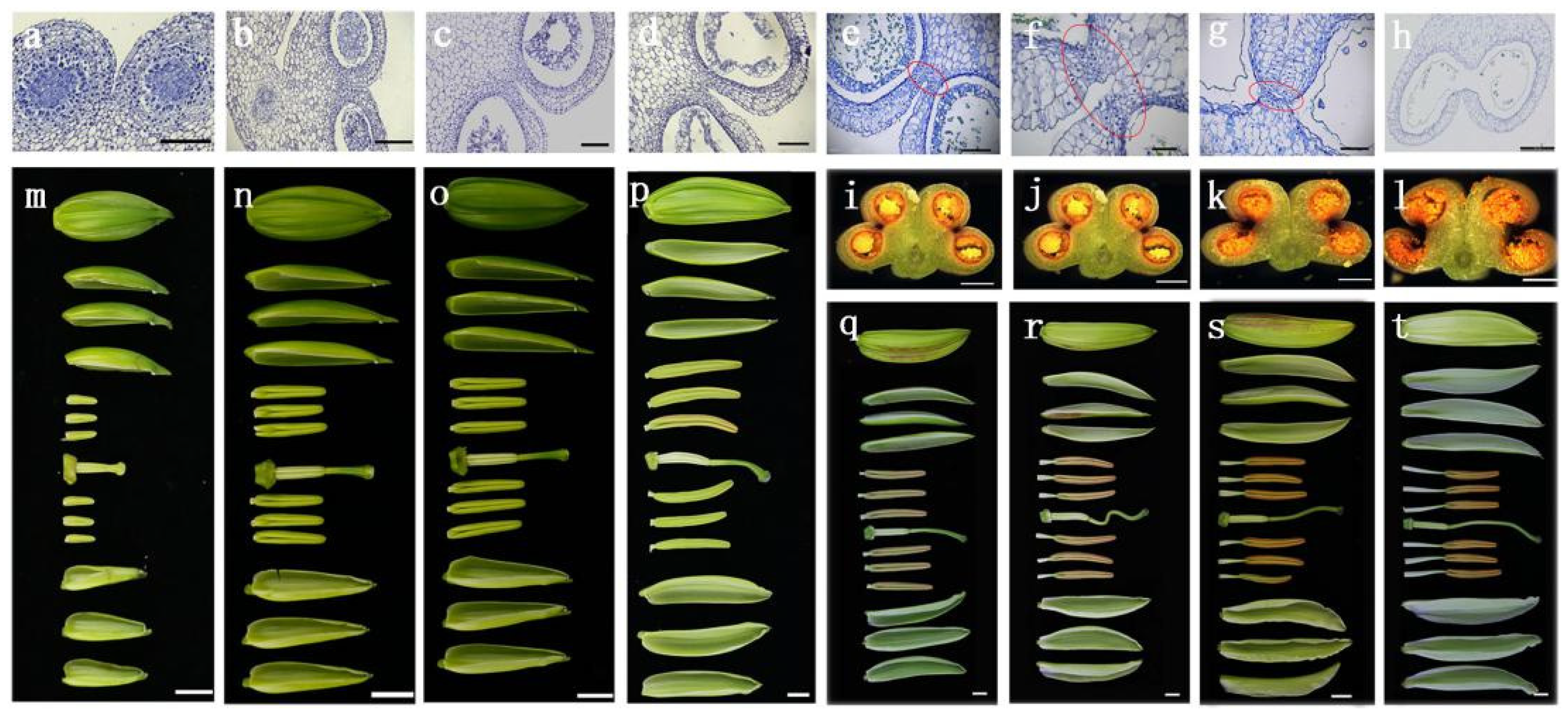
2.1. Occurrence of Anther Dehiscence in Lily Flower Buds 6–8 cm in Length
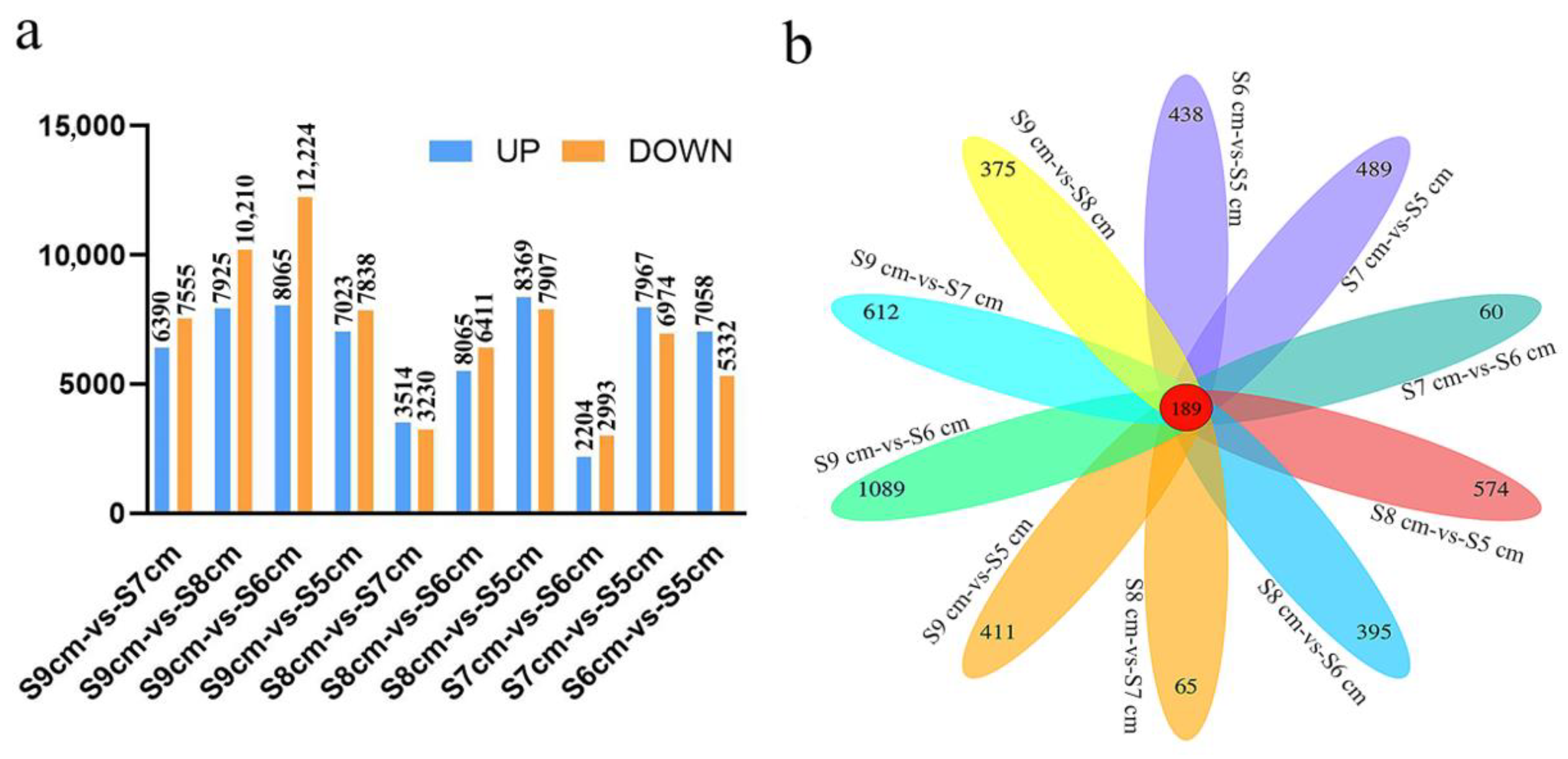
2.2. Regulation of Stomium Zone Degradation Showing Fewer Differentially Expressed Genes
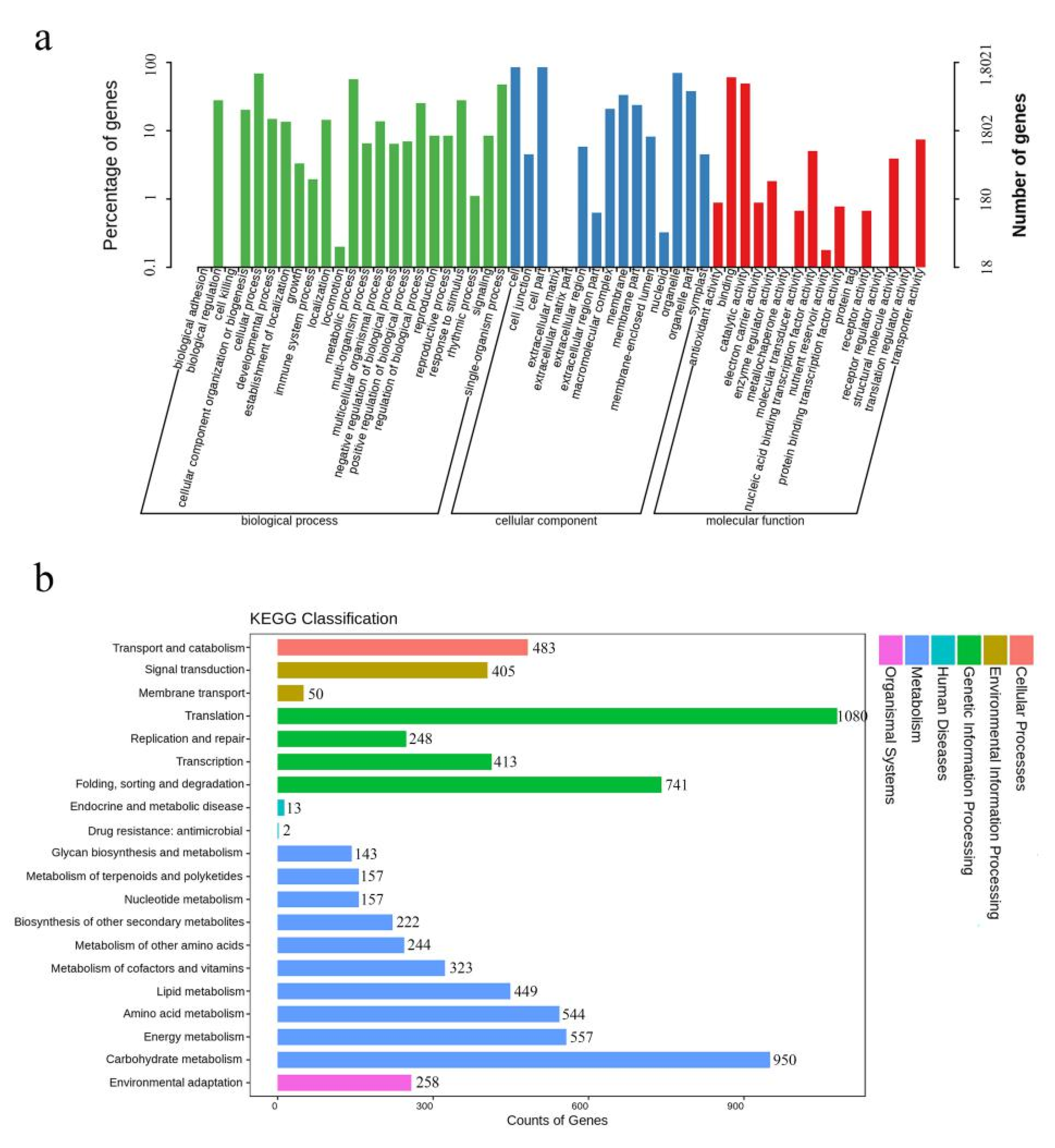
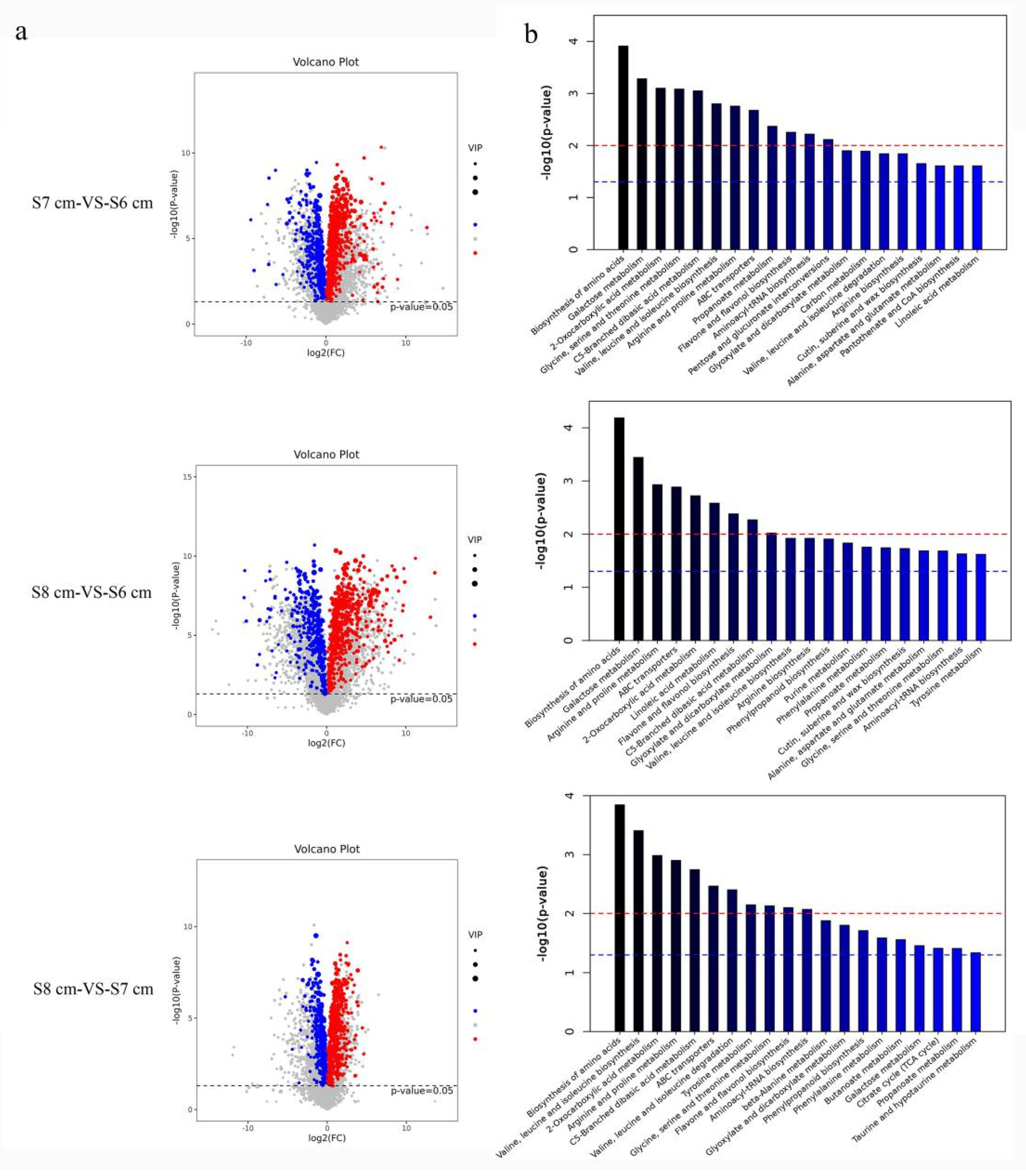
2.3. Classification Categories of Differentially Expressed Genes Involved in Anther Dehiscence
2.4. Jasmonate Biosynthesis and Signal Transduction Roles in Stomium Zone Degradation
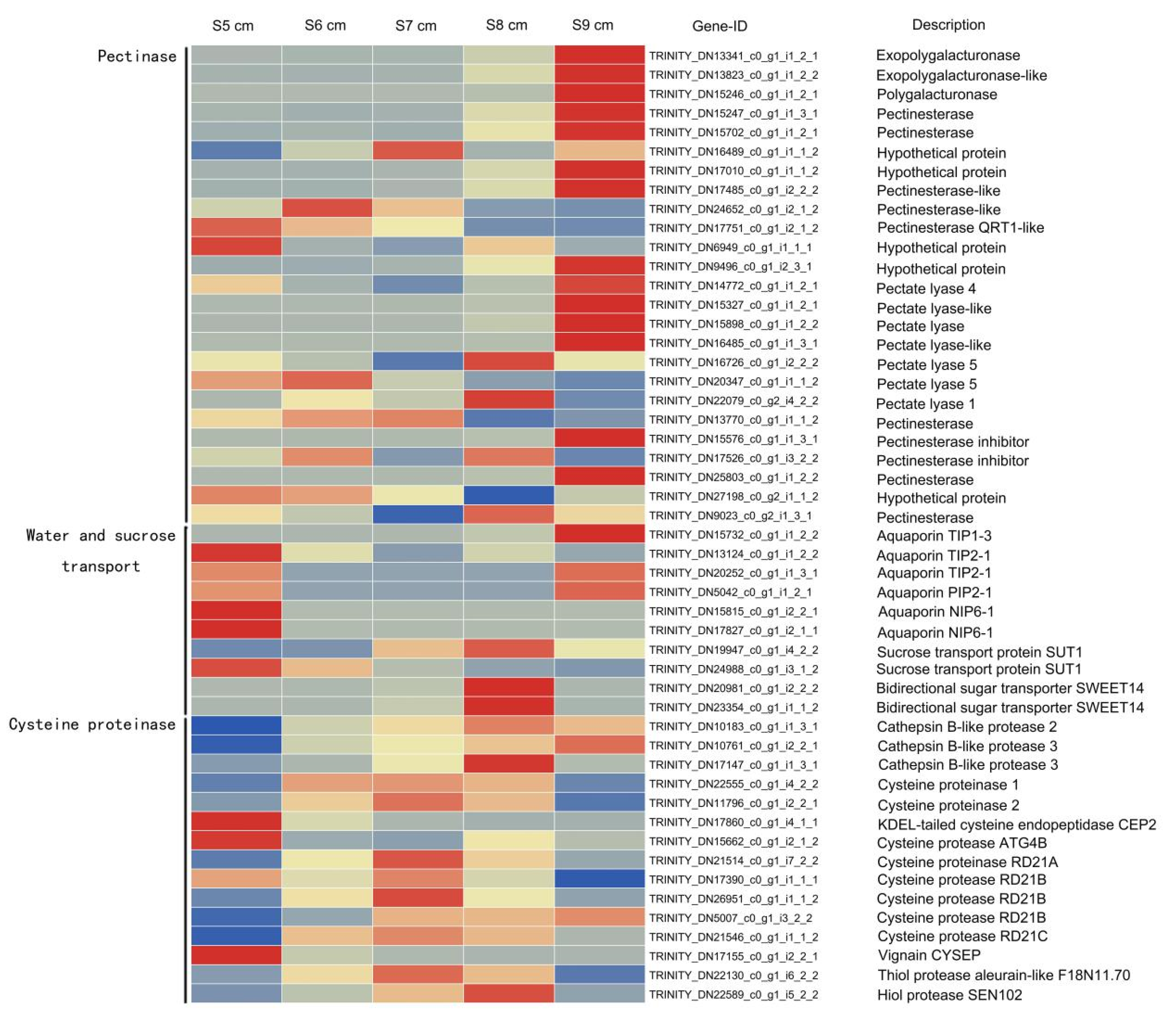
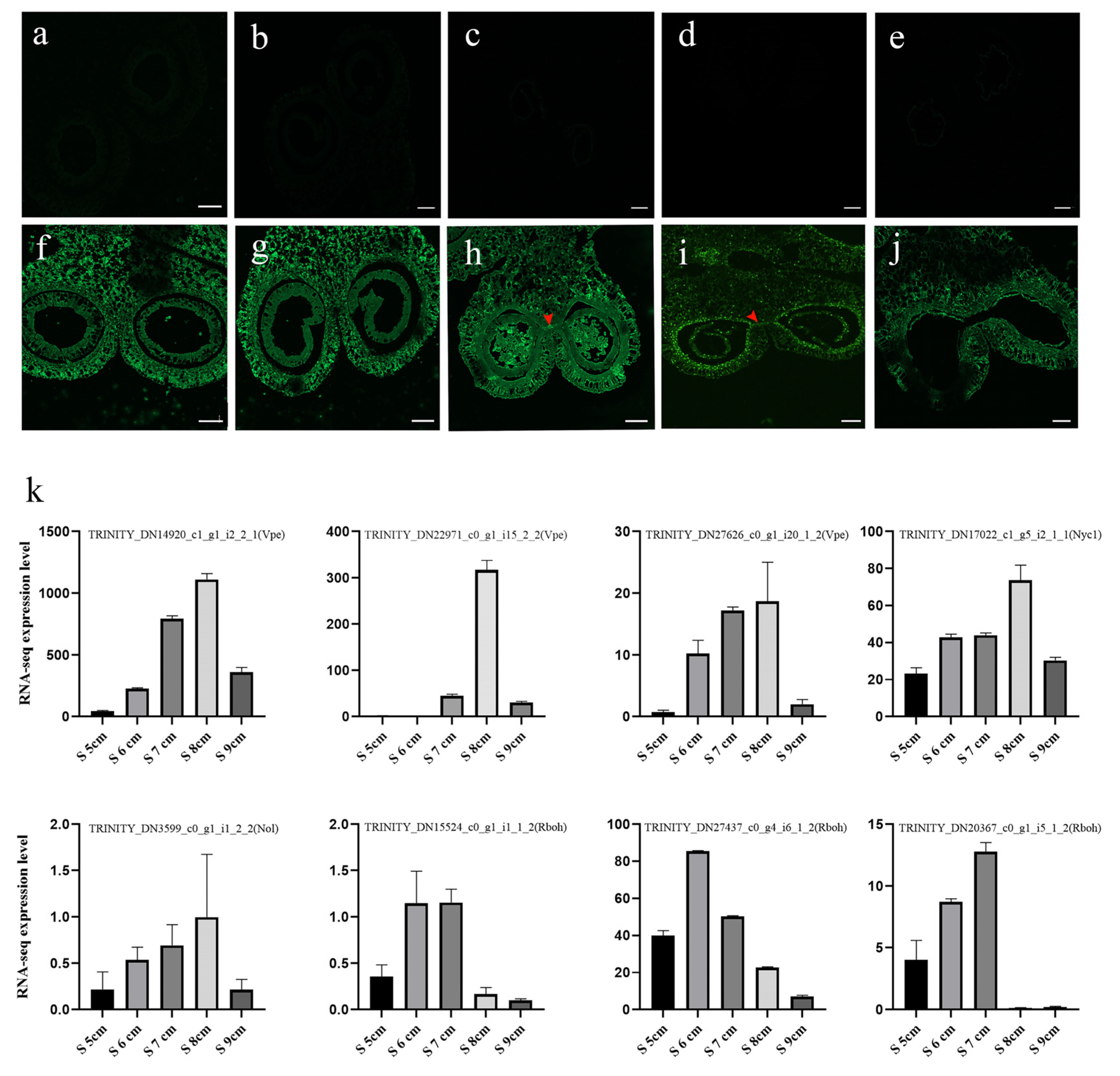
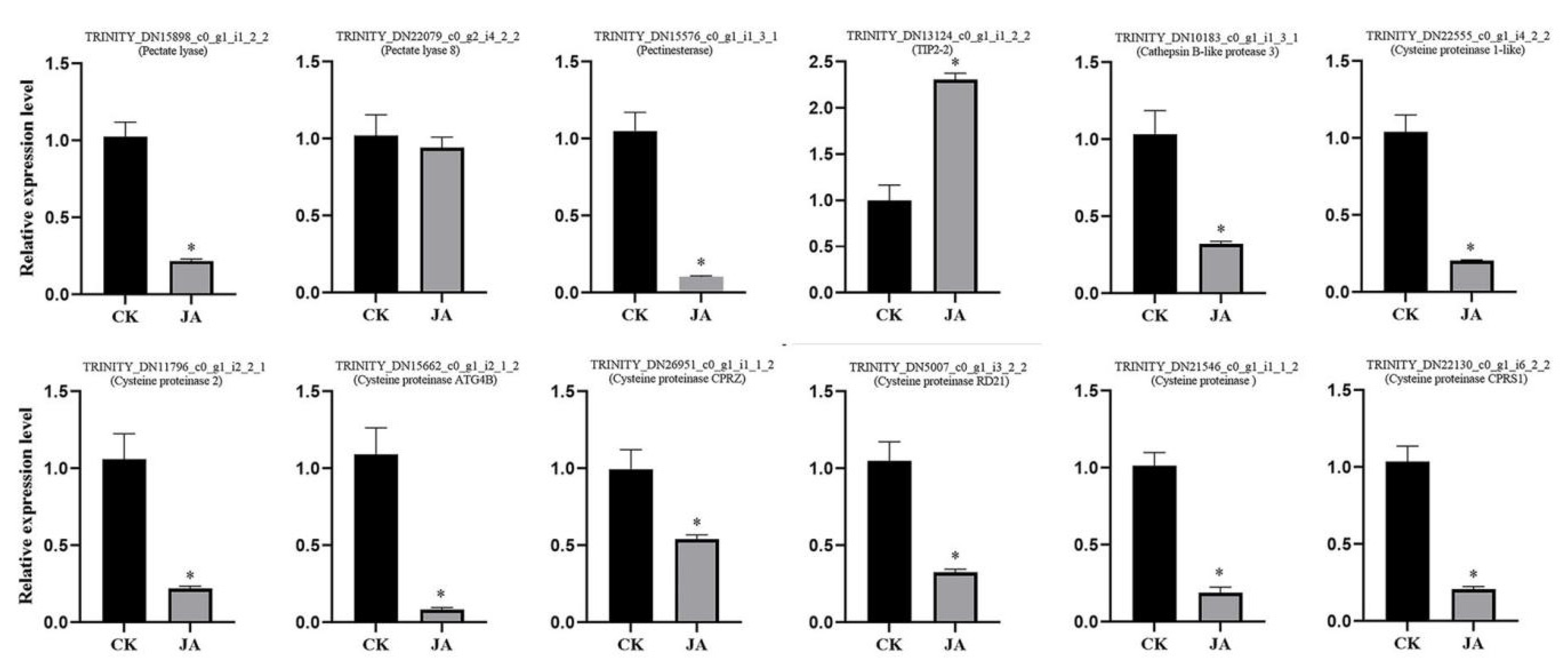
2.5. Genes Associated with Cysteine Proteinases during Stomium Zone Degradation
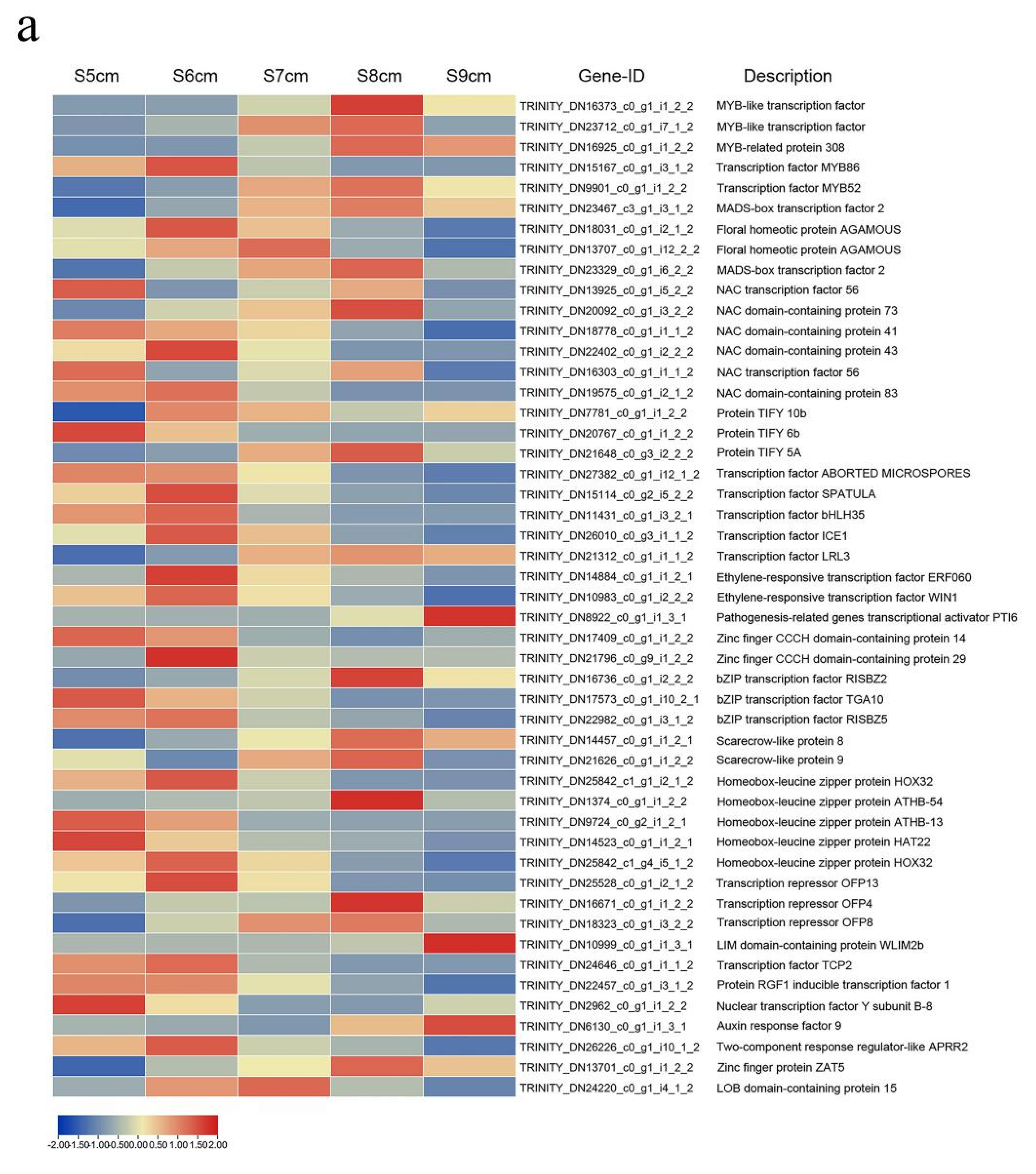
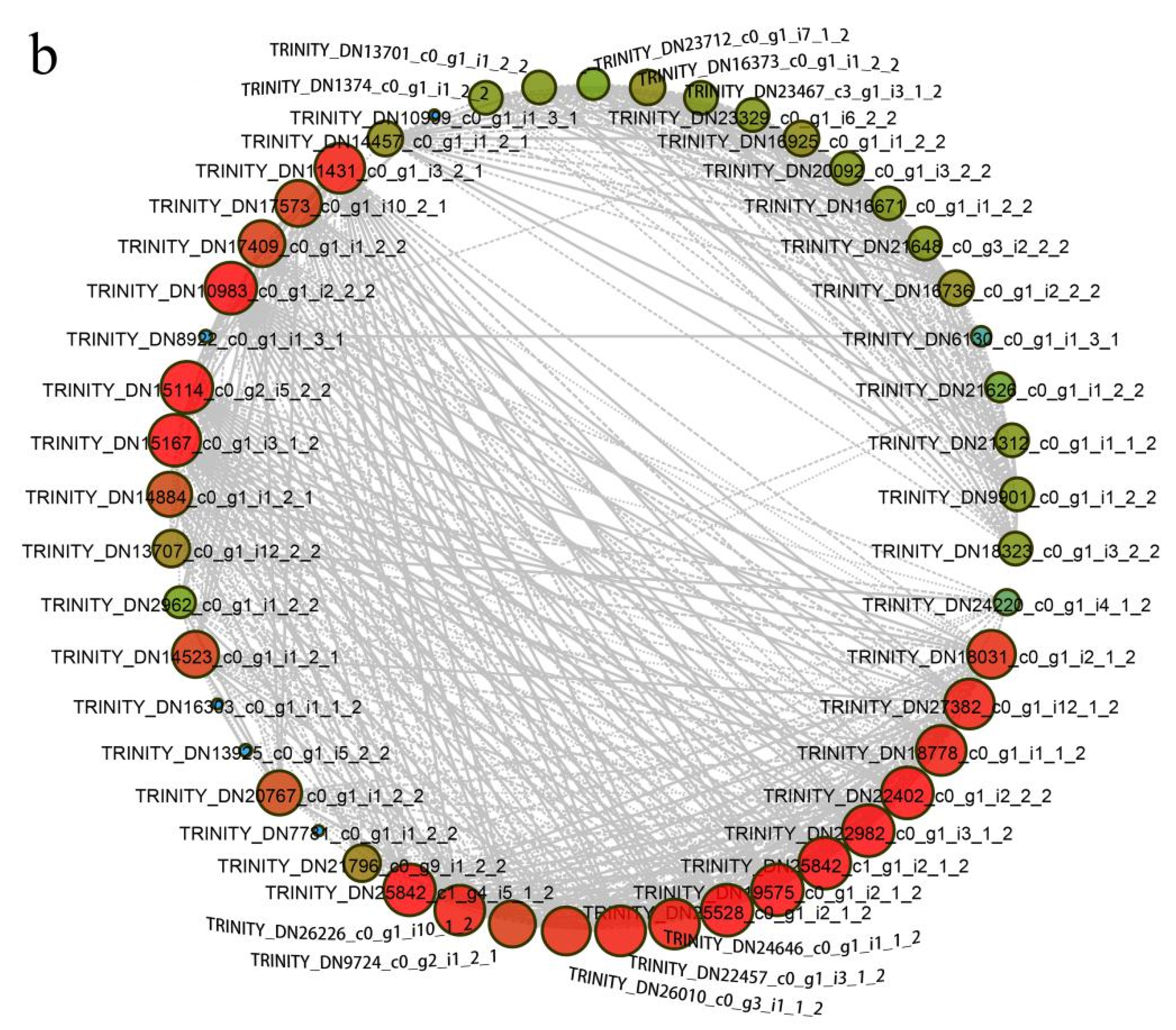
2.6. Transcription Factors Roles in Anther Dehiscence
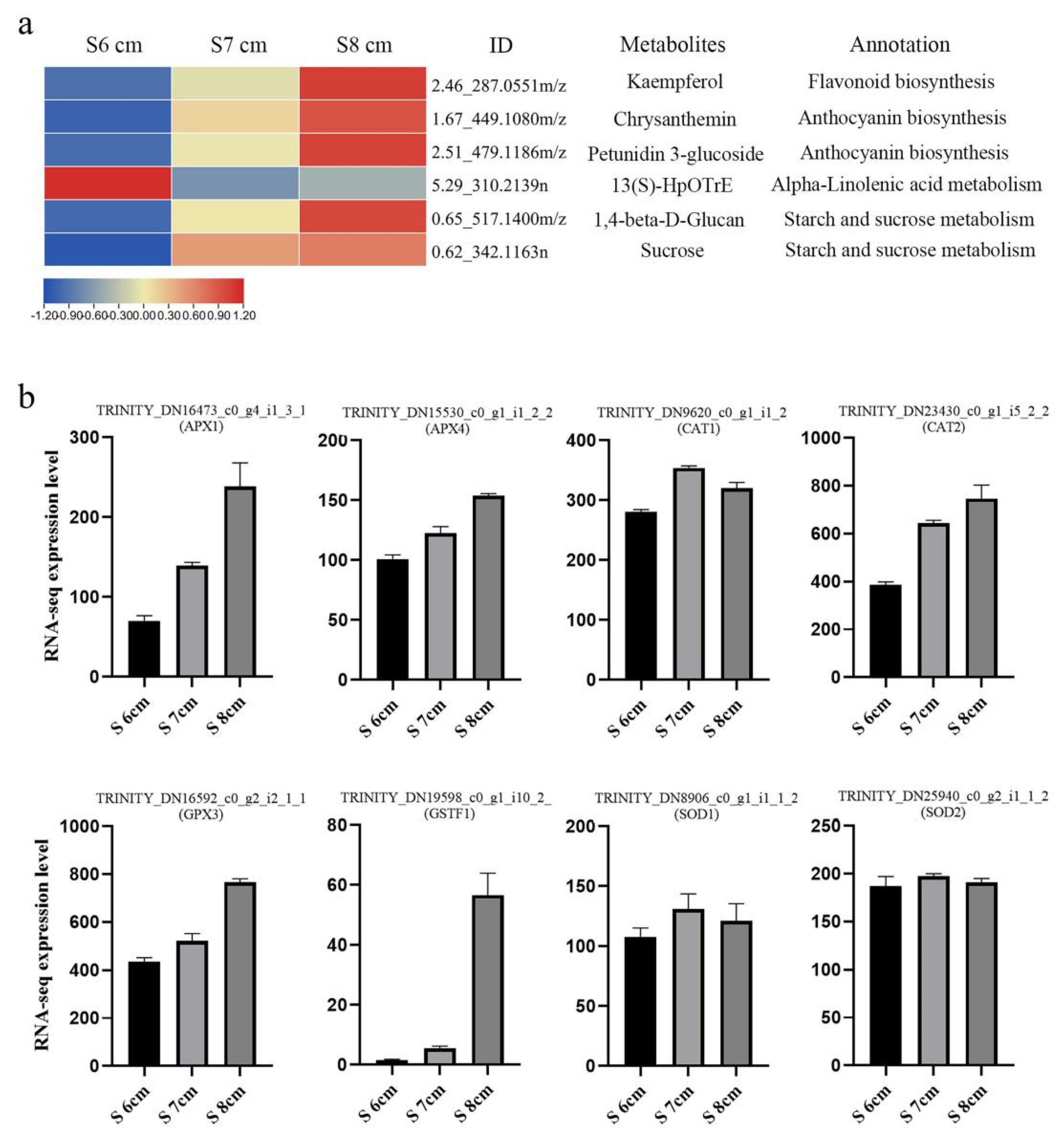
2.7. Flavone and Flavanol Biosynthesis Is Enriched in the Critical Degeneration Stage of the Stomium Zone
2.8. Reactive Oxygen Species Mediates Stomium Zone Degeneration
2.9. Stomium Zone Degeneration as a Process of Programmed Cell Death
2.10. Jasmonates and Regulation of the Expression of Genes Associated with Stomium Zone Degeneration
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Sampling
4.2. RNA-Seq
4.3. RNA Data Processing and Analysis
4.4. Heat Map and Weighted Network Analyses
4.5. Sample Preparation and Metabolite Extraction
4.6. Liquid Chromatography−Tandem Mass Spectrometry
4.7. Data Preprocessing and Statistical Analyses
4.8. TUNEL Assay
4.9. Reverse-Transcription Quantitative PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cao, X.; Yi, J.; Wu, Z.; Luo, X.; Zhong, X.; Wu, J.; Khan, M.A.; Zhao, Y.; Yi, M. Involvement of Ca2+ and CaM3 in Regulation of Thermotolerance in Lily (Lilium longiflorum). Plant Mol. Biol. Rep. 2013, 31, 1293–1304. [Google Scholar] [CrossRef]
- Gong, B.; Yi, J.; Wu, J.; Sui, J.; Khan, M.A.; Wu, Z.; Zhong, X.; Seng, S.; He, J.; Yi, M. LlHSFA1, a novel heat stress transcription factor in lily (Lilium longiflorum), can interact with LlHSFA2 and enhance the thermotolerance of transgenic Arabidopsis thaliana. Plant Cell Rep. 2014, 33, 1519–1533. [Google Scholar] [CrossRef]
- Wilson, Z.A.; Song, J.; Taylor, B.; Yang, C. The final split: The regulation of anther dehiscence. J. Exp. Bot. 2011, 62, 1633–1649. [Google Scholar] [CrossRef] [Green Version]
- Xue, L.; Wu, H.; Chen, Y.; Li, X.; Hou, J.; Lu, J.; Wei, S.; Dai, X.; Olson, M.S.; Liu, J. Evidences for a role of two Y-specific genes in sex determination in Populus deltoides. Nat. Commun. 2020, 11, 5893. [Google Scholar] [CrossRef]
- Yuan, G.; Wu, Z.; Liu, X.; Li, T.; Teng, N. Characterization and functional analysis of LoUDT1, a bHLH transcription fac-tor related to anther development in the lily oriental hybrid Siberia (Lilium spp.). Plant Physiol. Biochem. 2021, 166, 1087–1095. [Google Scholar] [CrossRef] [PubMed]
- Teng, R.; Wu, Z.; Xu, S.; Hou, H.; Teng, N. A Novel Lateral Organ Boundary-domain Factor CmLBD2 Positively Regulates Pollen Development via Activating CmACOS5 in Chrysanthemum morifolium. Plant Cell Physiol. 2021. [Google Scholar] [CrossRef]
- Podolyan, A.; Luneva, O.; Klimenko, E.; Breygina, M. Oxygen radicals and cytoplasm zoning in growing lily pollen tubes. Plant Reprod. 2021, 34, 103–115. [Google Scholar] [CrossRef]
- Cecchetti, V.; Altamura, M.M.; Brunetti, P.; Petrocelli, V.; Falasca, G.; Ljung, K.; Costantino, P.; Cardarelli, M. Auxin controls Arabidopsis anther dehiscence by regulating endothecium lignification and jasmonic acid biosynthesis. Plant J. 2013, 74, 411–422. [Google Scholar] [CrossRef]
- Sanders, P.M.; Bui, A.Q.; Weterings, K.; McIntire, K.; Hsu, Y.-C.; Lee, P.Y.; Truong, M.T.; Beals, T.; Goldberg, R. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 1991, 1, 297–322. [Google Scholar] [CrossRef]
- Yang, C.; Xu, Z.; Song, J.; Conner, K.; Barrena, G.V.; Wilson, Z.A. Arabidopsis MYB26/MALE STERILE35 Regulates Secondary Thickening in the Endothecium and Is Essential for Anther Dehiscence. Plant Cell 2007, 19, 534–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steiner-Lange, S.; Unte, U.S.; Eckstein, L.; Yang, C.; Wilson, Z.A.; Schmelzer, E.; Dekker, K.; Saedler, H. Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J. 2003, 34, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Song, J.; Ferguson, A.C.; Klisch, D.; Simpson, K.; Mo, R.; Taylor, B.; Mitsuda, N.; Wilson, Z.A. Transcription Factor MYB26 Is Key to Spatial Specificity in Anther Secondary Thickening Formation. Plant Physiol. 2017, 175, 333–350. [Google Scholar] [CrossRef] [PubMed]
- Mitsuda, N.; Iwase, A.; Yamamoto, H.; Yoshida, M.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 2007, 19, 270–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, R.; Richardson, E.A.; Ye, Z.-H. The MYB46 Transcription Factor Is a Direct Target of SND1 and Regulates Secondary Wall Biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2776–2792. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.-F.; Wang, B.; Feng, Y.-F.; Xue, J.-S.; Qian, X.-X.; Liu, S.-Q.; Zhou, J.; Yu, Y.-H.; Yang, N.-Y.; Xu, P.; et al. Auxin response factor17 directly regulates MYB108 for anther dehiscence. Plant Physiol. 2019, 181, 645–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cecchetti, V.; Altamura, M.M.; Falasca, G.; Costantino, P.; Cardarelli, M. Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 2008, 20, 1760–1774. [Google Scholar] [CrossRef] [Green Version]
- Caldelari, D.; Wang, G.; Farmer, E.E.; Dong, X. Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol. Biol. 2010, 75, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis gene required for jasmonate-regulated de-fense and fertility. Science 1998, 280, 1091–1094. [Google Scholar] [CrossRef]
- Song, S.; Qi, T.; Huang, H.; Ren, Q.; Wu, D.; Chang, C.; Peng, W.; Liu, Y.; Peng, J.; Xie, D. The jasmonate-zim domain proteins interact with the R2R3-MYB tran-scription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 2011, 23, 1000–1013. [Google Scholar] [CrossRef] [Green Version]
- Qi, T.; Huang, H.; Song, S.; Xie, D. Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell 2015, 27, 1620–1633. [Google Scholar] [CrossRef] [Green Version]
- Sui, J.; He, J.; Wu, J.; Gong, B.; Cao, X.; Seng, S.; Wu, Z.; Wu, C.; Liu, C.; Yi, M. Characterization and Functional Analysis of Transcription Factor LoMYB80 Related to Anther Development in Lily (Lilium Oriental Hybrids). J. Plant Growth Regul. 2015, 34, 545–557. [Google Scholar] [CrossRef]
- Sanders, P.M.; Bui, A.Q.; Le, B.H.; Goldberg, R.B. Differentiation and degeneration of cells that play a major role in tobacco anther dehiscence. Sex. Plant Reprod. 2005, 17, 219–241. [Google Scholar] [CrossRef]
- Dong, J.; Kim, S.T.; Lord, E.M. Plantacyanin Plays a Role in Reproduction in Arabidopsis. Plant Physiol. 2005, 138, 778–789. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, W.; Feng, Y.; Tu, M.; Wittich, P.E.; Bate, N.J.; Messing, J. Transcriptome and metabolome reveal distinct carbon allocation patterns during internode sugar accumulation in different sorghum genotypes. Plant Biotechnol. J. 2018, 17, 472–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wu, Z.; Wu, H.; Fang, W.; Chen, F.; Teng, N. Transcriptome Profiling Unravels a Vital Role of Pectin and Pectinase in Anther Dehiscence in Chrysanthemum. Int. J. Mol. Sci. 2019, 20, 5865. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wu, Z.; Wang, L.; Wu, M.; Zhang, D.; Fang, W.; Chen, F.; Teng, N. Cytological and Molecular Characteristics of Pollen Abortion in Lily with Dysplastic Tapetum. Hortic. Plant J. 2019, 5, 281–294. [Google Scholar] [CrossRef]
- Pichersky, E.; Lewinsohn, E. Convergent Evolution in Plant Specialized Metabolism. Annu. Rev. Plant Biol. 2011, 62, 549–566. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Wu, Y.; Wang, G.; Wang, T.; Cao, F. Integrated analysis of the transcriptome and metabolome in young and mature leaves of Ginkgo bilobal L. Ind. Crops Prod. 2020, 143, 111906. [Google Scholar] [CrossRef]
- Meihls, L.N.; Handrick, V.; Glauser, G.; Barbier, H.; Kaur, H.; Haribal, M.M.; Lipka, A.E.; Gershenzon, J.; Buckler, E.S.; Erb, M.; et al. Natural variation in maize aphid resistance is associated with 2,4-Dihydroxy-7-Methoxy-1,4-Benzoxazin-3-One Glucoside methyltransferase activity. Plant Cell 2013, 25, 2341–2355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quadrana, L.; Almeida, J.; Asís, R.; Duffy, T.; Dominguez, P.G.; Bermudez, L.F.; Conti, G.; Da Silva, J.V.C.; Peralta, I.E.; Colot, V.; et al. Natural occurring epialleles determine vitamin E accumulation in tomato fruits. Nat. Commun. 2014, 5, 4027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, J.; Philippe, R.N.; Noel, J.P. The rise of chemo diversity in plants. Science 2012, 336, 1667–1670. [Google Scholar] [CrossRef] [Green Version]
- Zheng, S.; Li, J.; Ma, L.; Wang, H.; Ni, E.; Jiang, D.; Zhuang, C. OsAGO2 controls ROS production and the initiation of tapetal PCD by epigenetically regulating OsHXK1 expression in rice anthers. Proc. Natl. Acad. Sci. USA 2019, 116, 7549–7558. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.Y.; He, Y.Q.; Li, L.Y.; Liu, H.R.; Hong, G.J. Involvement of the R2R3-MYB transcription factors MYB21 and its hom-ologs in regulating the stamen flavonols accumulation in Arabidopsis. J. Exp. Bot. 2021, 72, 12. [Google Scholar] [CrossRef]
- Cheng, H.; Zhang, Q.; Guo, D. Genes that Respond to H2O2 Are Also Evoked under Light in Arabidopsis. Mol. Plant 2013, 6, 226–228. [Google Scholar] [CrossRef] [Green Version]
- Denness, L.; McKenna, J.; Segonzac, C.; Wormit, A.; Madhou, P.; Bennett, M.; Mansfield, J.; Zipfel, C.; Hamann, T. Cell Wall Damage-Induced Lignin Biosynthesis Is Regulated by a Reactive Oxygen Species- and Jasmonic Acid-Dependent Process in Arabidopsis. Plant Physiol. 2011, 156, 1364–1374. [Google Scholar] [CrossRef] [Green Version]
- Song, S.; Qi, T.; Wasternack, C.; Xie, D. Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr. Opin. Plant Biol. 2014, 21, 112–119. [Google Scholar] [CrossRef]
- Okada, K.; Ueda, J.; Komaki, M.K.; Bell, C.J.; Shimura, Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 1991, 3, 677–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, R.J.; Spielman, M.; Dickinson, H.G. Stamen Structure and Function. Plant Cell 2004, 16, S46–S60. [Google Scholar] [CrossRef]
- Katsir, L.; Chung, H.S.; Koo, A.J.; A Howe, G. Jasmonate signaling: A conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 2008, 11, 428–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, S.; Qi, T.; Fan, M.; Zhang, X.; Gao, H.; Huang, H.; Wu, D.; Guo, H.; Xie, D. The bHLH Subgroup IIId Factors Negatively Regulate Jasmonate-Mediated Plant Defense and Development. PLoS Genet. 2013, 9, e1003653. [Google Scholar] [CrossRef] [Green Version]
- Ishiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, J.; Okada, K. The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 2013, 13, 2191–2209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Kumar, P.P. Characterization of two ethylene receptors PhERS1 and PhETR2 from petunia: PhETR2 regulates timing of anther dehiscence. J. Exp. Bot. 2006, 58, 533–544. [Google Scholar] [CrossRef] [Green Version]
- Savada, R.P.; Ozga, J.A.; Jayasinghege, C.P.A.; Waduthanthri, K.D.; Reinecke, D.M. Heat stress differentially modifies ethylene biosynthesis and signaling in pea floral and fruit tissues. Plant Mol. Biol. 2017, 95, 313–331. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Li, P.; Chen, M.; Lee, Y.; Yang, C. Forever young flower negatively regulates ethylene response DNA-binding factors by activating an ethylene-responsive factor to control Arabidopsis floral organ senescence and abscission. Plant Physiol. 2015, 168, 1666–1683. [Google Scholar] [CrossRef]
- Peng, J. Gibberellin and Jasmonate Crosstalk during Stamen Development. J. Integr. Plant Biol. 2009, 51, 1064–1070. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.A.; Elliott, K.A.; Gonzalez-Carranza, Z.H. Abscission, dehiscence, and other cell separation processes. Annu. Rev. Plant Biol. 2002, 53, 131–158. [Google Scholar] [CrossRef]
- Ormancey, M.; Thuleau, P.; van der Hoorn, R.A.; Grat, S.; Testard, A.; Kamal, K.Y.; Boudsocq, M.; Cotelle, V.; Mazars, C. Sphingolipid-induced cell death in Arabidopsis is negatively regulated by the papain-like cysteine protease RD21. Plant Sci. 2019, 280, 12–17. [Google Scholar] [CrossRef]
- Senatore, A.; Trobacher, C.P.; Greenwood, J.S. Ricinosomes Predict Programmed Cell Death Leading to Anther Dehiscence in Tomato. Plant Physiol. 2008, 149, 775–790. [Google Scholar] [CrossRef] [Green Version]
- Stadler, R.; Truernit, E.; Gahrtz, M.; Sauer, N. The AtSUC1 sucrose carrier may represent the osmotic driving force for anther dehiscence and pollen tube growth in Arabidopsis. Plant J. 1999, 19, 269–278. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, J.; Du, M.-L.; Li, L.; Wang, X.-L.; Li, X.-B. A cotton gene encoding MYB-Like transcription factor is specifically ex-pressed in pollen and is involved in regulation of late anther/pollen development. Plant Cell Physiol. 2013, 6, 893–906. [Google Scholar] [CrossRef]
- Huang, H.; Gong, Y.; Liu, B.; Wu, D.; Zhang, M.; Xie, D.; Song, S. The DELLA proteins interact with MYB21 and MYB24 to regulate filament elongation in Arabidopsis. BMC Plant Biol. 2020, 20, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, T.; Ng, K.-H.; Lim, T.-S.; Yu, H.; Meyerowitz, E.M. The Homeotic Protein AGAMOUS Controls Late Stamen Development by Regulating a Jasmonate Biosynthetic Gene in Arabidopsis. Plant Cell 2007, 19, 3516–3529. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Bian, J.; Liu, S.; Song, H.; Shi, N.; Tao, Y.; Wang, H. Flower-specific expression of Arabidopsis PCS1 driven by AGAMOUS second intron in tobacco decreases the fertility of transgenic plants. Mol. Breed. 2010, 27, 337–346. [Google Scholar] [CrossRef]
- Zhong, R.; Ye, Z.-H. The Arabidopsis NAC transcription factor NST2 functions together with SND1 and NST1 to regulate secondary wall biosynthesis in fibers of inflorescence stems. Plant Signal. Behav. 2015, 10, e989746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. The NAC Transcription Factors NST1 and NST2 of Arabidopsis Regulate Secondary Wall Thickenings and Are Required for Anther Dehiscence. Plant Cell 2005, 17, 2993–3006. [Google Scholar] [CrossRef] [Green Version]
- Lyu, X.; Chen, S.; Liao, N.; Liu, J.; Hu, Z.; Yang, J.; Zhang, M. Characterization of watermelon anther and its programmed cell death-associated events during dehiscence under cold stress. Plant Cell Rep. 2019, 38, 1551–1561. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, D.; Lv, X.; Wang, Y.; Xun, Z.; Liu, Z.; Li, F.; Lu, H. The Cysteine Protease CEP1, a Key Executor Involved in Tapetal Programmed Cell Death, Regulates Pollen Development in Arabidopsis. Plant Cell 2014, 26, 2939–2961. [Google Scholar] [CrossRef] [Green Version]
- Shindo, T.; Villamil, J.C.M.; Hörger, A.; Song, J.; Van Der Hoorn, R.A.L. A Role in Immunity for Arabidopsis Cysteine Protease RD21, the Ortholog of the Tomato Immune Protease C14. PLoS ONE 2012, 7, e29317. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Cui, K.; Hu, Q.; Wu, C.; Li, G.; Huang, J.; Peng, S. Response of spikelet water status to high temperature and its relationship with heat tolerance in rice. Crop J. 2021, 26, 2939. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Roberts, A.; Trapnell, C.; Donaghey, J.; Rinn, J.L.; Pachter, L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011, 12, R22. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Roberts, A.; Pachter, L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods 2013, 10, 71–73. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Liu, N.; Zhang, W.; Wu, C.; Jiang, Y.; Ma, J.; Li, M.; Sui, S. Integrated transcriptome and proteome analysis provides insight into chilling-induced dormancy breaking in Chimonanthus praecox. Hortic. Res. 2020, 7, 198. [Google Scholar] [CrossRef]
- Zhao, W.; Baldwin, E.A.; Bai, J.; Plotto, A.; Irey, M. Comparative analysis of the transcriptomes of the calyx abscission zone of sweet orange insights into the huanglongbing-associated fruit abscission. Hortic. Res. 2019, 6, 71. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Boccard, J.; Rutledge, D. A consensus orthogonal partial least squares discriminant analysis (OPLS-DA) strategy for multi-block Omics data fusion. Anal. Chim. Acta. 2013, 769, 30–39. [Google Scholar] [CrossRef]
- Zhang, S.; Zhao, Q.; Zeng, D.; Xu, J.; Zhou, H.; Wang, F.; Ma, N.; Li, Y. RhMYB108, an R2R3-MYB transcription factor, is involved in ethylene- and JA-induced petal senescence in rose plants. Hortic. Res. 2019, 6, 131. [Google Scholar] [CrossRef] [Green Version]
- Chechanovsky, N.; Hovav, R.; Frenkel, R.; Faigenboim, A.; Eselson, Y.; Petreikov, M.; Moy, M.; Shen, S.; Schaffer, A.A. Low temperature upregulates cwp expression and modifies alternative splicing patterns, increasing the severity of cwp-induced tomato fruit cuticular microfissures. Hortic. Res. 2019, 6, 122. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, L.; Liu, X.; Wu, Z.; Teng, N. Transcriptome and Metabolome Analyses Provide Insights into the Stomium Degeneration Mechanism in Lily. Int. J. Mol. Sci. 2021, 22, 12124. https://doi.org/10.3390/ijms222212124
He L, Liu X, Wu Z, Teng N. Transcriptome and Metabolome Analyses Provide Insights into the Stomium Degeneration Mechanism in Lily. International Journal of Molecular Sciences. 2021; 22(22):12124. https://doi.org/10.3390/ijms222212124
Chicago/Turabian StyleHe, Ling, Xinyue Liu, Ze Wu, and Nianjun Teng. 2021. "Transcriptome and Metabolome Analyses Provide Insights into the Stomium Degeneration Mechanism in Lily" International Journal of Molecular Sciences 22, no. 22: 12124. https://doi.org/10.3390/ijms222212124
APA StyleHe, L., Liu, X., Wu, Z., & Teng, N. (2021). Transcriptome and Metabolome Analyses Provide Insights into the Stomium Degeneration Mechanism in Lily. International Journal of Molecular Sciences, 22(22), 12124. https://doi.org/10.3390/ijms222212124





