Insights into Regulation of C2 and C4 Photosynthesis in Amaranthaceae/Chenopodiaceae Using RNA-Seq
Abstract
1. Introduction
2. Results
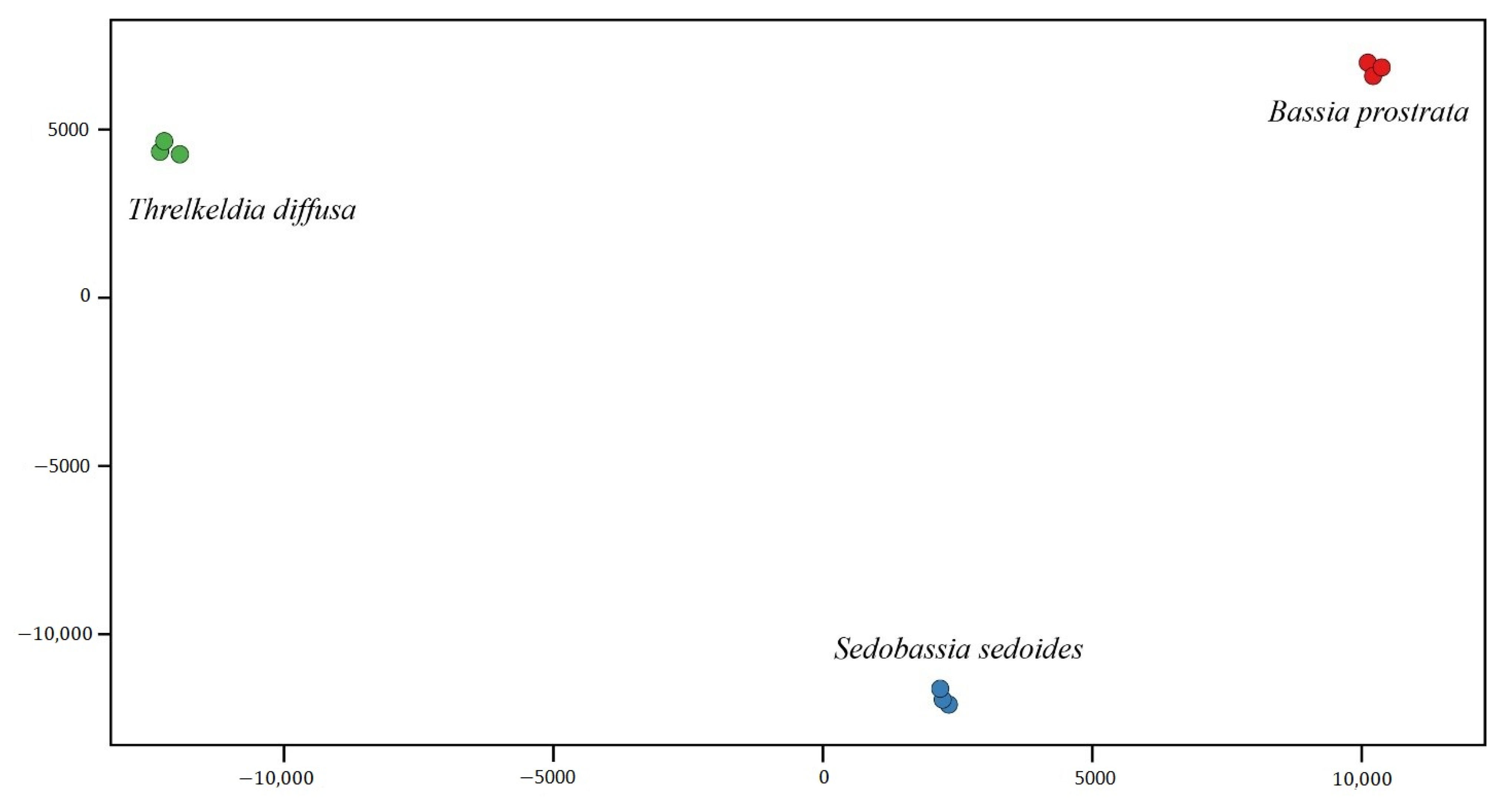
2.1. Descriptive Statistics of RNA Data and De Novo Assembly
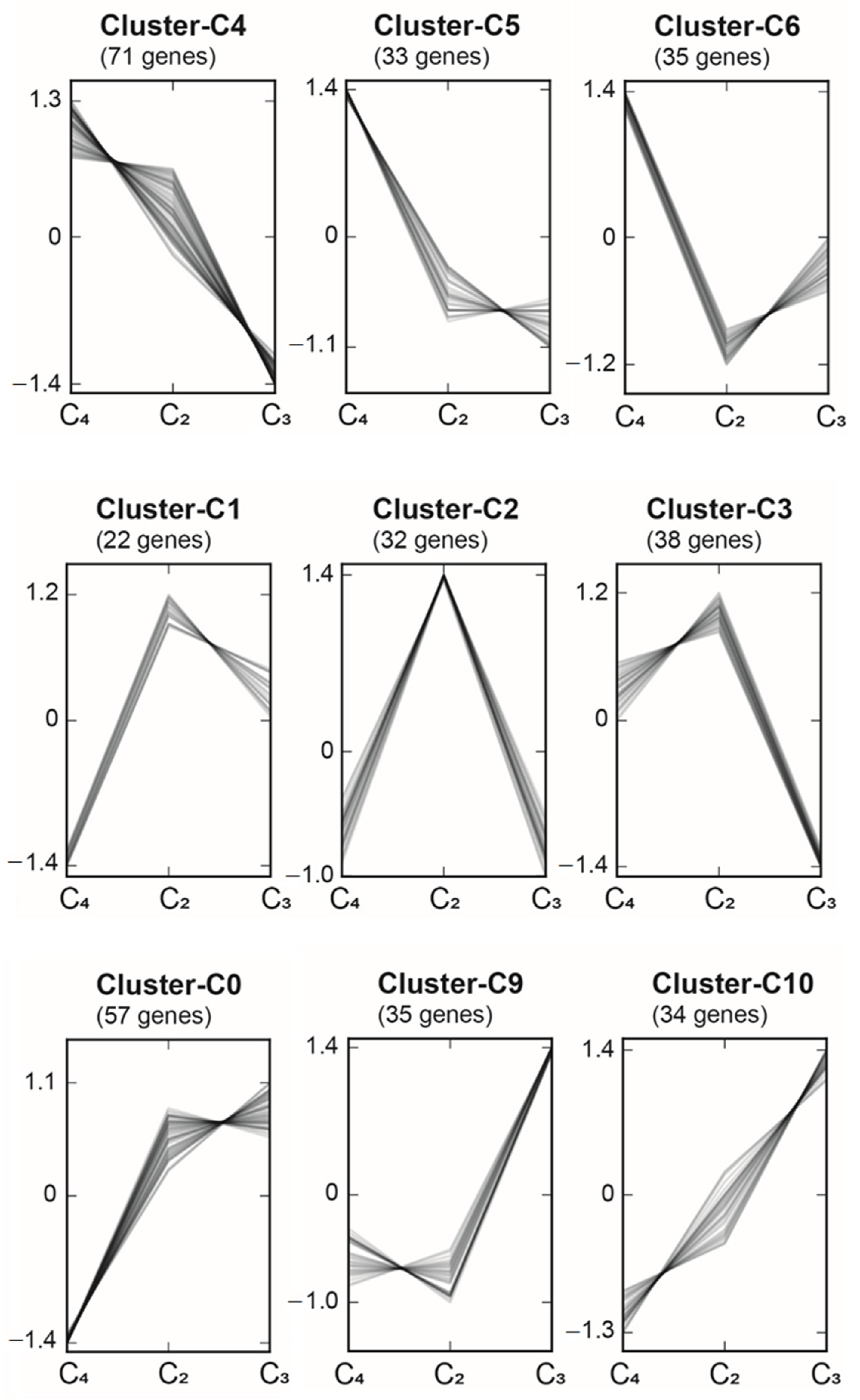
2.2. Differential Expression Genes (DEGs) within Camphorosmeae
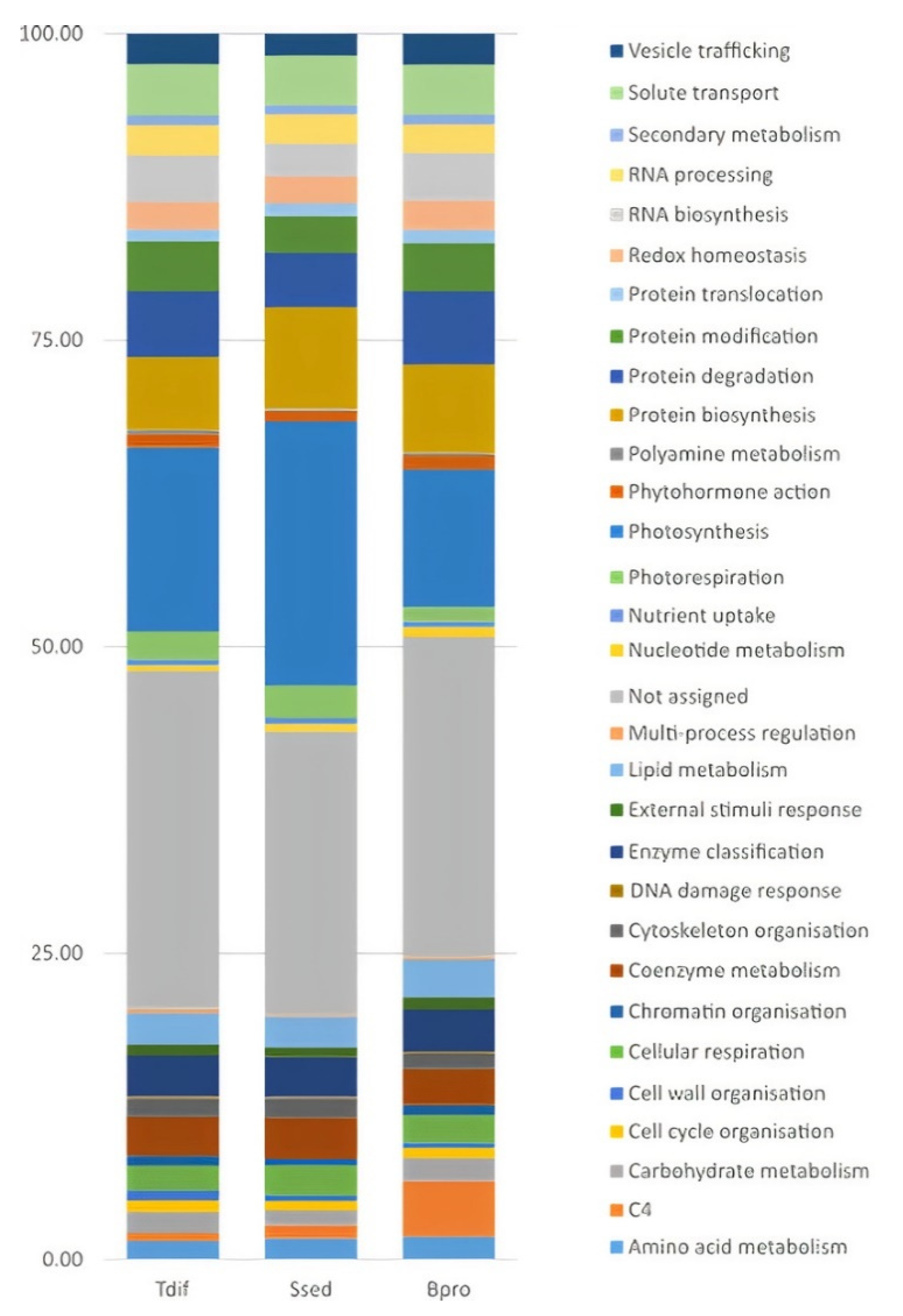
2.3. Functional Classification and Enrichment of DEGs within the Camphorosmeae
2.4. Differential Expression of C4-Related Genes in C3, C2 and C4 Camphorosmeae Species
2.5. Differential Expression of Key Photorespiration Genes in C3, C2 and C4 Camphorosmeae Species
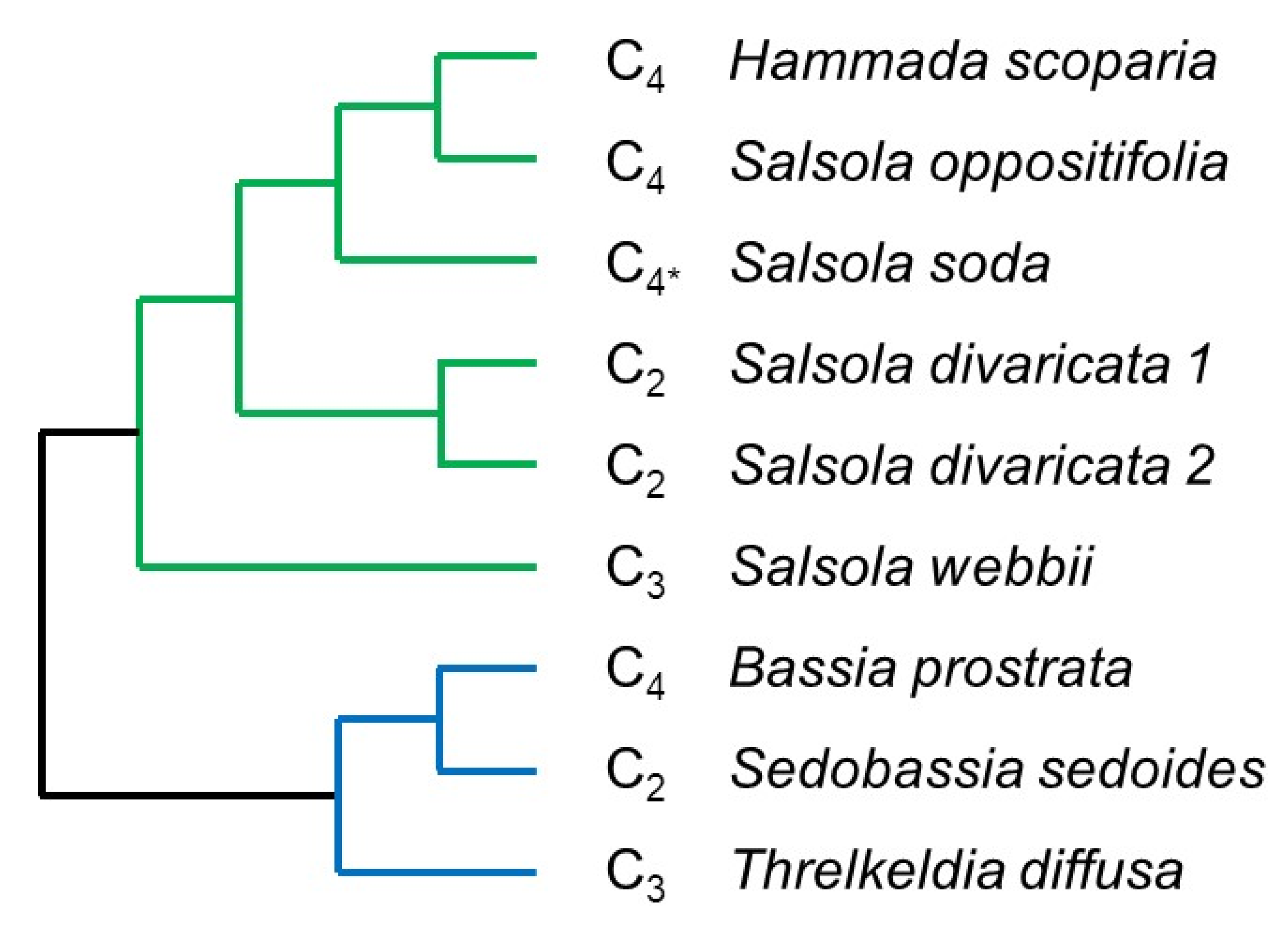
2.6. Regulatory Elements in C3, C2, and C4 Species of the Amaranthaceae/Chenopodiaceae Alliance
3. Discussion
3.1. Transcriptome Analysis in Camphorosmeae
3.2. C4 Key Enzymes in C4 and C2 Camphorosmeae Species
3.3. C4 Key Enzymes in C4 and C2 Camphorosmeae Species
3.4. Regulation of C3, C2 and C4 Photosynthesis in Amaranthaceae/Chenopodiaceae
4. Materials and Methods
4.1. Plant Material
4.2. RNA Isolation and Sequencing
4.3. Data Access
4.4. RNA-Seq Data Processing
4.5. Differential Gene Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bauwe, H.; Hagemann, M.; Fernie, A.R. Photorespiration: Players, partners and origin. Trends Plant Sci. 2010, 15, 330–336. [Google Scholar] [CrossRef]
- Gowik, U.; Westhoff, P. The Path from C3 to C4 photosynthesis. Plant Physiol. 2011, 155, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Christin, P.A.; Osborne, C.P.; Chatelet, D.S.; Columbus, J.T.; Besnard, G.; Hodkinson, T.R.; Garrison, L.M.; Vorontsova, M.S.; Edwards, E.J. Anatomical enablers and the evolution of C4 photosynthesis in grasses. Proc. Natl. Acad. Sci. USA 2013, 110, 1381–1386. [Google Scholar] [CrossRef]
- Kadereit, G.; Bohley, K.; Lauterbach, M.; Tefarikis, D.T.; Kadereit, J.W. C3–C4 intermediates may be of hybrid origin—A reminder. New Phytol. 2017, 215, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Sage, R.F.; Khoshravesh, R.; Sage, T.L. From proto-Kranz to C4 Kranz: Building the bridge to C 4 photosynthesis. J. Exp. Bot. 2014, 65, 3341–3356. [Google Scholar] [CrossRef]
- Bräutigam, A.; Gowik, U. Photorespiration connects C3 and C4 photosynthesis. J. Exp. Bot. 2016, 67, 2953–2962. [Google Scholar] [CrossRef]
- Schlüter, U.; Weber, A.P.M. Regulation and Evolution of C 4 Photosynthesis. Annu. Rev. Plant Biol. 2020, 71, 183–215. [Google Scholar] [CrossRef]
- Sage, R.F.; Monson, R.K.; Ehleringer, J.R.; Adachi, S.; Pearcy, R.W. Some like it hot: The physiological ecology of C4 plant evolution. Oecologia 2018, 187, 941–966. [Google Scholar] [CrossRef]
- Monson, R.K.; Edwards, G.E.; Ku, M.S.B. C3-C4 Intermediate Photosynthesi Plants. Bioscience 1984, 34, 563–566. [Google Scholar] [CrossRef]
- Edwards, G.E.; Ku, M.S.B. Biochemistry of C3–C4 Intermediates. In The Biochemistry of Plants; Hatch, M.D., Boardman, N.K., Eds.; Academic Press: London, UK, 1987; Volume 10, pp. 275–325. [Google Scholar]
- Oakley, J.C.; Sultmanis, S.; Stinson, C.R.; Sage, T.L.; Sage, R.F. Comparative studies of C3 and C4 Atriplex hybrids in the genomics era: Physiological assessments. J. Exp. Bot. 2014, 65, 3637–3647. [Google Scholar] [CrossRef]
- Sage, R.F. A portrait of the C4 photosynthetic family on the 50th anniversary of its discovery: Species number, evolutionary lineages, and Hall of Fame. J. Exp. Bot. 2016, 67, 4039–4056. [Google Scholar] [CrossRef]
- Pyankov, V.I.; Artyusheva, E.G.; Edwards, G.E.; Black, C.C.; Soltis, P.S. Phylogenetic analysis of tribe Salsoleae (Chenopodiaceae) based on ribosomal its sequences: Implications for the evolution of photosynthesis types. Am. J. Bot. 2001, 88, 1189–1198. [Google Scholar] [CrossRef]
- Kadereit, G.T.; Borsch, K.; Weising, H.F. Phylogeny of Amaranthaceae and Chenopodiaceae and the evolution of C4 photosynthesis. Int. J. Plant Sci. 2003, 164, 959–986. [Google Scholar] [CrossRef]
- Kadereit, G.; Ackerly, D.; Pirie, M.D. A broader model for C4 photosynthesis evolution in plants inferred from the goosefoot family (Chenopodiaceae s.s.). Proc. R. Soc. B Biol. Sci. 2012, 279, 3304–3311. [Google Scholar] [CrossRef]
- Kadereit, G.; Lauterbach, M.; Pirie, M.D.; Arafeh, R.; Freitag, H. When do different C4 leaf anatomies indicate independent C 4 origins? Parallel evolution of C4 leaf types in Camphorosmeae (Chenopodiaceae). J. Exp. Bot. 2014, 65, 3499–3511. [Google Scholar] [CrossRef][Green Version]
- Schütze, P.; Freitag, H.; Weising, K. An integrated molecular and morphological study of the subfamily Suaedoideae ulbr. (Chenopodiaceae). Plant Syst. Evol. 2003, 239, 257–286. [Google Scholar] [CrossRef]
- Kadereit, G.; Freitag, H. Molecular phylogeny of Camphorosmeae (Camphorosmoideae, Chenopodiaceae): Implications for biogeography, evolution of C4-photosynthesis and taxonomy. Taxon 2011, 60, 51–78. [Google Scholar] [CrossRef]
- Akhani, H.; Edwards, G.; Roalson, E.H. Diversification of the Old World Salsoleae s.l. (Chenopodiaceae): Molecular Phylogenetic Analysis of Nuclear and Chloroplast Data Sets and a Revised Classification Author (s): Hossein Akhani, Gerald Edwards, and Eric H. Roalson Diversification O. Int. J. Plant Sci. 2007, 168, 931–956. [Google Scholar] [CrossRef]
- Freitag, H.; Kadereit, G. C3 and C4 leaf anatomy types in Camphorosmeae (Camphorosmoideae, Chenopodiaceae). Plant Syst. Evol. 2014, 300, 665–687. [Google Scholar] [CrossRef]
- Voznesenskaya, E.V.; Koteyeva, N.K.; Akhani, H.; Roalson, E.H.; Edwards, G.E. Structural and physiological analyses in Salsoleae (Chenopodiaceae) indicate multiple transitions among C3, intermediate, and C4 photosynthesis. J. Exp. Bot. 2013, 64, 3583–3604. [Google Scholar] [CrossRef]
- Schüssler, C.; Freitag, H.; Koteyeva, N.; Schmidt, D.; Edwards, G.; Voznesenskaya, E.; Kadereit, G. Molecular phylogeny and forms of photosynthesis in tribe Salsoleae (Chenopodiaceae). J. Exp. Bot. 2017, 68, 207–223. [Google Scholar] [CrossRef]
- Lauterbach, M.; Billakurthi, K.; Kadereit, G.; Ludwig, M.; Westhoff, P.; Gowik, U. C3 cotyledons are followed by C4 leaves: Intra-individual transcriptome analysis of Salsola soda (Chenopodiaceae). J. Exp. Bot. 2017, 68, 161–176. [Google Scholar] [CrossRef]
- Lauterbach, M.; Schmidt, H.; Billakurthi, K.; Hankeln, T.; Westhoff, P.; Gowik, U.; Kadereit, G. De novo transcriptome assembly and comparison of C3, C3-C4, and C4 species of tribe Salsoleae (Chenopodiaceae). Front. Plant Sci. 2017, 8, 1939. [Google Scholar] [CrossRef]
- Monteiro, A.; Podlaha, O. Wings, horns, and butterfly eyespots: How do complex traits evolve? PLoS Biol. 2009, 7, 0209–0216. [Google Scholar] [CrossRef] [PubMed]
- Aubry, S.; Kelly, S.; Kümpers, B.M.C.; Smith-Unna, R.D.; Hibberd, J.M. Deep Evolutionary Comparison of Gene Expression Identifies Parallel Recruitment of Trans-Factors in Two Independent Origins of C4 Photosynthesis. PLoS Genet. 2016, 10, e1006087. [Google Scholar] [CrossRef]
- Windhövel, A.; Hein, I.; Dabrowa, R.; Stockhaus, J. Characterization of a novel class of plant homeodomain proteins that bind to the C4 phosphoenolpyruvate carboxylase gene of Flaveria trinervia. Plant Mol. Biol. 2001, 45, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Brutnell, T.P. A synthesis of transcriptomic surveys to dissect the genetic basis of C4 photosynthesis. Curr. Opin. Plant Biol. 2016, 31, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Schwacke, R.; Ponce-Soto, G.Y.; Krause, K.; Bolger, A.M.; Arsova, B.; Hallab, A.; Gruden, K.; Stitt, M.; Bolger, M.E.; Usadel, B. MapMan4: A Refined Protein Classification and Annotation Framework Applicable to Multi-Omics Data Analysis. Mol. Plant 2019, 12, 879–892. [Google Scholar] [CrossRef]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, 1182–1187. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Dohm, J.C.; Minoche, A.E.; Holtgräwe, D.; Capella-Gutiérrez, S.; Zakrzewski, F.; Tafer, H.; Rupp, O.; Sörensen, T.R.; Stracke, R.; Reinhardt, R.; et al. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 2014, 505, 546–549. [Google Scholar] [CrossRef]
- Abu-Jamous, B.; Kelly, S. Clust: Automatic extraction of optimal co-expressed gene clusters from gene expression data. Genome Biol. 2018, 19, 172. [Google Scholar] [CrossRef]
- Gowik, U.; Bräutigam, A.; Weber, K.L.; Weber, A.P.M.; Westhoff, P. Evolution of C 4 photosynthesis in the genus flaveria: How many and which genes does it take to make C 4? Plant Cell 2011, 23, 2087–2105. [Google Scholar] [CrossRef] [PubMed]
- Mallmann, J.; Heckmann, D.; Bräutigam, A.; Lercher, M.J.; Weber, A.P.M.; Westhoff, P.; Gowik, U. The role of photorespiration during the evolution of C4 photosynthesis in the genus Flaveria. eLife 2014, 2014, e02478. [Google Scholar] [CrossRef]
- Schulze, S.; Mallmann, J.; Burscheidt, J.; Koczor, M.; Streubel, M.; Bauwe, H.; Gowik, U.; Westhoff, P. Evolution of C4 photosynthesis in the genus flaveria: Establishment of a photorespiratory CO2 pump. Plant Cell 2013, 25, 2522–2535. [Google Scholar] [CrossRef] [PubMed]
- Sage, R.F. Russ Monson and the evolution of C4 photosynthesis. Oecologia 2021, 21, 1–18. [Google Scholar] [CrossRef]
- Bräutigam, A.; Kajala, K.; Wullenweber, J.; Sommer, M.; Gagneul, D.; Weber, K.L.; Carr, K.M.; Gowik, U.; Maß, J.; Lercher, M.J.; et al. An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiol. 2011, 155, 142–156. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bräutigam, A.; Weber, A.P.M.; Zhu, X.G. Three distinct biochemical subtypes of C4 photosynthesis? A modelling analysis. J. Exp. Bot. 2014, 65, 3567–3578. [Google Scholar] [CrossRef] [PubMed]
- Furumoto, T.; Yamaguchi, T.; Ohshima-Ichie, Y.; Nakamura, M.; Tsuchida-Iwata, Y.; Shimamura, M.; Ohnishi, J.; Hata, S.; Gowik, U.; Westhoff, P.; et al. A plastidial sodium-dependent pyruvate transporter. Nature 2011, 476, 472–476. [Google Scholar] [CrossRef]
- Walters, R.G.; Ibrahim, D.G.; Horton, P.; Kruger, N.J. A mutant of Arabidopsis lacking the triose-phosphate/phosphate translocator reveals metabolic regulation of starch breakdown in the light. Plant Physiol. 2004, 135, 891–906. [Google Scholar] [CrossRef]
- Sage, R.F. Photorespiratory compensation: A driver for biological diversity. Plant Biol. 2013, 15, 624–638. [Google Scholar] [CrossRef] [PubMed]
- Lundgren, M.R. C2 photosynthesis: A promising route towards crop improvement? New Phytol. 2020, 228, 1734–1740. [Google Scholar] [CrossRef] [PubMed]
- Döring, F.; Streubel, M.; Bräutigam, A.; Gowik, U. Most photorespiratory genes are preferentially expressed in the bundle sheath cells of the C4 grass Sorghum bicolor. J. Exp. Bot. 2016, 67, 3053–3064. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef]
- Kotak, S.; Port, M.; Ganguli, A.; Bicker, F.; Von Koskull-Döring, P. Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class a Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. 2004, 39, 98–112. [Google Scholar] [CrossRef]
- Pernas, M.; Ryan, E.; Dolan, L. Schizoriza Controls Tissue System Complexity in Plants. Curr. Biol. 2010, 20, 818–823. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.; Schippers, J.H.M.; Welker, A.; Mieulet, D.; Guiderdoni, E.; Mueller-Roeber, B. Transcription factor oshsfc1b regulates salt tolerance and development in oryza sativa ssp. japonica. AoB Plants 2012, 12, pls011. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef]
- Grimplet, J.; Pimentel, D.; Agudelo-Romero, P.; Martinez-Zapater, J.M.; Fortes, A.M. The Lateral Organ Boundaries Domain gene family in grapevine: Genome-wide characterization and expression analyses during developmental processes and stress responses. Sci. Rep. 2017, 7, 15968. [Google Scholar] [CrossRef]
- Slewinski, T.L.; Anderson, A.A.; Zhang, C.; Turgeon, R. Scarecrow plays a role in establishing Kranz anatomy in maize leaves. Plant Cell Physiol. 2012, 53, 2030–2037. [Google Scholar] [CrossRef]
- Slewinski, T.L. Using evolution as a guide to engineer Kranz-type C4 photosynthesis. Front. Plant Sci. 2013, 4, 212. [Google Scholar] [CrossRef]
- Wang, P.; Kelly, S.; Fouracre, J.P.; Langdale, J.A. Genome-wide transcript analysis of early maize leaf development reveals gene cohorts associated with the differentiation of C4 Kranz anatomy. Plant J. 2013, 75, 656–670. [Google Scholar] [CrossRef] [PubMed]
- Fouracre, J.P.; Ando, S.; Langdale, J.A. Cracking the Kranz enigma with systems biology. J. Exp. Bot. 2014, 65, 3327–3339. [Google Scholar] [CrossRef]
- Lori Tausta, S.; Li, P.; Si, Y.; Gandotra, N.; Liu, P.; Sun, Q.; Brutnell, T.P.; Nelson, T. Developmental dynamics of Kranz cell transcriptional specificity in maize leaf reveals early onset of C4-related processes. J. Exp. Bot. 2014, 65, 3543–3555. [Google Scholar] [CrossRef]
- Semiarti, E.; Ueno, Y.; Tsukaya, H.; Iwakawa, H.; Machida, C.; Machida, Y. The asymmetric leaves 2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 2001, 128, 1771–1783. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Frugis, G.; Colgan, D.; Chua, N.H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000, 14, 3024–3036. [Google Scholar] [CrossRef]
- Vroemen, C.W.; Mordhorst, A.P.; Albrecht, C.; Kwaaitaal, M.A.C.J.; De Vries, S.C. The Cup-Shaped Cotyledon 3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 2003, 15, 1563–1577. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Ohtani, M.; Mitsuda, N.; Kubo, M.; Ohme-Takagi, M.; Fukuda, H.; Demura, T. Vnd-Interacting 2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 2010, 22, 1249–1263. [Google Scholar] [CrossRef]
- Huang, Q.; Wang, Y.; Li, B.; Chang, J.; Chen, M.; Li, K.; Yang, G.; He, G. TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 2015, 15, 268. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, X.; Chao, J.; Zhang, Z.; Wang, W.; Guo, Y. NAC family transcription factors in tobacco and their potential role in regulating leaf senescence. Front. Plant Sci. 2018, 871, 1900. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Huang, G.Q.; Zou, D.; Yan, J.Q.; Li, Y.; Hu, S.; Li, X.B. The cotton (Gossypium hirsutum) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers. New Phytol. 2018, 217, 625–640. [Google Scholar] [CrossRef]
- Ebrahimian-Motlagh, S.; Ribone, P.A.; Thirumalaikumar, V.P.; Allu, A.D.; Chan, R.L.; Mueller-Roeber, B.; Balazadeh, S. Jungbrunnen 1 confers drought tolerance downstream of the HD-Zip I Transcription factor AtHB13. Front. Plant Sci. 2017, 8, 2118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, T.; Wang, X.; Wang, J.; Gu, K.; Yu, J.; Hu, D.; Hao, Y. Genome-wide Identification and Expression Analyses of Homeodomain-leucine Zipper Family Genes Reveal Their Involvement in Stress Response in Apple (Malus × domestica). Hortic. Plant J. 2021, in press. [Google Scholar] [CrossRef]
- Li, P.; Ponnala, L.; Gandotra, N.; Wang, L.; Si, Y.; Tausta, S.L.; Kebrom, T.H.; Provart, N.; Patel, R.; Myers, C.R.; et al. The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 2010, 42, 1060–1067. [Google Scholar] [CrossRef]
- Borba, A.R.; Serra, T.S.; Górska, A.; Gouveia, P.; Cordeiro, A.M.; Reyna-Llorens, I.; Kneřová, J.; Barros, P.M.; Abreu, I.A.; Oliveira, M.M.; et al. Synergistic binding of bHLH transcription factors to the promoter of the maize NADP-ME gene used in C4 photosynthesis is based on an ancient code found in the ancestral C3 state. Mol. Biol. Evol. 2018, 35, 1690–1705. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2013, 29, 644–652. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Kriventseva, E.V.; Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Dias, R.; Simão, F.A.; Zdobnov, E.M. OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019, 47, D807–D811. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
| C2 vs. C4* | C3 vs. C4* | C3 vs. C2* | |||||
|---|---|---|---|---|---|---|---|
| Transcripts | Genes | Log2FC | Padj | Log2FC | P | Log2FC | Padj |
| Bv_006710_gkqg.t1 | MDH | 1.29 | 1.01 × 10−10 | 2.84 | 9.92 × 10−42 | 1.55 | 1.74 × 10−14 |
| Bv1_004490_tyfq.t1 | PHT4 | 1.57 | 1.56 × 10−11 | 2.89 | 1.72 × 10−31 | 1.32 | 7.56 × 10−08 |
| Bv1_013550_fjqs.t1 | PPdK | 3.37 | 2.32 × 10−54 | 4.63 | 2.78 × 10−90 | 1.26 | 8.92 × 10−10 |
| Bv2_031080_twkf.t1 | AspAt | 1.32 | 2.84 × 10−06 | 2.15 | 1.44 × 10−13 | 0.82 | 0.003667 |
| Bv3_049110_qgnh.t1 | PHT1 | 3.16 | 8.05 × 10−08 | - | - | - | - |
| Bv4_072630_xjai.t1 | DIT | - | - | 1.30 | 4.88 × 10−06 | 2.61 | 3.46 × 10−19 |
| Bv5_117240_yhsk.t1 | PPT | 1.94 | 7.13 × 10−13 | 3.61 | 5.61 × 10−35 | 1.67 | 2.18 × 10−09 |
| Bv6_135140_uyxu.t1 | Asn Synthetase | 2.56 | 5.60 × 10−07 | 2.42 | 1.93 × 10−06 | - | - |
| Bv6_148840_uffy.t1 | CA | 3.80 | 6.66 × 10−38 | 3.81 | 4.08 × 10−38 | - | - |
| Bv7_169130_kwer.t1 | AlaAT | 2.61 | 4.89 × 10−17 | 5.17 | 4.74 × 10−48 | 2.56 | 2.34 × 10−15 |
| Bv8_182550_kstq.t1 | TPT | - | - | 1.48 | 1.20 × 10−10 | 2.29 | 2.71 × 10−22 |
| Bv8_194450_rkme.t1 | CA | - | - | - | - | 1.48 | 0.000263 |
| Bv8_195530_sxjq.t1 | BASS2 | 3.51 | 5.00 × 10−68 | 3.97 | 6.26 × 10−83 | 0.46 | 0.018625 |
| Bv8_200290_ujgk.t1 | TPT | 3.25 | 1.25 × 10−05 | 1.97 | 0.005513 | ||
| Bv9_209750_xeaz.t1 | PEPC | 2.95 | 3.29 × 10−17 | 4.12 | 6.98 × 10−29 | 1.18 | 0.000429 |
| Bv9_215520_prze.t1 | DIT | 1.48 | 1.23 × 10−08 | 2.63 | 5.15 × 10−22 | 1.16 | 2.27 × 10−05 |
| Bv9_224000_xpgi.t1 | NHD | 2.48 | 1.17 × 10−21 | 2.68 | 7.04 × 10−25 | - | - |
| Bv9_224840_zmjw.t1 | NADP-ME | 1.29 | 8.44 × 10−08 | 3.86 | 9.54 × 10−47 | 2.56 | 1.34 × 10−23 |
| C2* vs. C4 | C3 vs. C2* | C3* vs. C4 | |||||
|---|---|---|---|---|---|---|---|
| Transcripts | Genes | Log2FC | Padj | Log2FC | Padj | Log2FC | Padj |
| Bv5_106360_ipey.t1 | GDC-H | 1.862968 | 3.37 × 10−15 | 0.990549 | 1.80 × 10−05 | 0.872432 | 0.000167 |
| Bv_012000_yknj.t1 | GDC-P | 1.253616 | 9.20 × 10−08 | - | - | 2.068131 | 5.39 × 10−18 |
| Bv3_059720_tshd.t1 | GDC-L | 1.281024 | 1.13 × 10−11 | - | - | 0.953812 | 3.95 × 10−07 |
| Bv3_065510_eeis.t1 | SHMT | - | - | 1.133566 | 0.000228 | - | - |
| Bv4_073470_iswc.t1 | AGT/SGT | 1.254967 | 2.18 × 10−07 | - | - | 1.130439 | 2.83 × 10−06 |
| Bv4_074740_miaa.t1 | PGP | 1.831751 | 1.99 × 10−16 | 0.505181 | 0.019216 | 1.326602 | 1.77 × 10−09 |
| Bv4_094290_jgpp.t1 | GOX | 1.708751 | 1.51 × 10−18 | 0.993515 | 1.94 × 10−07 | 0.715241 | 0.000175 |
| Bv5_107350_ydma.t1 | 2.435018 | 1.25 × 10−13 | - | - | 2.32744 | 1.09 × 10−12 | |
| Bv6_127540_qdph.t1 | GDC-T | 2.464812 | 2.21 × 10−23 | 1.030248 | 1.32 × 10−05 | 1.434602 | 2.66 × 10−09 |
| Bv6_148110_nuir.t1 | GGT | 2.380809 | 3.21 × 10−21 | 1.223501 | 4.63 × 10−07 | 1.157324 | 2.02 × 10−06 |
| Bv6_152820_wtfn.t1 | SHMT | 1.721992 | 2.64 × 10−18 | 0.734934 | 0.000137 | 0.987067 | 3.75 × 10−07 |
| Bv8_184280_guso.t1 | 0.677745 | 0.006074 | - | - | 1.06935 | 3.85 × 10−06 | |
| Bv9_213980_zwen.t1 | HPR | 0.96884 | 0.000282 | - | - | 0.531187 | 0.045313 |
| Bv9_220360_xogt.t2 | GLYK | 0.689722 | 0.001207 | 0.708265 | 0.000885 | - | - |
| Cluster-4 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| BBX15 (CO-Like) | SHR (GRAS) | SCZ (HSF) | LBD41 (LBD) | ||||||
| Lineage | Species (C3 vs. C4*) | LF2C | Padj | LF2C | Padj | LF2C | Padj | LF2C | Padj |
| Salsoleae | Salweb vs. Hsco | 8.8 | 3.84 × 10−24 | 2.23 | 1.55 × 10−08 | 5.59 | 1.73 × 10−05 | 3.71 | 6.05 × 10−13 |
| Salweb vs. Salopp | 11.86 | 6.56 × 10−61 | 1.95 | 2.56 × 10−06 | 5.09 | 0.000374 | 4.26 | 1.64 × 10−17 | |
| Salweb vs. Salsod | 11.57 | 6.71 × 10−62 | 2.63 | 1.85 × 10−13 | 7.66 | 4.94 × 10−20 | 2.84 | 2.06 × 10−07 | |
| Camphorosmeae | Tdif vs. Bpro | 10.99 | 3.77 × 10−51 | 4.21 | 9.49 × 10−12 | 3.4 | 2.71 × 10−06 | 6.71 | 5.11 × 10−10 |
| Salweb vs. Bpro | 10.82 | 2.50 × 10−47 | 1.45 | 0.0011 | 6.12 | 1.58 × 10−07 | 2.22 | 0.00019 | |
| Salso. × Camph. | Tdif vs. Hsco | 8.96 | 2.75 × 10−27 | 4.99 | 2.89 × 10−19 | 2.84 | 0.000272 | 8.23 | 1.45 × 10−21 |
| Tdif vs. Salsod | 11.72 | 1.85 × 10−63 | 5.34 | 5.60 × 10−25 | 5.25 | 3.76 × 10−21 | 6.96 | 1.91 × 10−12 | |
| Tdif vs. Salopp | 12.03 | 6.61 × 10−65 | 4.7 | 1.56 × 10−15 | 2.44 | 0.00602 | 8.68 | 3.84 × 10−26 | |
| Cluster-9 | Cluster-10 | ||||||
|---|---|---|---|---|---|---|---|
| ATHB13 (HD-ZIP) | HSFA6B (HSF) | NAC083 (NAC) | |||||
| Lineage | Species (C4 vs. C3*) | LF2C | Padj | LF2C | Padj | LF2C | Padj |
| Salsoleae | Hsco vs. Salweb | 9.02 | 5.45 × 10−24 | 1.21 | 0.00017 | 0.73 | 0.00398 |
| Salopp vs. Salweb | 1.71 | 2.77 × 10−05 | 1.54 | 1.32 × 10−05 | 1.42 | 4.44 × 10−08 | |
| Salsod vs. Salweb | 1.14 | 0.00077 | 1.43 | 1.65 × 10−06 | 0.76 | 0.0004 | |
| Camphorosmeae | Bpro vs. Tdif | 8.89 | 2.80 × 10−23 | 1.7 | 1.08 × 10−06 | 11.74 | 3.28 × 10−82 |
| Bpro vs. Salweb | 8.99 | 8.45 × 10−24 | 1.91 | 5.76 × 10−08 | 11.28 | 1.31 × 10−72 | |
| Salso. × Camph. | Hsco vs. Tdif | 8.92 | 1.78 × 10−23 | 1 | 0.00169 | 1.01 | 5.11 × 10−05 |
| Salsod vs. Tdif | 0.87 | 0.00956 | 1.23 | 4.09 × 10−05 | 1.04 | 6.71 × 10−07 | |
| Salopp vs. Tdif | 1.45 | 0.00038 | 1.34 | 0.00011 | 1.71 | 4.00 × 10−11 | |
| Cluster-2 | |||
|---|---|---|---|
| bHLH106 (bHLH) | |||
| Lineage | Species (C3 vs. C2*) | LF2C | Padj |
| Salsoleae | Sweb vs. Saldi1 | 1.68 | 8.90 × 10−13 |
| Sweb vs. Saldi2 | 1.63 | 4.07 × 10−11 | |
| Camphorosmeae | Tdif vs. Sedsed | 0.45 | 0.04616 |
| Salso. × Camph. | Tdif vs. Sdi1 | 1.06 | 3.33 × 10−07 |
| Tdif vs. Sdi2 | 1.01 | 2.79 × 10−06 | |
| Sweb vs. Sedsed | 1.07 | 4.19 × 10−05 | |
| Species (C4 vs. C2*) | LF2C | Padj | |
| Salsoleae | Hsco vs. Saldi1 | 1.37 | 1.23 × 10−10 |
| Hsco vs. Saldi 2 | 1.32 | 3.58 × 10−09 | |
| Salopp vs. Saldi1 | 2.17 | 7.37 × 10−19 | |
| Salopp vs. Saldi2 | 2.12 | 1.03 × 10−16 | |
| Salsod vs. Saldi1 | 1.21 | 3.24 × 10−10 | |
| Salsod vs. Saldi2 | 1.16 | 7.30 × 10−09 | |
| Camphorosmeae | Bpro vs. Sedsed | 0.56 | 0.01398 |
| Salso. × Camph. | Bpro vs. Saldi1 | 1.17 | 2.14 × 10−08 |
| Bpro vs. Saldi2 | 1.12 | 2.21 × 10−07 | |
| Hsco vs. Sedsed | 0.76 | 0.001176 | |
| Salopp vs. Sedsed | 1.56 | 9.00 × 10−09 | |
| Salsod vs. Sedsed | 0.6 | 0.00483 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siadjeu, C.; Lauterbach, M.; Kadereit, G. Insights into Regulation of C2 and C4 Photosynthesis in Amaranthaceae/Chenopodiaceae Using RNA-Seq. Int. J. Mol. Sci. 2021, 22, 12120. https://doi.org/10.3390/ijms222212120
Siadjeu C, Lauterbach M, Kadereit G. Insights into Regulation of C2 and C4 Photosynthesis in Amaranthaceae/Chenopodiaceae Using RNA-Seq. International Journal of Molecular Sciences. 2021; 22(22):12120. https://doi.org/10.3390/ijms222212120
Chicago/Turabian StyleSiadjeu, Christian, Maximilian Lauterbach, and Gudrun Kadereit. 2021. "Insights into Regulation of C2 and C4 Photosynthesis in Amaranthaceae/Chenopodiaceae Using RNA-Seq" International Journal of Molecular Sciences 22, no. 22: 12120. https://doi.org/10.3390/ijms222212120
APA StyleSiadjeu, C., Lauterbach, M., & Kadereit, G. (2021). Insights into Regulation of C2 and C4 Photosynthesis in Amaranthaceae/Chenopodiaceae Using RNA-Seq. International Journal of Molecular Sciences, 22(22), 12120. https://doi.org/10.3390/ijms222212120






