Mechanisms of Nucleosome Reorganization by PARP1
Abstract
:1. Introduction
2. Results
2.1. Experimental Approaches for Analysis of PARP1-Dependent Changes in Nucleosome Structure
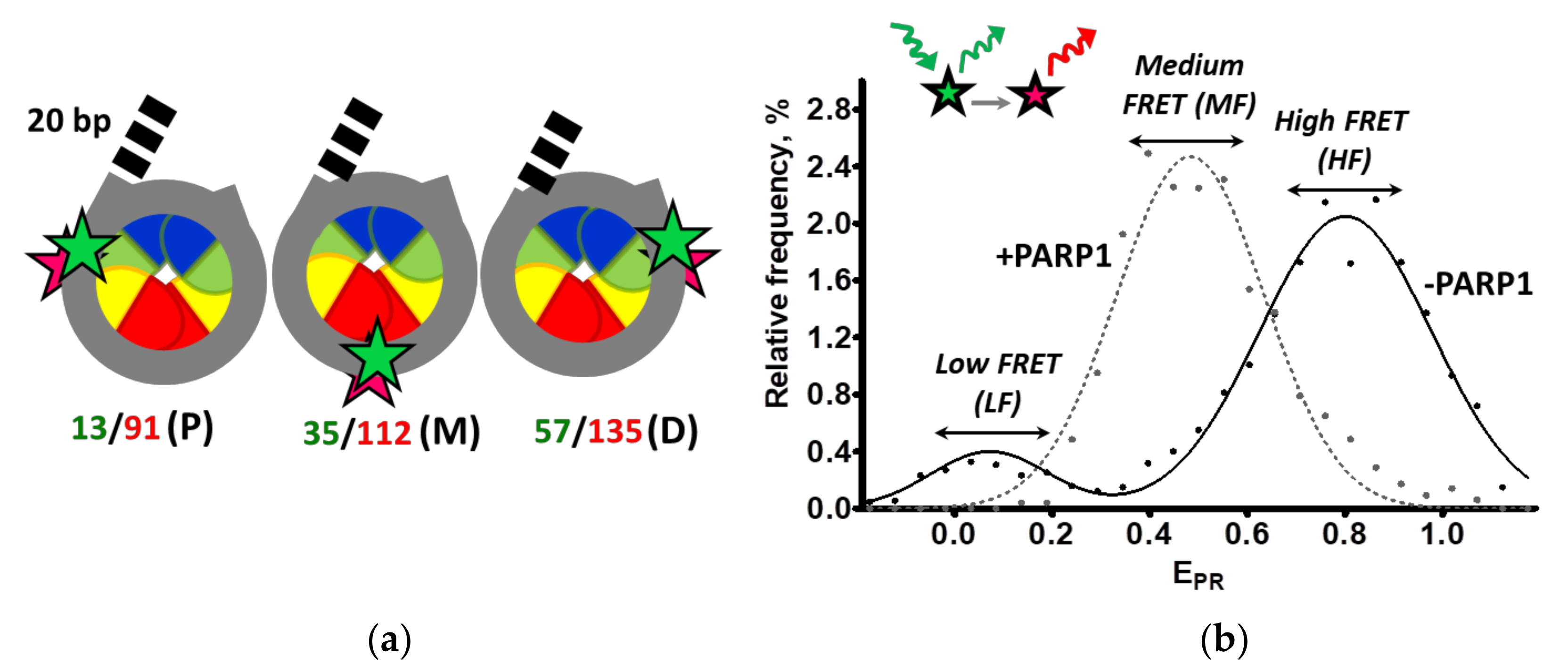
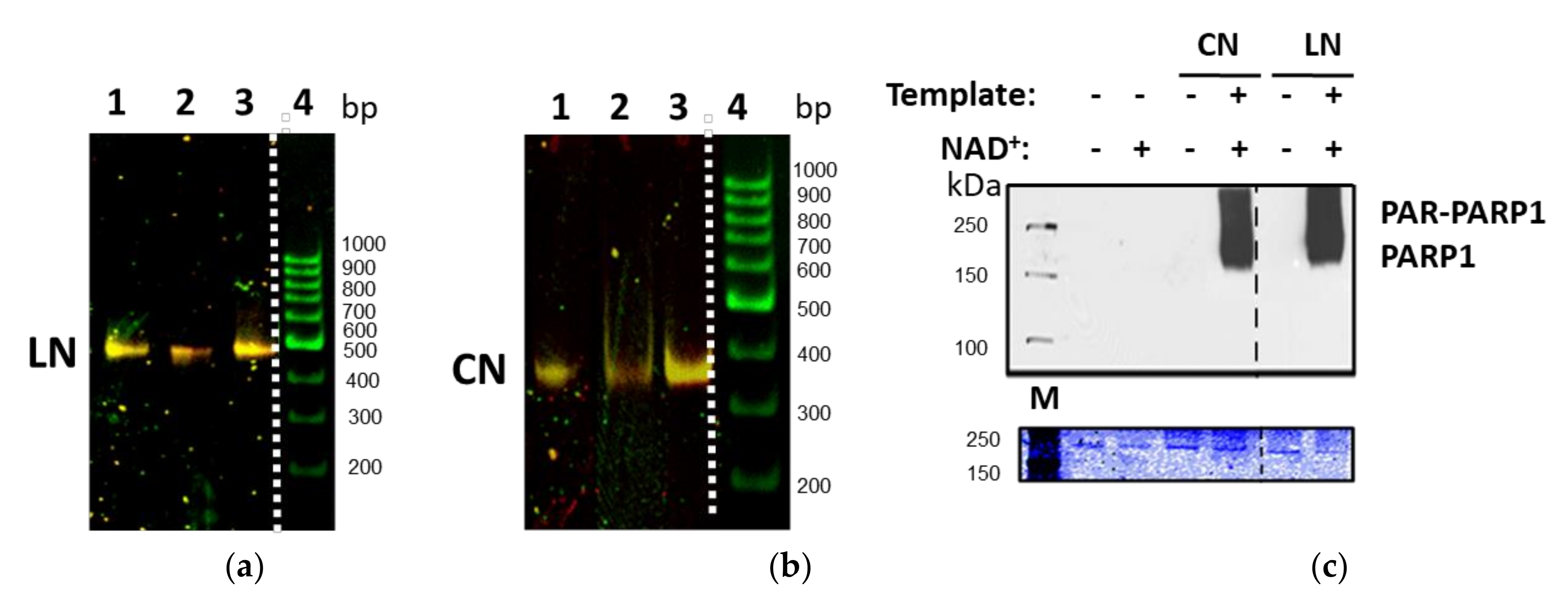
2.2. Characterization of the Nucleosomes and PARP1–Nucleosome Complexes
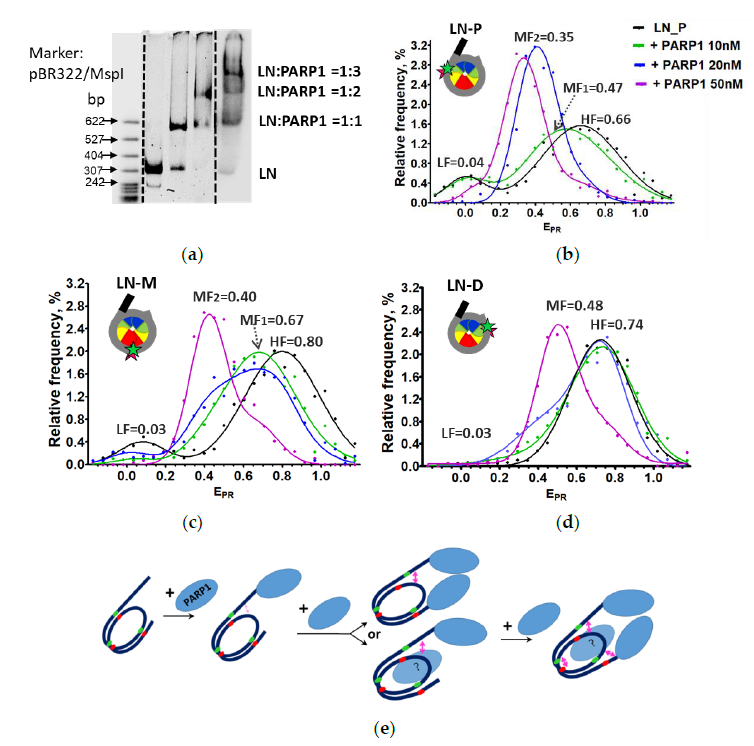
2.3. PARP1 Forms Structurally Distinct Complexes with Core Nucleosomes
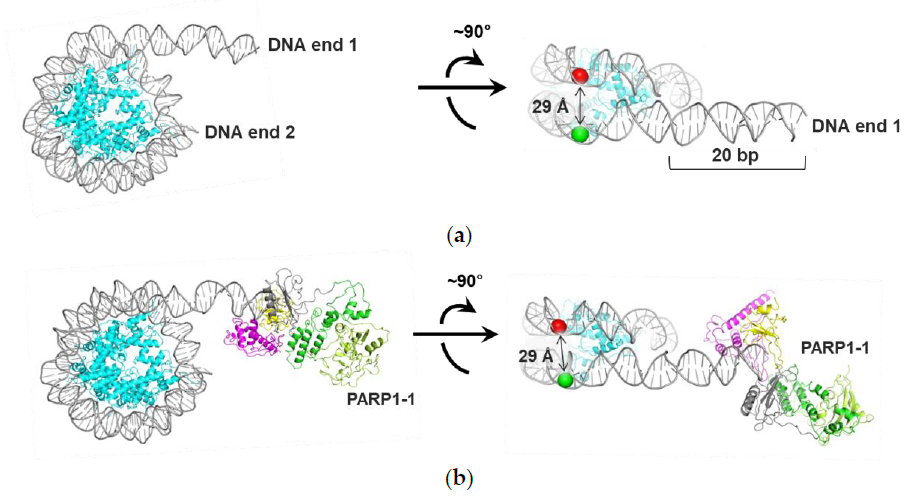
2.4. Nucleosomes Containing an Extended DNA Region Are Fully Reorganized Only after Binding of Three Molecules of PARP1
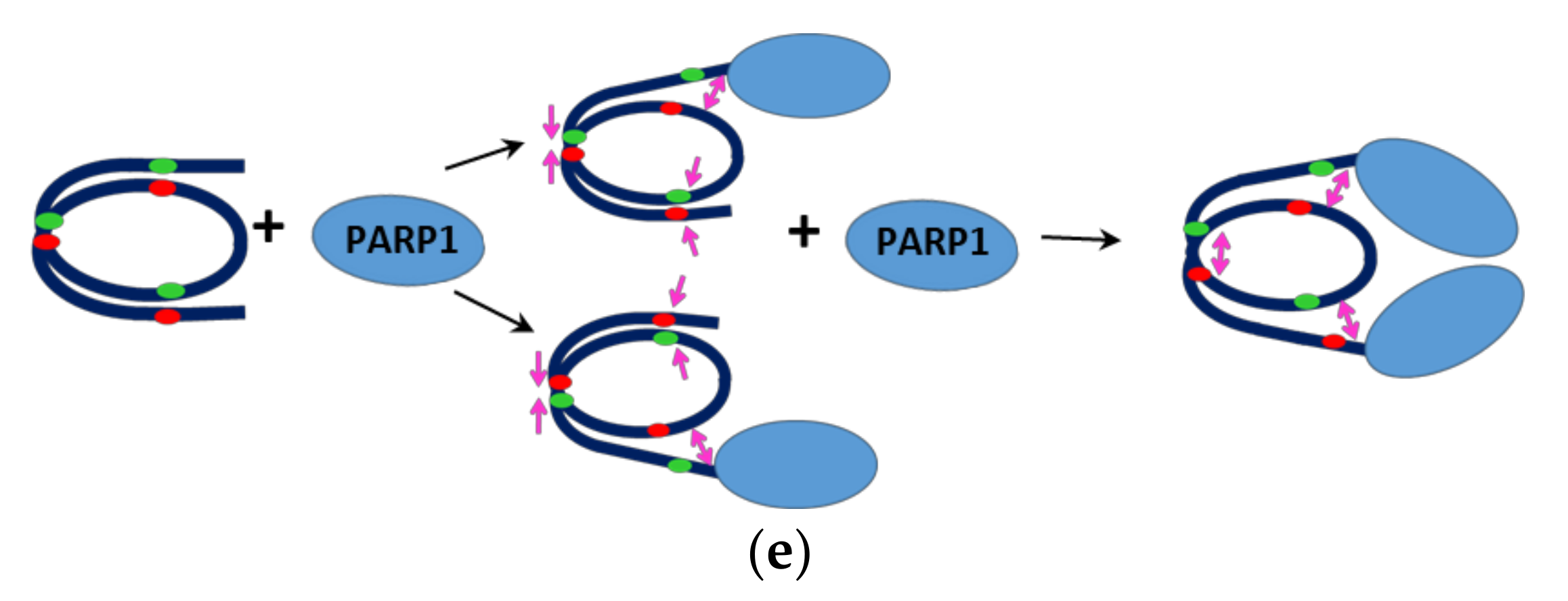
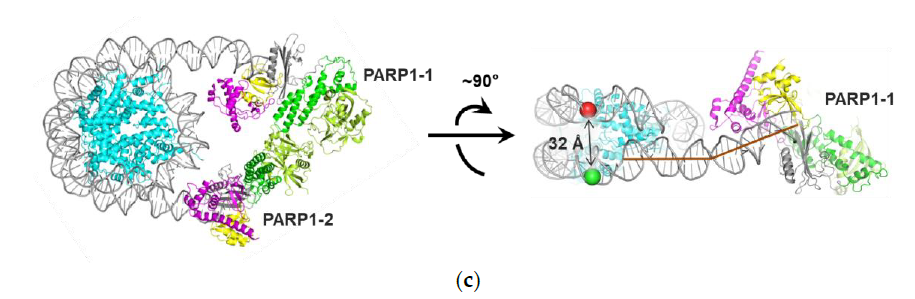
2.5. Interaction between Two PARP1 Molecules Bound to Double Strand DNA Breaks Can Induce Changes in the Structure of Nucleosomes: MD Simulations
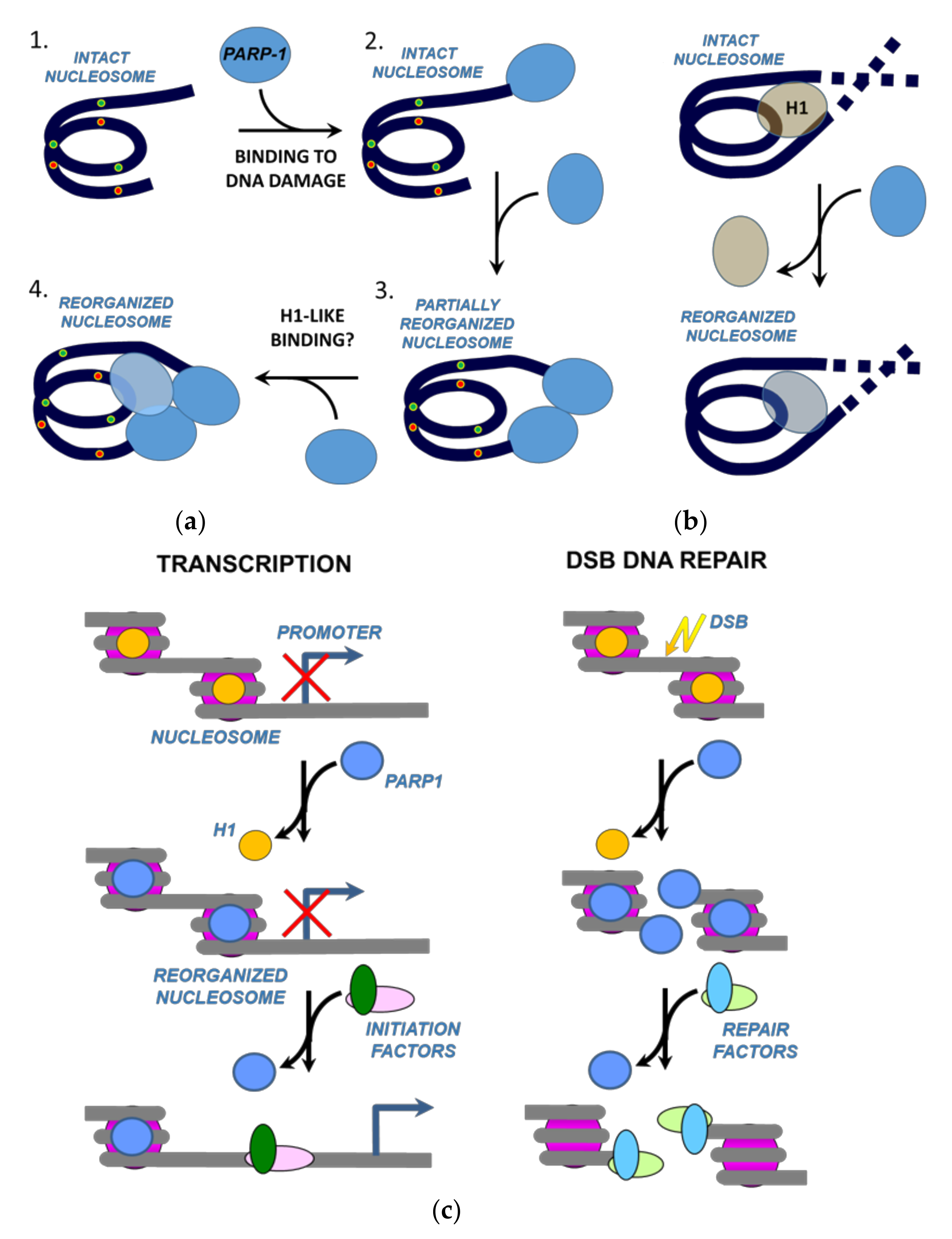
3. Discussion
4. Materials and Methods
4.1. Purification of Proteins, DNA Templates and Nucleosomes
4.2. spFRET Experiments in Solution
4.3. EMSA (Electrophoretic Mobility Shift Assay) Experiments
4.4. Single Particle Fluorescence Intensity Analysis of Nucleosomes in the Gel
4.5. Western Blot (WB) Experiments
4.6. DNaseI Footprinting
4.7. Molecular Modeling
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PARP1 | poly(ADP-ribose)polymerase 1 |
| PAR | poly(ADP-ribose) |
| MD | molecular dynamics |
| FRET | Förster resonance energy transfer |
| spFRET | single particle Förster resonance energy transfer |
| EMSA | electrophoretic mobility shift assay |
| Cy3 and Cy5 | fluorescent dyes cyanine 3 and cyanine 5 |
| EPR | proximity ratio (FRET efficiency without correction for detection sensitivity and quantum yields of fluorophores) |
| LN_P, LN_M, LN_D | nucleosomes with one 20 bp linker labeled with Cy3/Cy5 at positions 13/91 bp, 35/112 bp or 57/135 bp, respectively |
| CN_P, CN_M, CN_D | core nucleosomes labeled with Cy3/Cy5 at positions 13/91 bp, 35/112 bp or 57/135 bp, respectively |
References
- D’Amours, D.; Desnoyers, S.; D’Silva, I.; Poirier, G.G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999, 342 Pt 2, 249–268. [Google Scholar] [CrossRef]
- Kim, M.Y.; Mauro, S.; Gevry, N.; Lis, J.T.; Kraus, W.L. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 2004, 119, 803–814. [Google Scholar] [CrossRef] [Green Version]
- Nilov, D.K.; Pushkarev, S.V.; Gushchina, I.V.; Manasaryan, G.A.; Kirsanov, K.I.; Švedas, V.K. Modeling of the Enzyme-Substrate Complexes of Human Poly(ADP-Ribose) Polymerase 1. Biochemistry 2020, 85, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Manasaryan, G.; Suplatov, D.; Pushkarev, S.; Drobot, V.; Kuimov, A.; Švedas, V.; Nilov, D. Bioinformatic Analysis of the Nicotinamide Binding Site in Poly(ADP-Ribose) Polymerase Family Proteins. Cancers 2021, 13, 1201. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Kraus, W. Catalytic-Independent Functions of PARP-1 Determine Sox2 Pioneer Activity at Intractable Genomic Loci. Mol. Cell. 2017, 65, 589–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnakumar, R.; Kraus, W.L. The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol. Cell 2010, 39, 8–24. [Google Scholar] [CrossRef] [Green Version]
- Hottiger, M.O. Nuclear ADP-Ribosylation and Its Role in Chromatin Plasticity, Cell Differentiation, and Epigenetics. Annu. Rev. Biochem 2015, 84, 227–263. [Google Scholar] [CrossRef] [PubMed]
- Ogden, T.E.H.; Yang, J.C.; Schimpl, M.; Easton, L.E.; Underwood, E.; Rawlins, P.B.; McCauley, M.M.; Langelier, M.F.; Pascal, J.M.; Embrey, K.J.; et al. Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition. Nucleic Acids Res. 2021, 49, 2266–2288. [Google Scholar] [CrossRef] [PubMed]
- Langelier, M.F.; Planck, J.L.; Roy, S.; Pascal, J.M. Structural Basis for DNA Damage-Dependent Poly(ADP-ribosyl)ation by Human PARP-1. Science 2012, 336, 728–732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dawicki-McKenna, J.M.; Langelier, M.F.; DeNizio, J.E.; Riccio, A.A.; Cao, C.D.; Karch, K.R.; McCauley, M.; Steffen, J.D.; Black, B.E.; Pascal, J.M. PARP-1 Activation Requires Local Unfolding of an Autoinhibitory Domain. Mol. Cell 2015, 60, 755–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, C.; Ji, Y.; Wu, C.; Datz, H.; Boyle, C.; MacLeod, B.; Patel, S.; Ampofo, M.; Currie, M.; Harbin, J.; et al. Hit and run versus long-term activation of PARP-1 by its different domains fine-tunes nuclear processes. Proc. Natl. Acad. Sci. USA 2019, 116, 9941–9946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schreiber, V.; Dantzer, F.; Ame, J.C.; de Murcia, G. Poly(ADP-ribose): Novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006, 7, 517–528. [Google Scholar] [CrossRef]
- Pazzaglia, S.; Pioli, C. Multifaceted Role of PARP-1 in DNA Repair and Inflammation: Pathological and Therapeutic Implications in Cancer and Non-Cancer Diseases. Cells 2019, 9, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinnola, A.; Naumova, N.; Shah, M.; Tulin, A.V. Nucleosomal core histones mediate dynamic regulation of poly(ADP-ribose) polymerase 1 protein binding to chromatin and induction of its enzymatic activity. J. Biol. Chem. 2007, 282, 32511–32519. [Google Scholar] [CrossRef] [Green Version]
- Kraus, W.L.; Lis, J.T. PARP goes transcription. Cell 2003, 113, 677–683. [Google Scholar] [CrossRef] [Green Version]
- Krishnakumar, R.; Kraus, W.L. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell 2010, 39, 736–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupte, R.; Liu, Z.; Kraus, W.L. PARPs and ADP-ribosylation: Recent advances linking molecular functions to biological outcomes. Genes Dev. 2017, 31, 101–126. [Google Scholar] [CrossRef] [Green Version]
- Krishnakumar, R.; Gamble, M.; Frizzell, K.; Berrocal, J.; Kininis, M.; Kraus, W.L. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 2008, 319, 819–821. [Google Scholar] [CrossRef] [PubMed]
- Nalabothula, N.; Al-Jumaily, T.; Eteleeb, A.M.; Flight, R.M.; Xiaorong, S.; Moseley, H.; Rouchka, E.C.; Fondufe-Mittendorf, Y.N. Genome-Wide Profiling of PARP1 Reveals an Interplay with Gene Regulatory Regions and DNA Methylation. PLoS ONE 2015, 10, e0135410. [Google Scholar] [CrossRef]
- Muthurajan, U.M.; Hepler, M.R.; Hieb, A.R.; Clark, N.J.; Kramer, M.; Yao, T.; Luger, K. Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone. Proc. Natl. Acad. Sci. USA 2014, 111, 12752–12757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sultanov, D.C.; Gerasimova, N.S.; Kudryashova, K.S.; Maluchenko, N.V.; Kotova, E.Y.; Langelier, M.F.; Pascal, J.M.; Kirpichnikov, M.P.; Feofanov, A.V.; Studitsky, V.M. Unfolding of core nucleosomes by PARP-1 revealed by spFRET microscopy. Aims Genet. 2017, 4, 21–31. [Google Scholar] [CrossRef]
- Kulaeva, O.I.; Gaykalova, D.A.; Pestov, N.A.; Golovastov, V.V.; Vassylyev, D.G.; Artsimovitch, I.; Studitsky, V.M. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat. Struct. Mol. Biol. 2009, 16, 1272–1278. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Berrocal, J.G.; Yao, J.; DuMond, M.E.; Krishnakumar, R.; Ruhl, D.D.; Ryu, K.W.; Gamble, M.J.; Kraus, W.L. Regulation of poly(ADP-ribose) polymerase-1-dependent gene expression through promoter-directed recruitment of a nuclear NAD+ synthase. J. Biol. Chem. 2012, 287, 12405–12416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valieva, M.E.; Armeev, G.A.; Kudryashova, K.S.; Gerasimova, N.S.; Shaytan, A.K.; Kulaeva, O.I.; McCullough, L.L.; Formosa, T.; Georgiev, P.G.; Kirpichnikov, M.P.; et al. Large-scale ATP-independent nucleosome unfolding by a histone chaperone. Nat. Struct. Mol. Biol. 2016, 23, 1111–1116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilokapic, S.; Strauss, M.; Halic, M. Histone octamer rearranges to adapt to DNA unwrapping. Nat. Struct. Mol. Biol. 2018, 25, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Nir, E.; Michalet, X.; Hamadani, K.M.; Laurence, T.A.; Neuhauser, D.; Kovchegov, Y.; Weiss, S. Shot-noise limited single-molecule FRET histograms: Comparison between theory and experiments. J. Phys. Chem. B 2006, 110, 22103–22124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maluchenko, N.V.; Sultanov, D.S.; Kotova, E.Y.; Kirpichnikov, M.P.; Studitsky, V.M.; Feofanov, A.V. Histone Tails Promote PARP1-Dependent Structural Rearrangements in Nucleosomes. Dokl. Biochem. Biophys. 2019, 489, 377–379. [Google Scholar] [CrossRef]
- Bondarenko, V.A.; Steele, L.M.; Ujvari, A.; Gaykalova, D.A.; Kulaeva, O.I.; Polikanov, Y.S.; Luse, D.S.; Studitsky, V.M. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol. Cell 2006, 24, 469–479. [Google Scholar] [CrossRef]
- Vasudevan, D.; Chua, E.Y.; Davey, C.A. Crystal structures of nucleosome core particles containing the ‘601’ strong positioning sequence. J. Mol. Biol. 2010, 403, 1–10. [Google Scholar] [CrossRef]
- Clark, N.J.; Kramer, M.; Muthurajan, U.M.; Luger, K. Alternative modes of binding of Poly(ADP-ribose) polymerase 1 to free DNA and nucleosomes. J. Biol. Chem. 2012, 287, 32430–32439. [Google Scholar] [CrossRef] [Green Version]
- Langelier, M.F.; Ruhl, D.D.; Planck, J.L.; Kraus, W.L.; Pascal, J.M. The Zn3 domain of human Poly(ADP-ribose) polymerase-1 (PARP1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J. Biol. Chem. 2010, 285, 18877–18887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaykalova, D.A.; Kulaeva, O.I.; Bondarenko, V.A.; Studitsky, V.M. Preparation and analysis of uniquely positioned mononucleosomes. Methods Mol. Biol. 2009, 523, 109–123. [Google Scholar] [PubMed] [Green Version]
- Lu, X.J.; Olson, W.K. 3DNA: A versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 2008, 3, 1213–1227. [Google Scholar] [CrossRef] [PubMed]
- Nilov, D.; Maluchenko, N.; Kurgina, T.; Pushkarev, S.; Lys, A.; Kutuzov, M.; Gerasimova, N.; Feofanov, A.; Švedas, V.; Lavrik, O.; et al. Molecular Mechanisms of PARP-1 Inhibitor 7-Methylguanine. Int. J. Mol. Sci. 2020, 21, 2159. [Google Scholar] [CrossRef] [Green Version]
- Case, D.A.; Belfon, K.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., 3rd; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Giambasu, G. AMBER 2020; University of California: San Francisco, CA, USA, 2020; pp. 1–918. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivani, I.; Dans, P.D.; Noy, A.; Perez, A.; Faustino, I.; Hospital, A.; Walther, J.; Andrio, P.; Goni, R.; Balaceanu, A.; et al. Parmbsc1: A refined force field for DNA simulations. Nat. Methods 2016, 13, 55–58. [Google Scholar] [CrossRef] [Green Version]

| Name | Nucleotide Sequence |
|---|---|
| CN_P_forward | 5′-CCCGGTTCGCGC[Cy3-dT]CCCGCCTTCCGTGTGTTGTCGTCTCTCGG-3′ |
| CN_P_reverse | 5′-ACCCCAGGGACTTGAAGTAATAAGGACGGAGGGCCTCTTTCAACATCGATGCACGG[Cy5-dT]GGTTAG-3′ |
| CN_M_forward | 5′-CCCGGTTCGCGCTCCCGCCTTCCGTGTGTTGTCG[Cy5-dT]CTCTCGG-3′ |
| CN_M_reverse | 5′-ACCCCAGGGACTTGAAGTAATAAGGACGGAGGGCC[Cy3-dT]CTTTCAACATCGAT-3′ |
| CN_D_forward | 5′-CCCGGTTCGCGCTCCCGCCTTCCGTGTGTTGTCGTCTCTCGGGCGTCTAAGTACGC[Cy3-dT]TAGGC-3′ |
| CN_D_reverse | 5’-ACCCCAGGGACT[Cy5-dT]GAAGTAATAAGGACGGAGGGCCTCTTTC-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maluchenko, N.V.; Nilov, D.K.; Pushkarev, S.V.; Kotova, E.Y.; Gerasimova, N.S.; Kirpichnikov, M.P.; Langelier, M.-F.; Pascal, J.M.; Akhtar, M.S.; Feofanov, A.V.; et al. Mechanisms of Nucleosome Reorganization by PARP1. Int. J. Mol. Sci. 2021, 22, 12127. https://doi.org/10.3390/ijms222212127
Maluchenko NV, Nilov DK, Pushkarev SV, Kotova EY, Gerasimova NS, Kirpichnikov MP, Langelier M-F, Pascal JM, Akhtar MS, Feofanov AV, et al. Mechanisms of Nucleosome Reorganization by PARP1. International Journal of Molecular Sciences. 2021; 22(22):12127. https://doi.org/10.3390/ijms222212127
Chicago/Turabian StyleMaluchenko, Natalya V., Dmitry K. Nilov, Sergey V. Pushkarev, Elena Y. Kotova, Nadezhda S. Gerasimova, Mikhail P. Kirpichnikov, Marie-France Langelier, John M. Pascal, Md. Sohail Akhtar, Alexey V. Feofanov, and et al. 2021. "Mechanisms of Nucleosome Reorganization by PARP1" International Journal of Molecular Sciences 22, no. 22: 12127. https://doi.org/10.3390/ijms222212127
APA StyleMaluchenko, N. V., Nilov, D. K., Pushkarev, S. V., Kotova, E. Y., Gerasimova, N. S., Kirpichnikov, M. P., Langelier, M.-F., Pascal, J. M., Akhtar, M. S., Feofanov, A. V., & Studitsky, V. M. (2021). Mechanisms of Nucleosome Reorganization by PARP1. International Journal of Molecular Sciences, 22(22), 12127. https://doi.org/10.3390/ijms222212127








