The Escherichia coli Outer Membrane β-Barrel Assembly Machinery (BAM) Crosstalks with the Divisome
Abstract
:1. Introduction
2. Results
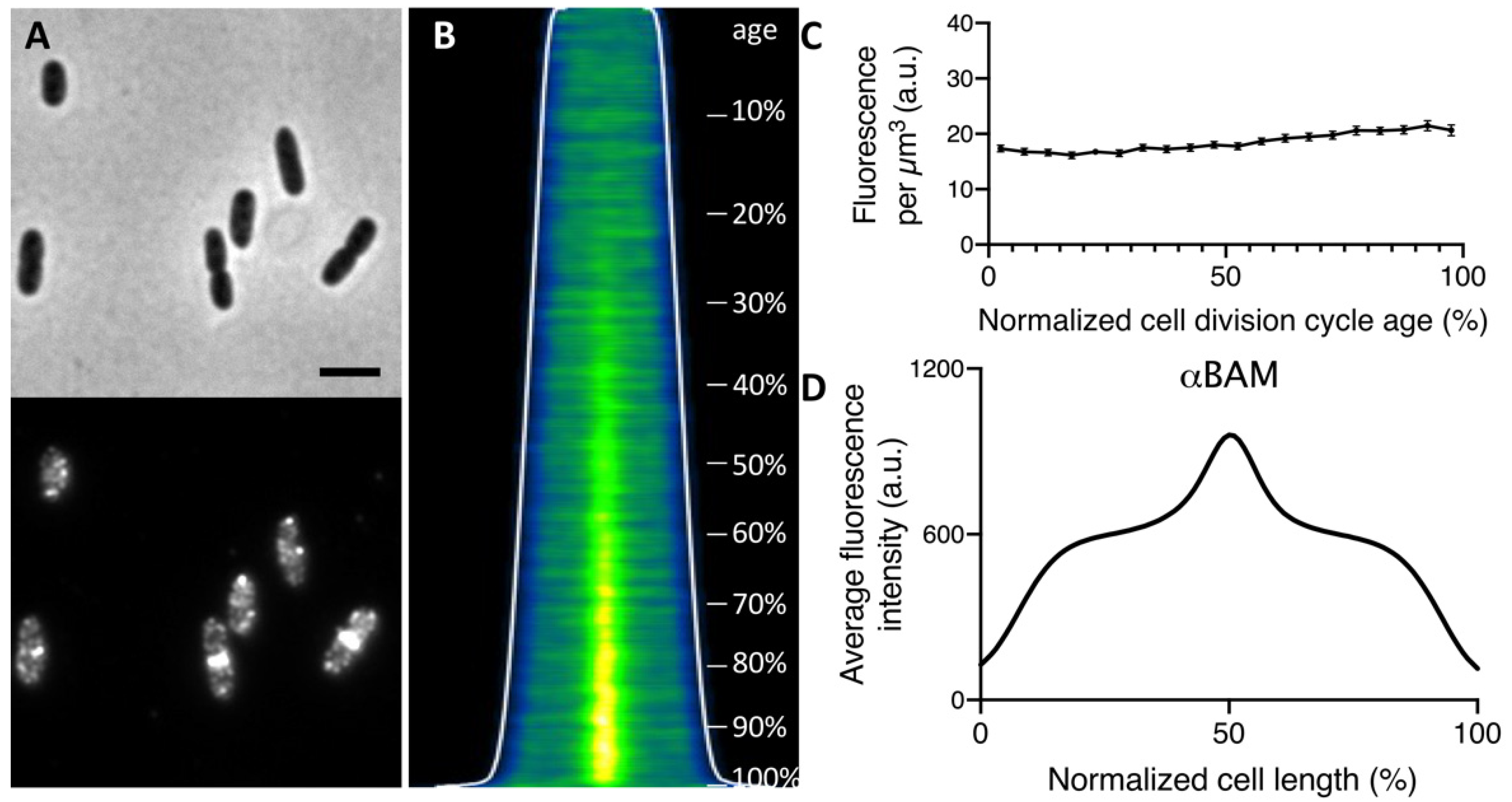
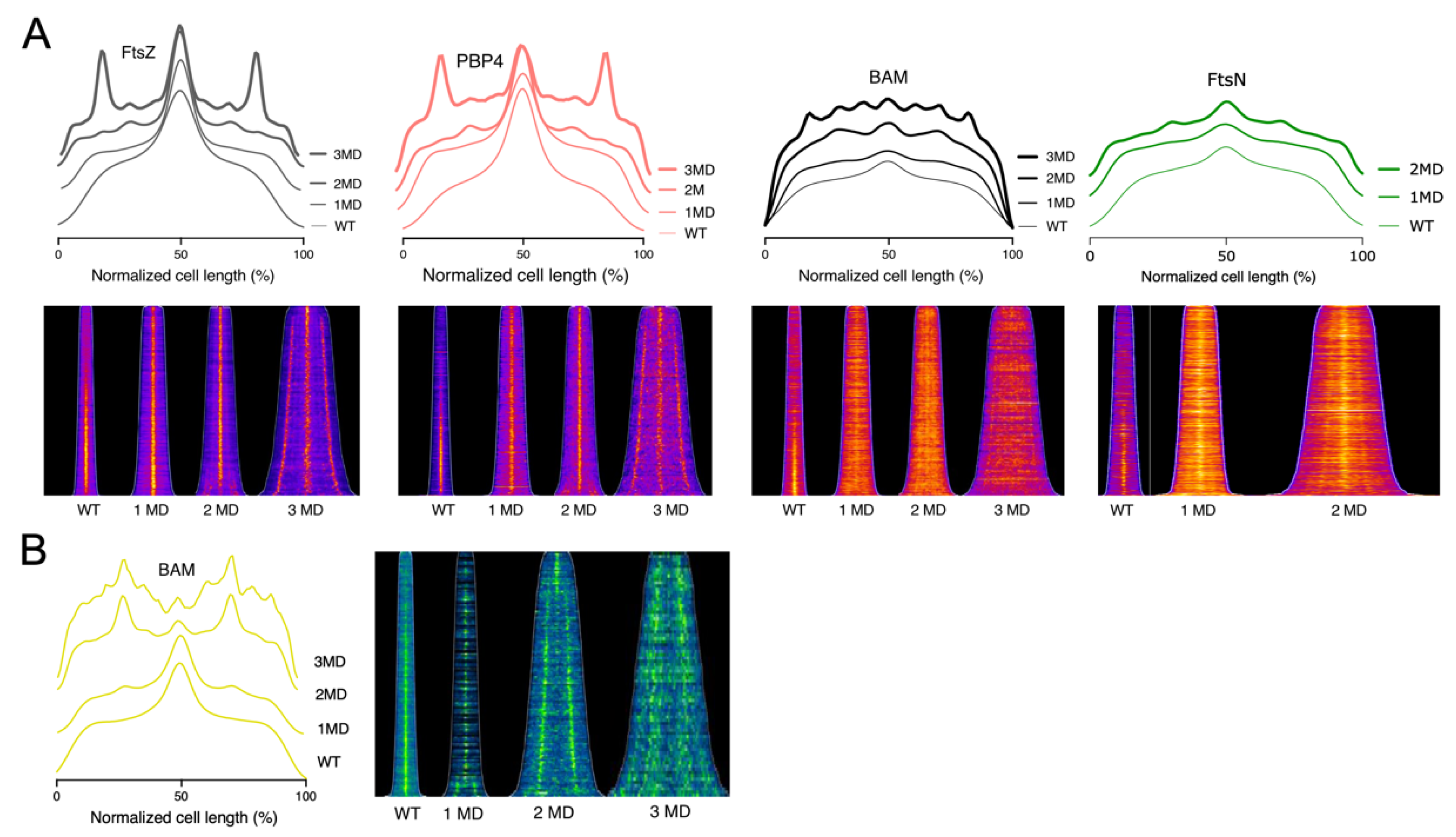
2.1. The Bam Complex Localizes at Midcell during Constriction
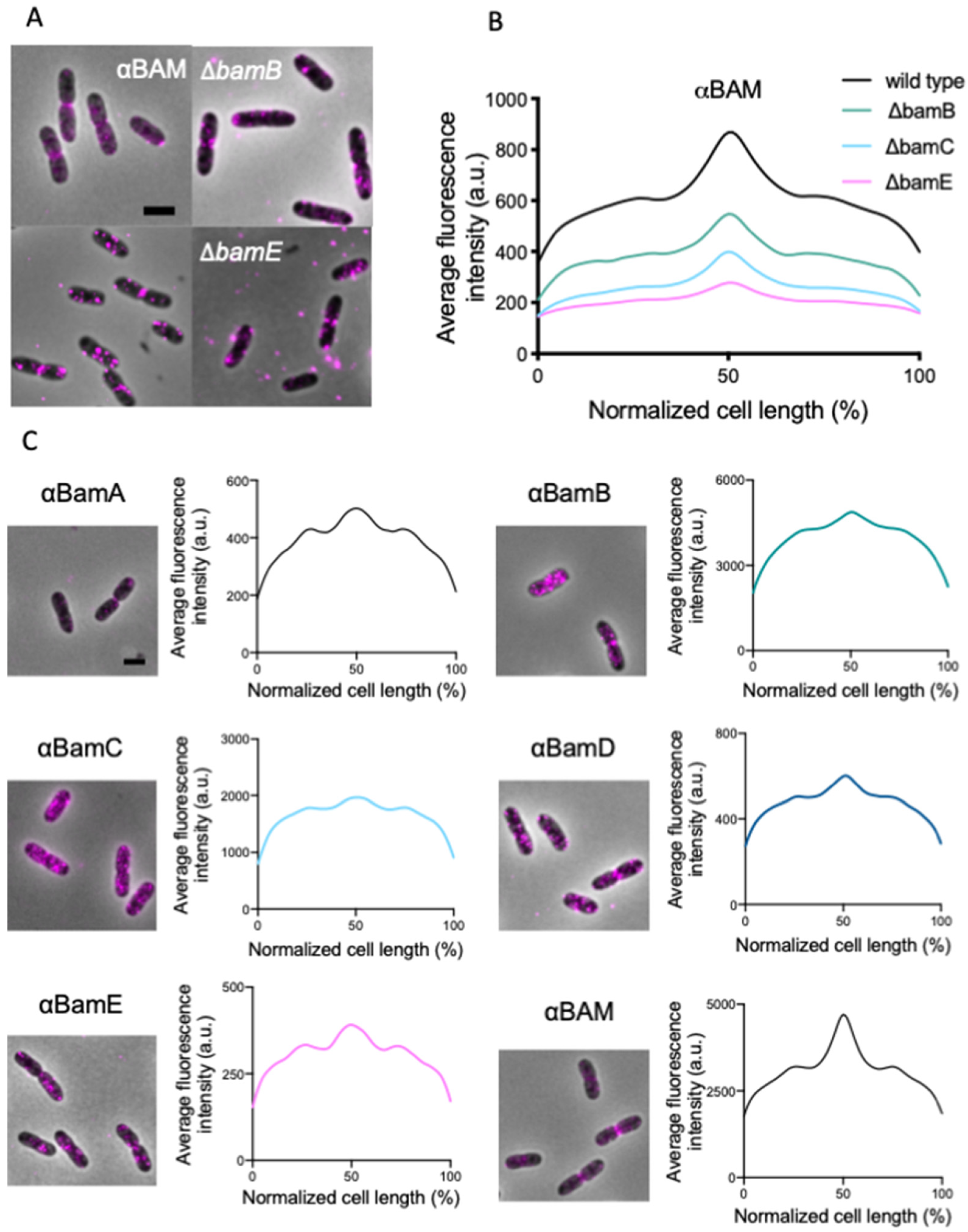
2.2. All BAM Subunits Localize at Midcell
2.3. Not All OM Proteins Exhibit a Midcell Localization like BAM
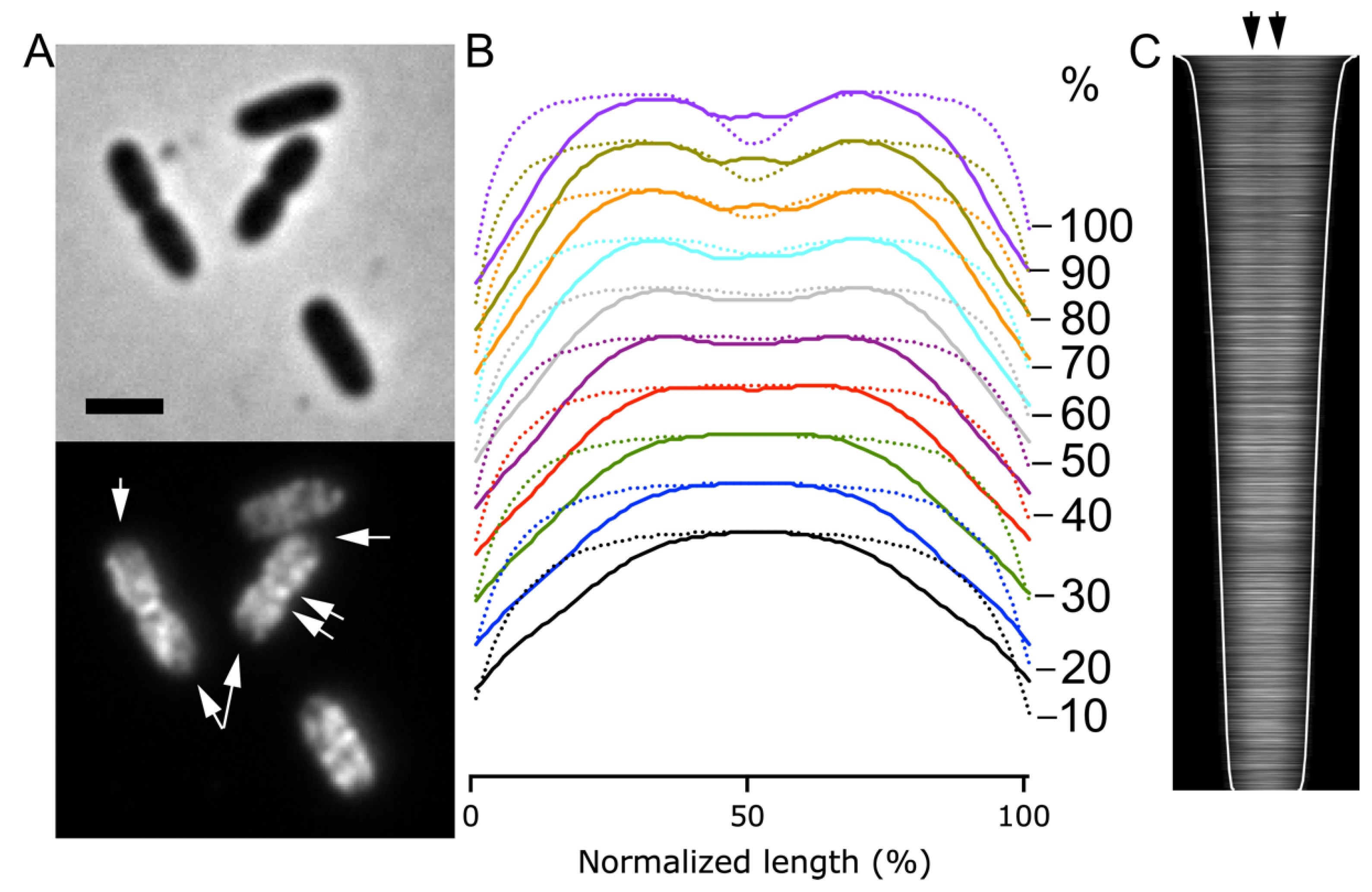
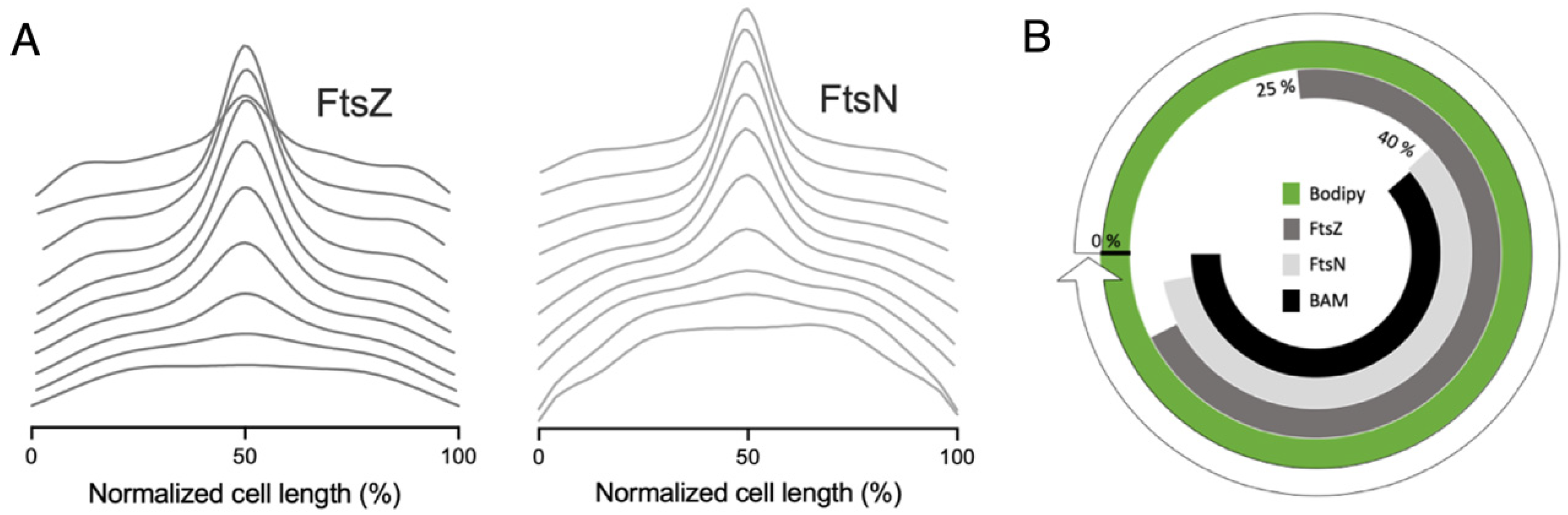
2.4. The BAM Complex Localizes at Midcell Simultaneously with Divisome Maturation
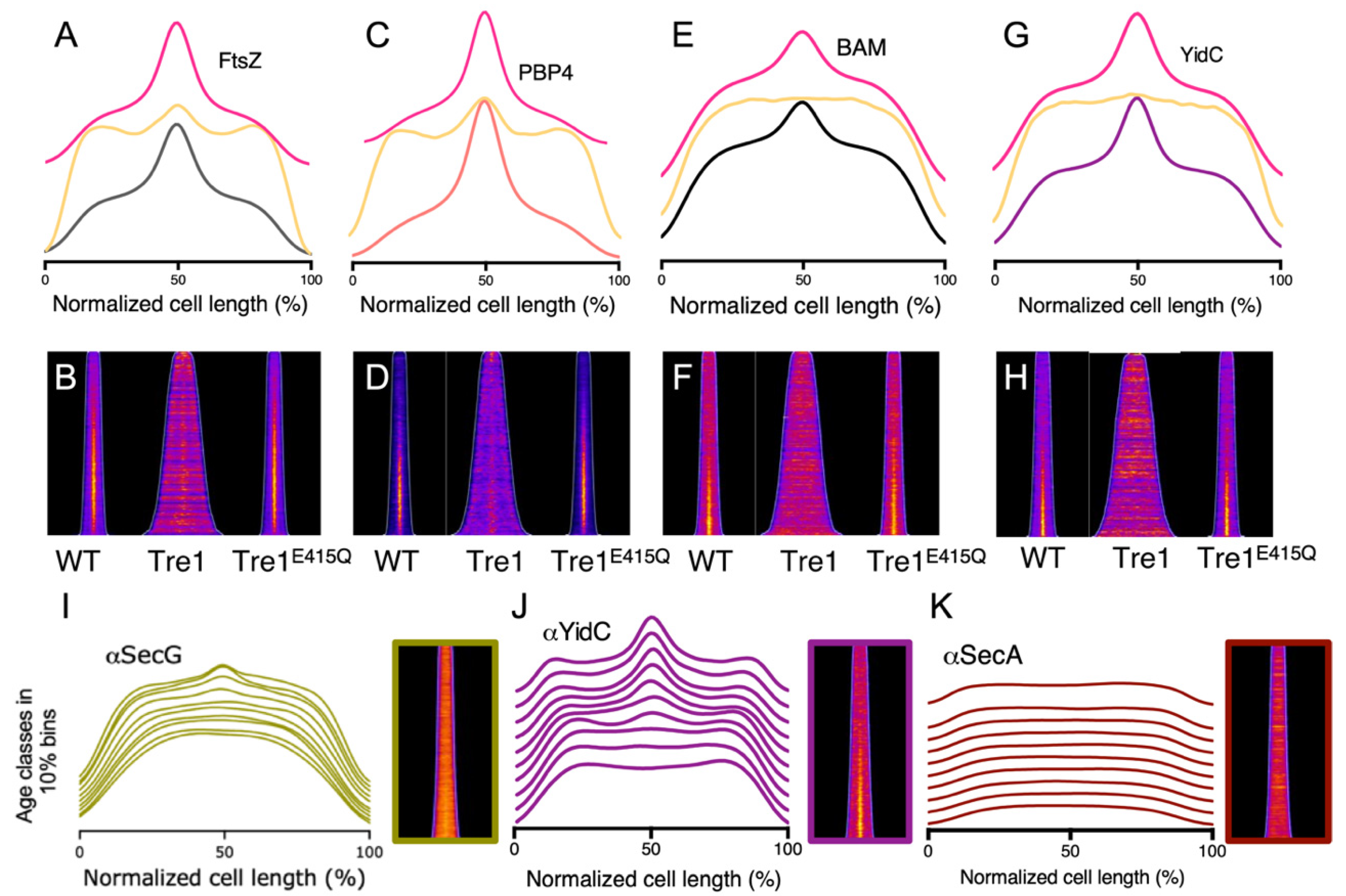
2.5. The BAM Midcell Localization Does Not Require Active Septation
2.6. The BAM Midcell Localization Does Not Depend on the Late Division Proteins
2.7. The BAM Complex Midcell Localization Depends on the Z-Ring Formation
2.8. YidC Colocalize at Midcell at the Same Time as the BAM Complex
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Fixation and Immunolabelling
4.3. Microscopy
4.4. Image Analysis
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nikaido, H. Advances in Enzymology and Related Areas of Molecular Biology. In Advances in Enzymology and Related Areas of Molecular Biology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011; Volume 77, pp. 1–60. [Google Scholar] [CrossRef] [Green Version]
- Glauert, A.M.; Thornley, M.J. The Topography of the Bacterial Cell Wall. Annu. Rev. Microbiol. 1969, 23, 159–198. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The Bacterial Cell Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef] [PubMed]
- Oswald, J.; Njenga, R.; Natriashvili, A.; Sarmah, P.; Koch, H.-G. The Dynamic SecYEG Translocon. Front. Mol. Biosci. 2021, 8, 664241. [Google Scholar] [CrossRef]
- Koch, H.-G.; Müller, M. Dissecting the Translocase and Integrase Functions of the Escherichia coli SecYEG Translocon. J. Cell Biol. 2000, 150, 689–694. [Google Scholar] [CrossRef] [Green Version]
- Xie, K.; Dalbey, R.E. Inserting Proteins into the Bacterial Cytoplasmic Membrane Using the Sec and YidC Translocases. Nat. Rev. Microbiol. 2008, 6, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, A. From the Sec Complex to the Membrane Insertase YidC. Biol. Chem. 2009, 390, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Paetzel, M.; Karla, A.; Strynadka, N.C.J.; Dalbey, R.E. Signal Peptidases. Chem. Rev. 2002, 102, 4549–4580. [Google Scholar] [CrossRef]
- Dalbey, R.E.; Wickner, W. Leader Peptidase Catalyzes the Release of Exported Proteins from the Outer Surface of the Escherichia coli Plasma Membrane. J. Biol. Chem. 1985, 260, 15925–15931. [Google Scholar] [CrossRef]
- Hussain, S.; Peterson, J.H.; Bernstein, H.D. Bam Complex-Mediated Assembly of Bacterial Outer Membrane Proteins Synthesized in an in Vitro Translation System. Sci. Rep. 2020, 10, 4557. [Google Scholar] [CrossRef]
- Hagan, C.L.; Kim, S.; Kahne, D. Reconstitution of Outer Membrane Protein Assembly from Purified Components. Science 2010, 328, 890–892. [Google Scholar] [CrossRef] [Green Version]
- Sklar, J.G.; Wu, T.; Kahne, D.; Silhavy, T.J. Defining the Roles of the Periplasmic Chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007, 21, 2473–2484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charlson, E.S.; Werner, J.N.; Misra, R. Differential Effects of YfgL Mutation on Escherichia coli Outer Membrane Proteins and Lipopolysaccharide. J. Bacteriol. 2006, 188, 7186–7194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ureta, A.R.; Endres, R.G.; Wingreen, N.S.; Silhavy, T.J. Kinetic Analysis of the Assembly of the Outer Membrane Protein LamB in Escherichia coli Mutants Each Lacking a Secretion or Targeting Factor in a Different Cellular Compartment. J. Bacteriol. 2006, 189, 446–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuong, P.; Bennion, D.; Mantei, J.; Frost, D.; Misra, R. Analysis of YfgL and YaeT Interactions through Bioinformatics, Mutagenesis, and Biochemistry. J. Bacteriol. 2008, 190, 1507–1517. [Google Scholar] [CrossRef] [Green Version]
- Webb, C.T.; Selkrig, J.; Perry, A.J.; Noinaj, N.; Buchanan, S.K.; Lithgow, T. Dynamic Association of BAM Complex Modules Includes Surface Exposure of the Lipoprotein BamC. J. Mol. Biol. 2012, 422, 545–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malinverni, J.C.; Werner, J.; Kim, S.; Sklar, J.G.; Kahne, D.; Misra, R.; Silhavy, T.J. YfiO Stabilizes the YaeT Complex and Is Essential for Outer Membrane Protein Assembly in Escherichia Coli. Mol. Microbiol. 2006, 61, 151–164. [Google Scholar] [CrossRef]
- Ricci, D.P.; Silhavy, T.J. The Bam Machine: A Molecular Cooper. Biochim. Biophys. Acta 2012, 1818, 1067–1084. [Google Scholar] [CrossRef] [Green Version]
- Chng, S.-S.; Gronenberg, L.S.; Kahne, D. Proteins Required for Lipopolysaccharide Assembly in Escherichia coli Form a Transenvelope Complex. Biochemistry 2010, 49, 4565–4567. [Google Scholar] [CrossRef] [Green Version]
- Gunasinghe, S.D.; Shiota, T.; Stubenrauch, C.J.; Schulze, K.E.; Webb, C.T.; Fulcher, A.J.; Dunstan, R.A.; Hay, I.D.; Naderer, T.; Whelan, D.R.; et al. The WD40 Protein BamB Mediates Coupling of BAM Complexes into Assembly Precincts in the Bacterial Outer Membrane. Cell Rep. 2018, 23, 2782–2794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mamou, G.; Inns, P.G.; Sun, D.; Kaminska, R.; Housden, N.G.; Cohen-Khait, R.; Miller, H.; Storek, K.M.; Rutherford, S.T.; Payandeh, J.; et al. Spatiotemporal Organization of BamA Governs the Pattern of Outer Membrane Protein Distribution in Growing Escherichia coli Cells. bioRxiv 2021. [Google Scholar] [CrossRef]
- Consoli, E.; Collet, J.-F.; den Blaauwen, T. The Escherichia coli Outer Membrane β-Barrel Assembly Machinery (BAM) Anchors the Peptidoglycan Layer by Spanning It with All Subunits. Int. J. Mol. Sci. 2021, 22, 1853. [Google Scholar] [CrossRef] [PubMed]
- Egan, A.J.F.; Vollmer, W. The Physiology of Bacterial Cell Division. Ann. N. Y. Acad. Sci. 2013, 1277, 8–28. [Google Scholar] [CrossRef] [PubMed]
- Aarsman, M.E.G.; Piette, A.; Fraipont, C.; Vinkenvleugel, T.M.F.; Nguyen-Distèche, M.; Blaauwen, T. den Maturation of the Escherichia coli Divisome Occurs in Two Steps. Mol. Microbiol. 2005, 55, 1631–1645. [Google Scholar] [CrossRef] [PubMed]
- Bi, E.; Lutkenhaus, J. FtsZ Ring Structure Associated with Division in Escherichia coli. Nature 1991, 354, 161–164. [Google Scholar] [CrossRef] [PubMed]
- Pichoff, S.; Shen, B.; Sullivan, B.; Lutkenhaus, J. FtsA Mutants Impaired for Self-Interaction Bypass ZipA Suggesting a Model in Which FtsA’s Self-Interaction Competes with Its Ability to Recruit Downstream Division Proteins. Mol. Microbiol. 2012, 83, 151–167. [Google Scholar] [CrossRef] [Green Version]
- Erickson, H.P.; Anderson, D.E.; Osawa, M. FtsZ in Bacterial Cytokinesis: Cytoskeleton and Force Generator All in One. Microbiol. Mol. Biol. Rev. 2010, 74, 504–528. [Google Scholar] [CrossRef] [Green Version]
- Addinall, S.G.; Cao, C.; Lutkenhaus, J. FtsN, a Late Recruit to the Septum in Escherichia coli. Mol. Microbiol. 1997, 25, 303–309. [Google Scholar] [CrossRef]
- Busiek, K.K.; Margolin, W. A Role for FtsA in SPOR-Independent Localization of the Essential Escherichia coli Cell Division Protein FtsN. Mol. Microbiol. 2014, 92, 1212–1226. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Persons, L.; Lee, L.; Boer, P.A.J.D. Roles for Both FtsA and the FtsBLQ Subcomplex in FtsN-Stimulated Cell Constriction in Escherichia coli. Mol. Microbiol. 2015, 95, 945–970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsang, M.-J.; Bernhardt, T.G. A Role for the FtsQLB Complex in Cytokinetic Ring Activation Revealed by an FtsL Allele That Accelerates Division. Mol. Microbiol. 2015, 95, 925–944. [Google Scholar] [CrossRef] [Green Version]
- Fraipont, C.; Alexeeva, S.; Wolf, B.; van der Ploeg, R.; Schloesser, M.; den Blaauwen, T.; Nguyen-Distèche, M. The Integral Membrane FtsW Protein and Peptidoglycan Synthase PBP3 Form a Subcomplex in Escherichia coli. Microbiology 2011, 157, 251–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taguchi, A.; Welsh, M.A.; Marmont, L.S.; Lee, W.; Sjodt, M.; Kruse, A.C.; Kahne, D.; Bernhardt, T.G.; Walker, S. FtsW Is a Peptidoglycan Polymerase That Is Functional Only in Complex with Its Cognate Penicillin-Binding Protein. Nat. Microbiol. 2019, 4, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Gray, A.N.; Egan, A.J.F.; Veer, I.L.V.; Verheul, J.; Colavin, A.; Koumoutsi, A.; Biboy, J.; Altelaar, A.F.M.; Damen, M.J.; Huang, K.C.; et al. Coordination of Peptidoglycan Synthesis and Outer Membrane Constriction during Escherichia coli Cell Division. eLife 2015, 4, e07118. [Google Scholar] [CrossRef] [Green Version]
- Botta, G.A.; Park, J.T. Evidence for Involvement of Penicillin-Binding Protein 3 in Murein Synthesis during Septation but Not during Cell Elongation. J. Bacteriol. 1981, 145, 333–340. [Google Scholar] [CrossRef] [Green Version]
- Spratt, B.G. Distinct Penicillin Binding Proteins Involved in the Division, Elongation, and Shape of Escherichia coli K12. Proc. Natl. Acad. Sci. USA 1975, 72, 2999–3003. [Google Scholar] [CrossRef] [Green Version]
- Pisabarro, A.G.; Prats, R.; Váquez, D.; Rodríguez-Tébar, A. Activity of Penicillin-Binding Protein 3 from Escherichia coli. J. Bacteriol. 1986, 168, 199–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scotti, P.A.; Urbanus, M.L.; Brunner, J.; de Gier, J.W.; von Heijne, G.; van der Does, C.; Driessen, A.J.; Oudega, B.; Luirink, J. YidC, the Escherichia coli Homologue of Mitochondrial Oxa1p, Is a Component of the Sec Translocase. EMBO J. 2000, 19, 542–549. [Google Scholar] [CrossRef] [Green Version]
- Urbanus, M.L.; Scotti, P.A.; Fröderberg, L.; Sääf, A.; de Gier, J.L.; Brunner, J.; Samuelson, J.C.; Dalbey, R.E.; Oudega, B.; Luirink, J. Sec-dependent Membrane Protein Insertion: Sequential Interaction of Nascent FtsQ with SecY and YidC. EMBO Rep. 2001, 2, 524–529. [Google Scholar] [CrossRef]
- Van der Laan, M.; Houben, E.N.G.; Nouwen, N.; Luirink, J.; Driessen, A.J.M. Reconstitution of Sec-dependent Membrane Protein Insertion: Nascent FtsQ Interacts with YidC in a SecYEG-dependent Manner. EMBO Rep. 2001, 2, 519–523. [Google Scholar] [CrossRef] [Green Version]
- Vischer, N.O.E.; Verheul, J.; Postma, M.; van den Berg van Saparoea, B.; Galli, E.; Natale, P.; Gerdes, K.; Luirink, J.; Vollmer, W.; Vicente, M.; et al. Cell Age Dependent Concentration of Escherichia coli Divisome Proteins Analyzed with ImageJ and ObjectJ. Front. Microbiol. 2015, 6, 586. [Google Scholar] [CrossRef] [Green Version]
- Banzhaf, M.; Yau, H.C.L.; Verheul, J.; Lodge, A.; Kritikos, G.; Mateus, A.; Cordier, B.; Hov, A.-K.; Stein, F.; Wartel, M.; et al. Outer Membrane Lipoprotein NlpI Scaffolds Peptidoglycan Hydrolases within Multi-Enzyme Complexes in Escherichia coli. EMBO J. 2020, 39, e102246. [Google Scholar] [CrossRef]
- Den Blaauwen, T.; Buddelmeijer, N.; Aarsman, M.E.; Hameete, C.M.; Nanninga, N. Timing of FtsZ Assembly in Escherichia coli. J. Bacteriol. 1999, 181, 5167–5175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pogliano, J.; Pogliano, K.; Weiss, D.S.; Losick, R.; Beckwith, J. Inactivation of FtsI Inhibits Constriction of the FtsZ Cytokinetic Ring and Delays the Assembly of FtsZ Rings at Potential Division Sites. Proc. Natl. Acad. Sci. USA 1997, 94, 559–564. [Google Scholar] [CrossRef] [Green Version]
- Den Blaauwen, T.; Aarsman, M.E.G.; Vischer, N.O.E.; Nanninga, N. Penicillin-Binding Protein PBP2 of Escherichia coli Localizes Preferentially in the Lateral Wall and at Mid-Cell in Comparison with the Old Cell Pole. Mol. Microbiol. 2003, 47, 539–547. [Google Scholar] [CrossRef]
- Verheul, J.; Lodge, A.; Yau, H.C.L.; Liu, X.; Liu, X.; Solovyova, A.S.; Typas, A.; Banzhaf, M.; Vollmer, W.; den Blaauwen, T. Midcell Localization of PBP4 of Escherichia coli Modulates the Timing of Divisome Assembly. bioRxiv 2021. [Google Scholar] [CrossRef]
- Taschner, P.E.; Huls, P.G.; Pas, E.; Woldringh, C.L. Division Behavior and Shape Changes in Isogenic FtsZ, FtsQ, FtsA, PbpB, and FtsE Cell Division Mutants of Escherichia coli during Temperature Shift Experiments. J. Bacteriol. 1988, 170, 1533–1540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wissel, M.C.; Weiss, D.S. Genetic Analysis of the Cell Division Protein FtsI (PBP3): Amino Acid Substitutions That Impair Septal Localization of FtsI and Recruitment of FtsN. J. Bacteriol. 2004, 186, 490–502. [Google Scholar] [CrossRef] [Green Version]
- Ting, S.-Y.; Bosch, D.E.; Mangiameli, S.M.; Radey, M.C.; Huang, S.; Park, Y.-J.; Kelly, K.A.; Filip, S.K.; Goo, Y.A.; Eng, J.K.; et al. Bifunctional Immunity Proteins Protect Bacteria against FtsZ-Targeting ADP-Ribosylating Toxins. Cell 2018, 175, 1380–1392.e14. [Google Scholar] [CrossRef] [Green Version]
- Koppelman, C.-M.; Aarsman, M.E.G.; Postmus, J.; Pas, E.; Muijsers, A.O.; Scheffers, D.-J.; Nanninga, N.; den Blaauwen, T. R174 of Escherichia coli FtsZ Is Involved in Membrane Interaction and Protofilament Bundling, and Is Essential for Cell Division. Mol. Microbiol. 2004, 51, 645–657. [Google Scholar] [CrossRef] [Green Version]
- Brandon, L.D.; Goehring, N.; Janakiraman, A.; Yan, A.W.; Wu, T.; Beckwith, J.; Goldberg, M.B. IcsA, a Polarly Localized Autotransporter with an Atypical Signal Peptide, Uses the Sec Apparatus for Secretion, Although the Sec Apparatus Is Circumferentially Distributed. Mol. Microbiol. 2003, 50, 45–60. [Google Scholar] [CrossRef] [Green Version]
- Weiss, D.S. Last but Not Least: New Insights into How FtsN Triggers Constriction during Escherichia Coli Cell Division. Mol. Microbiol. 2015, 95, 903–909. [Google Scholar] [CrossRef]
- Peters, N.T.; Dinh, T.; Bernhardt, T.G. A Fail-Safe Mechanism in the Septal Ring Assembly Pathway Generated by the Sequential Recruitment of Cell Separation Amidases and Their Activators. J. Bacteriol. 2011, 193, 4973–4983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerding, M.A.; Ogata, Y.; Pecora, N.D.; Niki, H.; Boer, P.A.J.D. Thetrans-Envelope Tol-Pal Complex Is Part of the Cell Division Machinery and Required for Proper Outer-Membrane Invagination during Cell Constriction in E. Coli. Mol. Microbiol. 2007, 63, 1008–1025. [Google Scholar] [CrossRef] [Green Version]
- Vassen, V.; Valotteau, C.; Feuillie, C.; Dague, C.F.; Dufrêne, Y.F.; Bolle, X.D. Localized Incorporation of Outer Membrane Components in the Pathogen Brucella Abortus. EMBO J. 2019, 38, e100323. [Google Scholar] [CrossRef] [PubMed]
- Pazos, M.; Peters, K.; Casanova, M.; Palacios, P.; VanNieuwenhze, M.; Breukink, E.; Vicente, M.; Vollmer, W. Z-Ring Membrane Anchors Associate with Cell Wall Synthases to Initiate Bacterial Cell Division. Nat. Commun. 2018, 9, 5090. [Google Scholar] [CrossRef] [Green Version]
- Banzhaf, M.; van den Berg van Saparoea, B.; Terrak, M.; Fraipont, C.; Egan, A.; Philippe, J.; Zapun, A.; Breukink, E.; Nguyen-Distèche, M.; den Blaauwen, T.; et al. Cooperativity of Peptidoglycan Synthases Active in Bacterial Cell Elongation. Mol. Microbiol. 2012, 85, 179–194. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.; Uehara, T.; Bernhardt, T.G. Beta-Lactam Antibiotics Induce a Lethal Malfunctioning of the Bacterial Cell Wall Synthesis Machinery. Cell 2014, 159, 1300–1311. [Google Scholar] [CrossRef] [Green Version]
- Uehara, T.; Park, J.T. Growth of Escherichia coli: Significance of Peptidoglycan Degradation during Elongation and Septation. J. Bacteriol. 2008, 190, 3914–3922. [Google Scholar] [CrossRef] [Green Version]
- Verhoeven, G.S.; Dogterom, M.; den Blaauwen, T. Absence of Long-Range Diffusion of OmpA in E. Coli Is Not Caused by Its Peptidoglycan Binding Domain. BMC Microbiol. 2013, 13, 66. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, A.S.; Young, K.D. Helical Disposition of Proteins and Lipopolysaccharide in the Outer Membrane of Escherichia coli. J. Bacteriol. 2005, 187, 1913–1922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Datsenko, K.A.; Wanner, B.L. One-Step Inactivation of Chromosomal Genes in Escherichia coli K-12 Using PCR Products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taschner, P.E.; Verest, J.G.; Woldringh, C.L. Genetic and Morphological Characterization of FtsB and NrdB Mutants of Escherichia coli. J. Bacteriol. 1987, 169, 19–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossiter, A.E.; Leyton, D.L.; Tveen-Jensen, K.; Browning, D.F.; Sevastsyanovich, Y.; Knowles, T.J.; Nichols, K.B.; Cunningham, A.F.; Overduin, M.; Schembri, M.A.; et al. The Essential β-Barrel Assembly Machinery Complex Components BamD and BamA Are Required for Autotransporter Biogenesis. J. Bacteriol. 2011, 193, 4250–4253. [Google Scholar] [CrossRef] [Green Version]
- Buddelmeijer, N.; Aarsman, M.; den Blaauwen, T. Immunolabeling of Proteins in Situ in Escherichia coli K12 Strains. Bio-Protocol 2013, 3, e852. [Google Scholar] [CrossRef] [Green Version]
- Edelstein, A.; Amodaj, N.; Hoover, K.; Vale, R.; Stuurman, N. Computer Control of Microscopes Using ΜManager. Curr. Protoc. Mol. Biol. 2010, 92, 14.20.1–14.20.17. [Google Scholar] [CrossRef] [Green Version]


| Strain | Genotype | Reference |
|---|---|---|
| BW25113 | lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | [62] |
| BW25113ΔompA | lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78 ompA::kan | [62] |
| LMC500 | F- araD139 Δ(argF-lac)U169 deoC1 flbB5301 ptsF25 rbsR relA1 rpsL150 lysA1 | [63] |
| ΔbamB | BW25113 ΔbamB::cam | [64] |
| ΔbamC | BW25113 ΔbamC::cam | [64] |
| ΔbamE | BW25113 ΔbamE::cam | [64] |
| PBP3ts | LMC500 ftsI(ts) | [47] |
| FtsQts | LMC500 ftsQ(ts) | [47] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Consoli, E.; Luirink, J.; den Blaauwen, T. The Escherichia coli Outer Membrane β-Barrel Assembly Machinery (BAM) Crosstalks with the Divisome. Int. J. Mol. Sci. 2021, 22, 12101. https://doi.org/10.3390/ijms222212101
Consoli E, Luirink J, den Blaauwen T. The Escherichia coli Outer Membrane β-Barrel Assembly Machinery (BAM) Crosstalks with the Divisome. International Journal of Molecular Sciences. 2021; 22(22):12101. https://doi.org/10.3390/ijms222212101
Chicago/Turabian StyleConsoli, Elisa, Joen Luirink, and Tanneke den Blaauwen. 2021. "The Escherichia coli Outer Membrane β-Barrel Assembly Machinery (BAM) Crosstalks with the Divisome" International Journal of Molecular Sciences 22, no. 22: 12101. https://doi.org/10.3390/ijms222212101
APA StyleConsoli, E., Luirink, J., & den Blaauwen, T. (2021). The Escherichia coli Outer Membrane β-Barrel Assembly Machinery (BAM) Crosstalks with the Divisome. International Journal of Molecular Sciences, 22(22), 12101. https://doi.org/10.3390/ijms222212101








