GPI-80 Augments NF-κB Activation in Tumor Cells
Abstract
:1. Introduction
2. Results
2.1. GPI-80 Expression Is Detected in Several Urologic Cancer Cell Lines, but the Expression Level Is Lower Than That of Neutrophils
2.2. Establishment of GPI-80-Overexpressing PC3 Cells and GPI-80 Gene-Deleted PC3 Cells
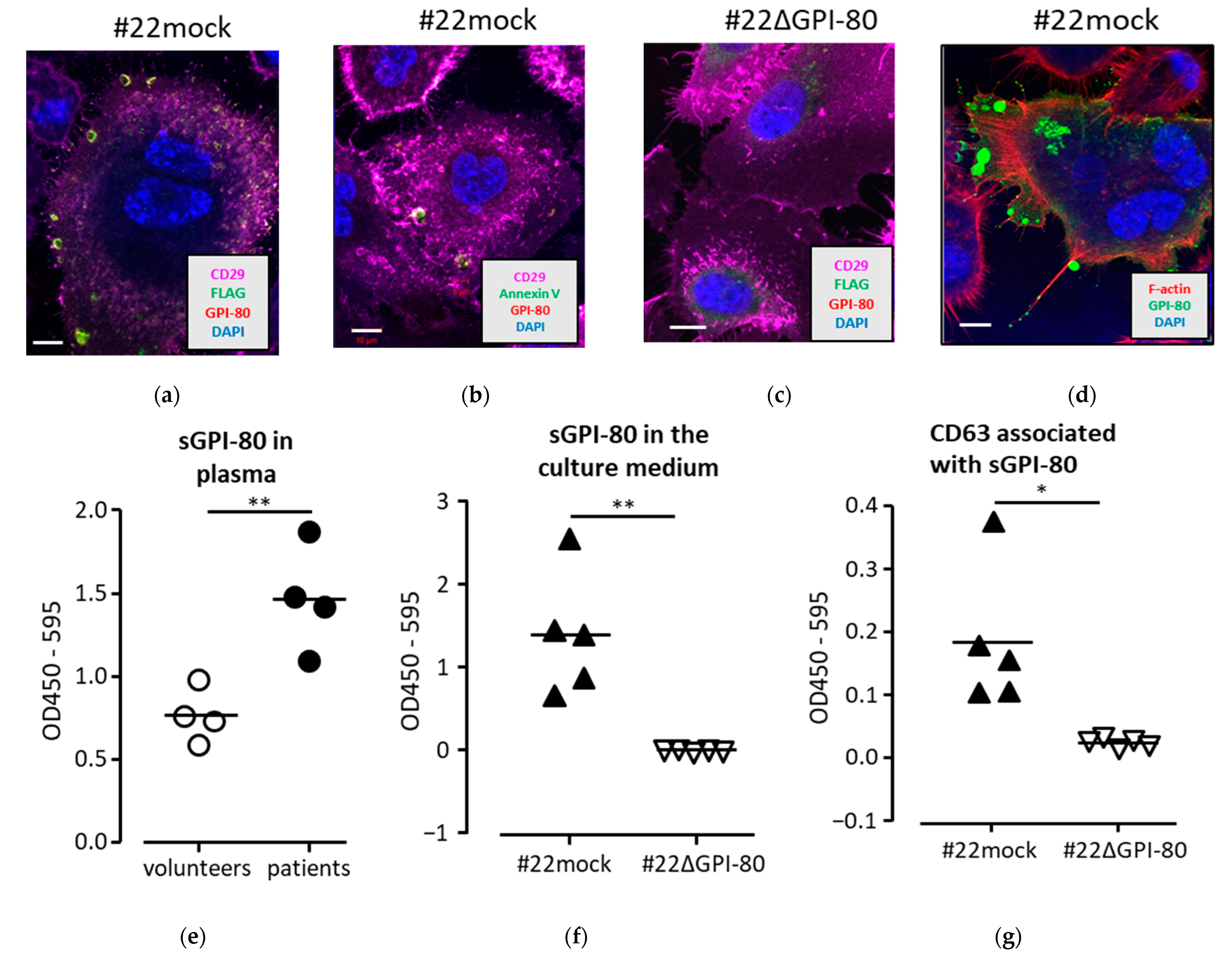
2.3. GPI-80 Localized in Vesicles and Was Detected in Conditioned Medium with Exosome Marker, CD63
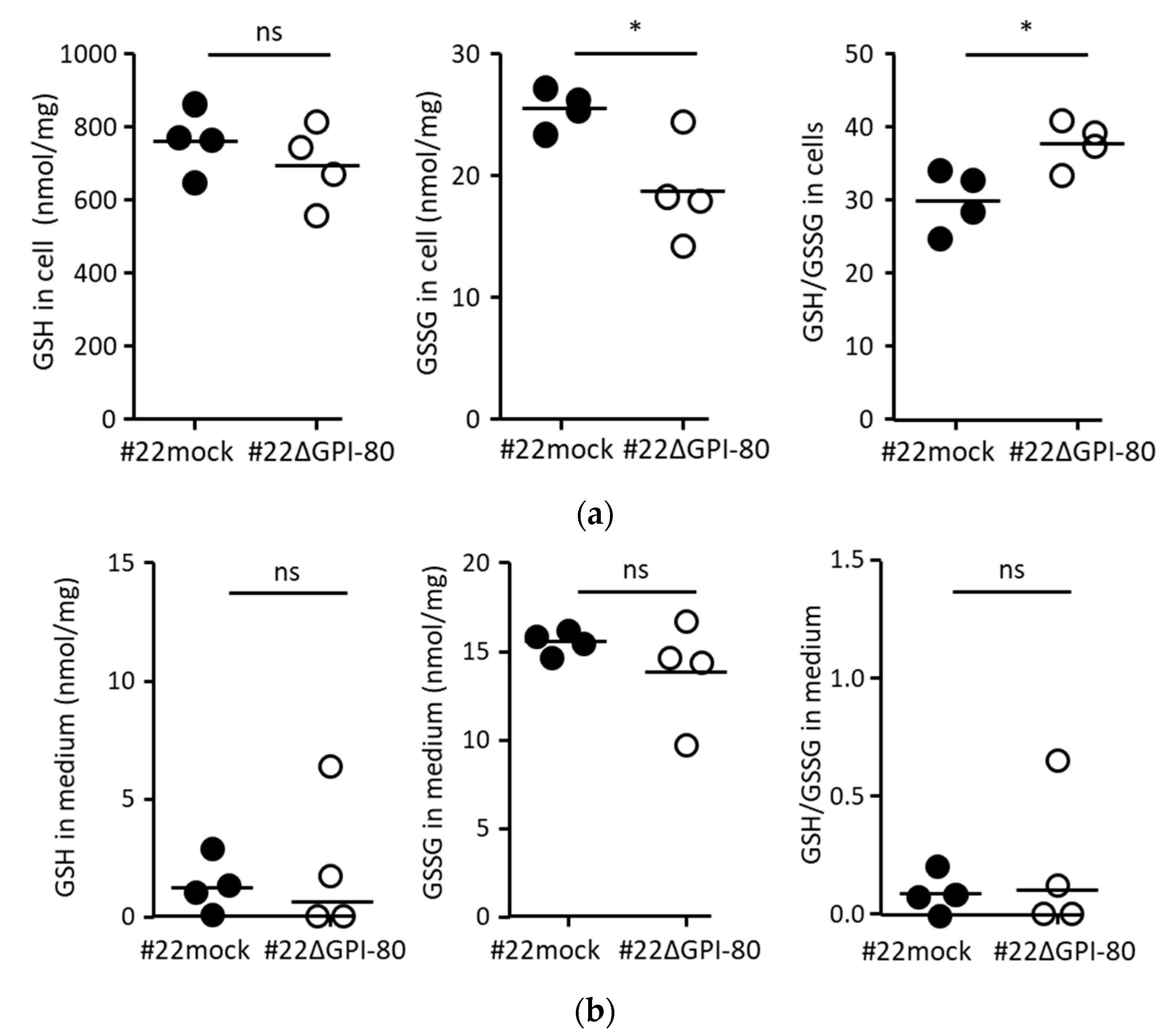
2.4. GPI-80 Tends to Undergo Intracellular Oxidization
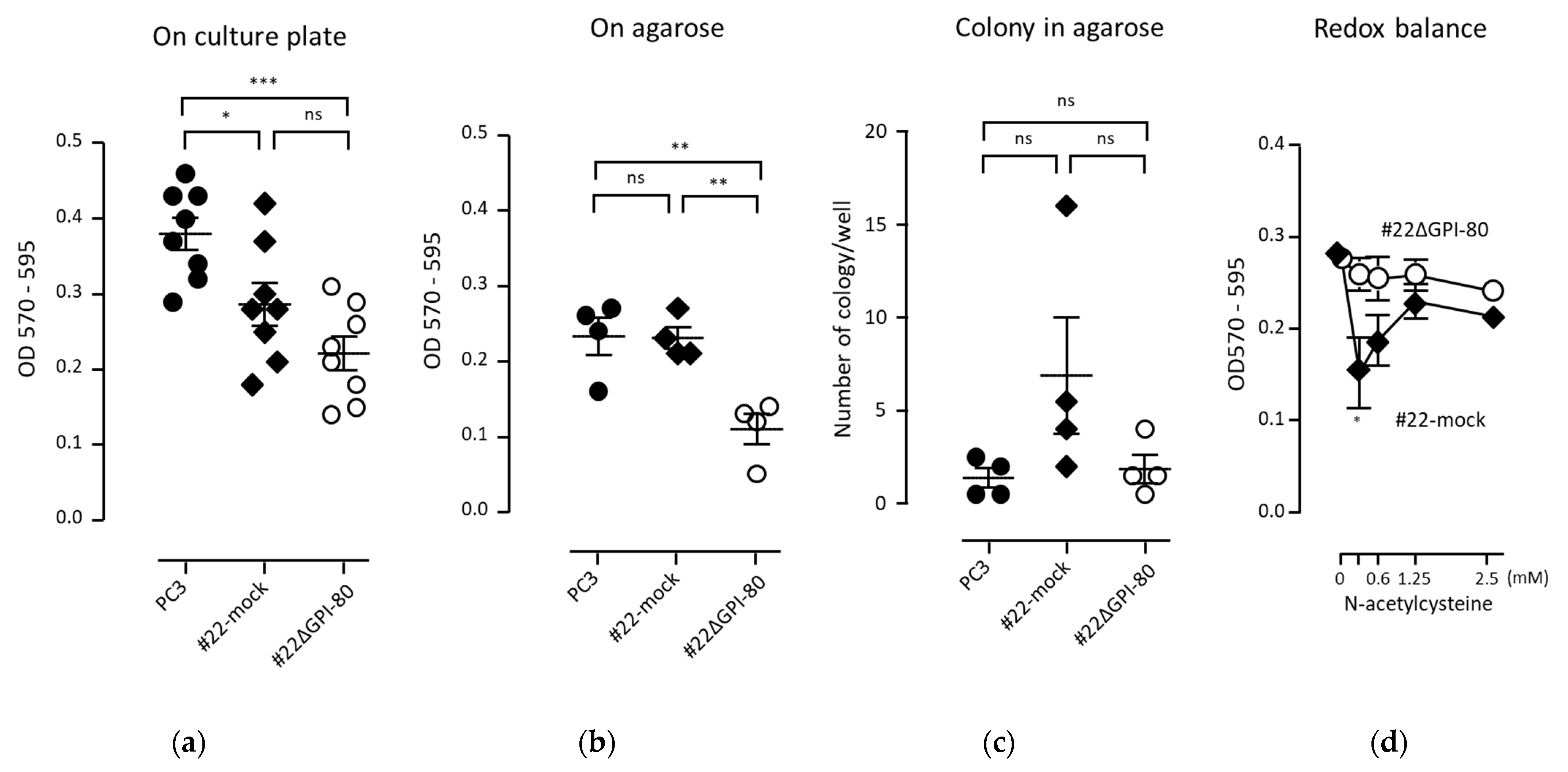
2.5. GPI-80 Expression Does Not Augment the Growth of PC3 Cells
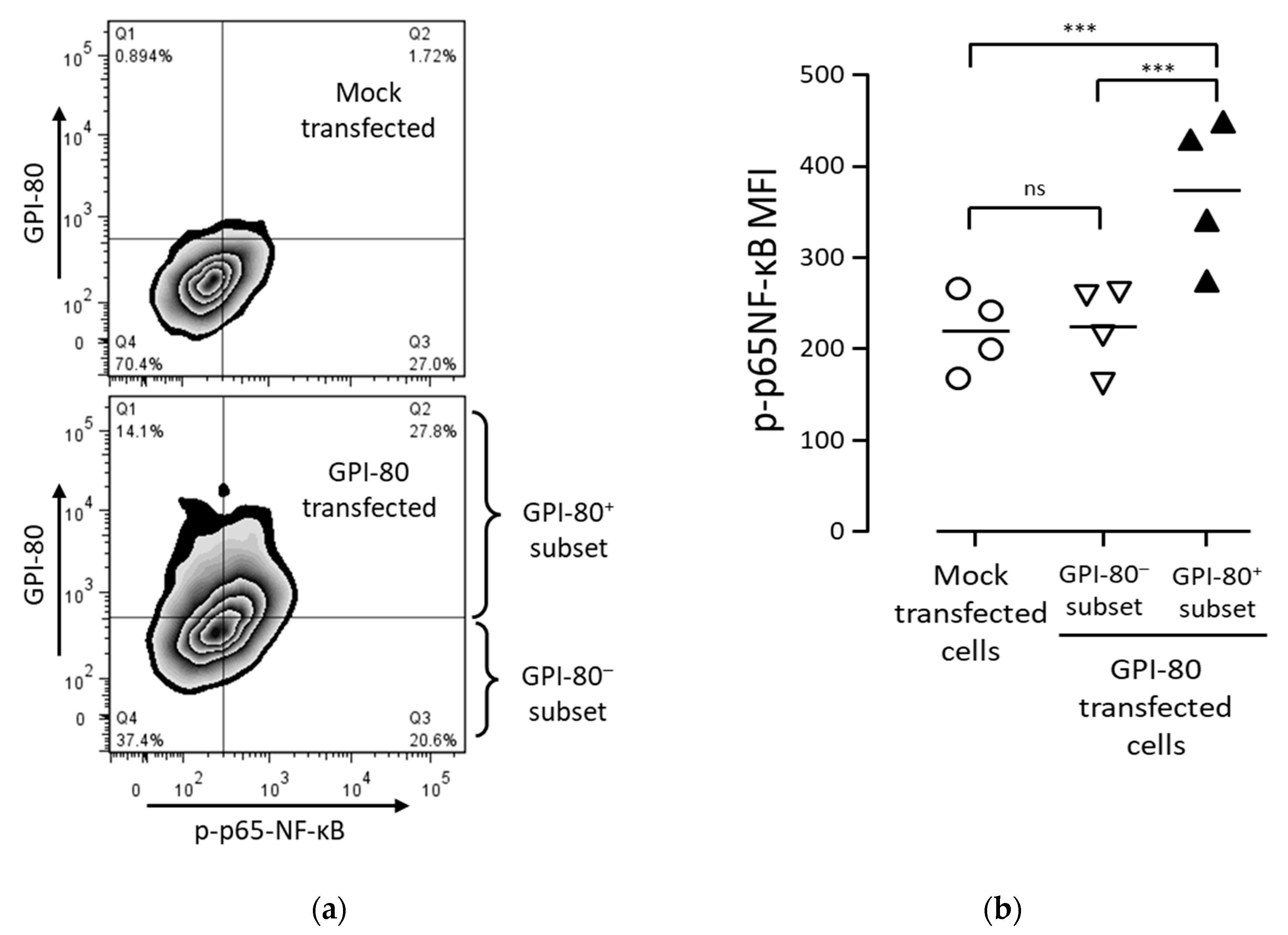
2.6. GPI-80 Augments NF-κB Activation
2.7. GPI-80 Expression Does Not Control Cell Adhesion and Migration in PC3 Cells
2.8. GPI-80 Has Weak Pantetheinase Activity in PC3 Cells
3. Discussion
4. Materials and Methods
4.1. Human Peripheral Blood and Plasma
4.2. Measurement of Reduced Glutathione (GSH) and Oxidized Glutathione (GSSG)
4.3. Measurement of Phosphorylated p65-NF-κB
4.4. Cell Growth Assay on Agarose and Colony Formation Assay in Soft Agarose
4.5. IL-1β Measurement
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martins, F.; Sofiya, L.; Sykiotis, G.P.; Lamine, F.; Maillard, M.; Fraga, M.; Shabafrouz, K.; Ribi, C.; Cairoli, A.; Guex-Crosier, Y.; et al. Adverse effects of immune-checkpoint inhibitors: Epidemiology, management and surveillance. Nat. Rev. Clin. Oncol. 2019, 16, 563–580. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bornhauser, B.; Cario, G.; Rinaldi, A.; Risch, T.; Rodriguez Martinez, V.; Schutte, M.; Warnatz, H.J.; Scheidegger, N.; Mirkowska, P.; Temperli, M.; et al. The hematopoietic stem cell marker VNN2 is associated with chemoresistance in pediatric B-cell precursor ALL. Blood Adv. 2020, 4, 4052–4064. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Watanabe, T.; Sakurai, S.; Ohtake, K.; Kinoshita, T.; Araki, A.; Fujita, T.; Takei, H.; Takeda, Y.; Sato, Y.; et al. A novel glycosylphosphatidyl inositol-anchored protein on human leukocytes: A possible role for regulation of neutrophil adherence and migration. J. Immunol. 1999, 162, 4277–4284. [Google Scholar] [PubMed]
- Nakamura-Sato, Y.; Sasaki, K.; Watanabe, H.; Araki, Y.; Sendo, F. Clustering on the forward surfaces of migrating neutrophils of a novel GPI-anchored protein that may regulate neutrophil adherence and migration. J. Leukoc. Biol. 2000, 68, 650–654. [Google Scholar] [PubMed]
- Dahlgren, C.; Karlsson, A.; Sendo, F. Neutrophil secretory vesicles are the intracellular reservoir for GPI-80, a protein with adhesion-regulating potential. J. Leukoc. Biol. 2001, 69, 57–62. [Google Scholar] [PubMed]
- Huang, J.; Takeda, Y.; Watanabe, T.; Sendo, F. A sandwich ELISA for detection of soluble GPI-80, a glycosylphosphatidyl-inositol (GPI)-anchored protein on human leukocytes involved in regulation of neutrophil adherence and migration—Its release from activated neutrophils and presence in synovial fluid of rheumatoid arthritis patients. Microbiol. Immunol. 2001, 45, 467–471. [Google Scholar]
- Chen, Y.; Huang, T.; Yang, X.; Liu, C.; Li, P.; Wang, Z.; Zhi, S. MicroRNA106a regulates the proliferation and invasion of human osteosarcoma cells by targeting VNN2. Oncol. Rep. 2018, 40, 2251–2259. [Google Scholar]
- D’Antonio, J.M.; Ma, C.; Monzon, F.A.; Pflug, B.R. Longitudinal analysis of androgen deprivation of prostate cancer cells identifies pathways to androgen independence. Prostate 2008, 68, 698–714. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, H.; Furihata, T.; Omotehara, F.; Sakai, T.; Horiuchi, H.; Shinagawa, Y.; Imura, J.; Ohkura, Y.; Tachibana, M.; Kubota, K.; et al. Identification of genes differentially expressed in a newly isolated human metastasizing esophageal cancer cell line, T.Tn-AT1, by cDNA microarray. Cancer Sci. 2003, 94, 699–706. [Google Scholar] [CrossRef] [Green Version]
- Nitto, T.; Onodera, K. Linkage between coenzyme a metabolism and inflammation: Roles of pantetheinase. J. Pharmacol. Sci. 2013, 123, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giessner, C.; Millet, V.; Mostert, K.J.; Gensollen, T.; Vu Manh, T.P.; Garibal, M.; Dieme, B.; Attaf-Bouabdallah, N.; Chasson, L.; Brouilly, N.; et al. Vnn1 pantetheinase limits the Warburg effect and sarcoma growth by rescuing mitochondrial activity. Life Sci. Alliance 2018, 1, e201800073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef] [Green Version]
- Arraud, N.; Gounou, C.; Linares, R.; Brisson, A.R. A simple flow cytometry method improves the detection of phosphatidylserine-exposing extracellular vesicles. J. Thromb. Haemost. 2015, 13, 237–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hugel, B.; Martinez, M.C.; Kunzelmann, C.; Freyssinet, J.M. Membrane microparticles: Two sides of the coin. Physiology 2005, 20, 22–27. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Sumide, K.; Kawamura, H.; Nakatsuka, R.; Fujioka, T.; Sonoda, Y. GPI-80 expression highly purifies human cord blood-derived primitive CD34-negative hematopoietic stem cells. Blood 2016, 128, 2258–2260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddig, P.J.; Juliano, R.L. Clinging to life: Cell to matrix adhesion and cell survival. Cancer Metastasis Rev. 2005, 24, 425–439. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R. Redox outside the box: Linking extracellular redox remodeling with intracellular redox metabolism. J. Biol. Chem. 2012, 287, 4397–4402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sart, S.; Song, L.; Li, Y. Controlling Redox Status for Stem Cell Survival, Expansion, and Differentiation. Oxid. Med. Cell Longev. 2015, 2015, 105135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aldini, G.; Altomare, A.; Baron, G.; Vistoli, G.; Carini, M.; Borsani, L.; Sergio, F. N-Acetylcysteine as an antioxidant and disulphide breaking agent: The reasons why. Free Radic. Res. 2018, 52, 751–762. [Google Scholar] [CrossRef]
- Spagnuolo, G.; D’Anto, V.; Cosentino, C.; Schmalz, G.; Schweikl, H.; Rengo, S. Effect of N-acetyl-L-cysteine on ROS production and cell death caused by HEMA in human primary gingival fibroblasts. Biomaterials 2006, 27, 1803–1809. [Google Scholar] [CrossRef]
- Finn, N.A.; Kemp, M.L. Pro-oxidant and antioxidant effects of N-acetylcysteine regulate doxorubicin-induced NF-kappa B activity in leukemic cells. Mol. Biosyst. 2012, 8, 650–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paszek, P.; Jackson, D.A.; White, M.R. Oscillatory control of signalling molecules. Curr. Opin. Genet. Dev. 2010, 20, 670–676. [Google Scholar] [CrossRef] [PubMed]
- Zambrano, S.; Bianchi, M.E.; Agresti, A. High-throughput analysis of NF-kappaB dynamics in single cells reveals basal nuclear localization of NF-kappaB and spontaneous activation of oscillations. PLoS ONE 2014, 9, e90104. [Google Scholar] [CrossRef] [PubMed]
- Hiscott, J.; Marois, J.; Garoufalis, J.; D’Addario, M.; Roulston, A.; Kwan, I.; Pepin, N.; Lacoste, J.; Nguyen, H.; Bensi, G.; et al. Characterization of a functional NF-kappa B site in the human interleukin 1 beta promoter: Evidence for a positive autoregulatory loop. Mol. Cell Biol. 1993, 13, 6231–6240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tak, P.P.; Firestein, G.S. NF-kappaB: A key role in inflammatory diseases. J. Clin. Invest. 2001, 107, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Greten, F.R.; Grivennikov, S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Tang, Y.; Peng, X.; Cai, X.; Wa, Q.; Ren, D.; Li, Q.; Luo, J.; Li, L.; Zou, X. Acidic extracellular pH promotes prostate cancer bone metastasis by enhancing PC-3 stem cell characteristics, cell invasiveness and VEGF-induced vasculogenesis of BM-EPCs. Oncol. Rep. 2016, 36, 2025–2032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, X.; Falzon, M. PTH-related protein modulates PC-3 prostate cancer cell adhesion and integrin subunit profile. Mol. Cell Endocrinol. 2003, 199, 165–177. [Google Scholar] [CrossRef]
- Canli, O.; Nicolas, A.M.; Gupta, J.; Finkelmeier, F.; Goncharova, O.; Pesic, M.; Neumann, T.; Horst, D.; Lower, M.; Sahin, U.; et al. Myeloid Cell-Derived Reactive Oxygen Species Induce Epithelial Mutagenesis. Cancer Cell 2017, 32, 869–883.e5. [Google Scholar] [CrossRef] [Green Version]
- Murata, Y.; Shimamura, T.; Hamuro, J. The polarization of T(h)1/T(h)2 balance is dependent on the intracellular thiol redox status of macrophages due to the distinctive cytokine production. Int. Immunol. 2002, 14, 201–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kokura, S.; Wolf, R.E.; Yoshikawa, T.; Granger, D.N.; Aw, T.Y. Molecular mechanisms of neutrophil-endothelial cell adhesion induced by redox imbalance. Circ. Res. 1999, 84, 516–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaiswing, L.; Zhong, W.; Liang, Y.; Jones, D.P.; Oberley, T.D. Regulation of prostate cancer cell invasion by modulation of extra- and intracellular redox balance. Free Radic. Biol. Med. 2012, 52, 452–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giannoni, E.; Fiaschi, T.; Ramponi, G.; Chiarugi, P. Redox regulation of anoikis resistance of metastatic prostate cancer cells: Key role for Src and EGFR-mediated pro-survival signals. Oncogene 2009, 28, 2074–2086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, F.; Haass, N.K. Microenvironment-Driven Dynamic Heterogeneity and Phenotypic Plasticity as a Mechanism of Melanoma Therapy Resistance. Front. Oncol. 2018, 8, 173. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Banerjee, R. Redox remodeling as an immunoregulatory strategy. Biochemistry 2010, 49, 1059–1066. [Google Scholar] [CrossRef] [Green Version]
- Takeda, Y.; Nara, H.; Araki, A.; Asao, H. Human peripheral neutrophils express functional IL-21 receptors. Inflammation 2014, 37, 1521–1532. [Google Scholar] [CrossRef] [PubMed]
- Horibata, S.; Vo, T.V.; Subramanian, V.; Thompson, P.R.; Coonrod, S.A. Utilization of the Soft Agar Colony Formation Assay to Identify Inhibitors of Tumorigenicity in Breast Cancer Cells. J. Vis. Exp. 2015, 99, e52727. [Google Scholar] [CrossRef] [Green Version]
- Nara, H.; Onoda, T.; Rahman, M.; Araki, A.; Juliana, F.M.; Tanaka, N.; Asao, H. WSB-1, a novel IL-21 receptor binding molecule, enhances the maturation of IL-21 receptor. Cell Immunol. 2011, 269, 54–59. [Google Scholar] [CrossRef]
- Takeda, Y.; Kato, T.; Ito, H.; Kurota, Y.; Yamagishi, A.; Sakurai, T.; Araki, A.; Nara, H.; Tsuchiya, N.; Asao, H. The pattern of GPI-80 expression is a useful marker for unusual myeloid maturation in peripheral blood. Clin. Exp. Immunol. 2016, 186, 373–386. [Google Scholar] [CrossRef] [Green Version]
- Nitto, T.; Takeda, Y.; Yoshitake, H.; Sendo, F.; Araki, Y. Structural divergence of GPI-80 in activated human neutrophils. Biochem. Biophys. Res. Commun. 2007, 359, 227–233. [Google Scholar] [CrossRef]
- Horton, R.M.; Cai, Z.L.; Ho, S.N.; Pease, L.R. Gene splicing by overlap extension: Tailor-made genes using the polymerase chain reaction. Biotechniques 1990, 8, 528–535. [Google Scholar] [CrossRef] [Green Version]
- Ho, S.N.; Hunt, H.D.; Horton, R.M.; Pullen, J.K.; Pease, L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 1989, 77, 51–59. [Google Scholar] [CrossRef]
- Ruan, B.H.; Cole, D.C.; Wu, P.; Quazi, A.; Page, K.; Wright, J.F.; Huang, N.; Stock, J.R.; Nocka, K.; Aulabaugh, A.; et al. A fluorescent assay suitable for inhibitor screening and vanin tissue quantification. Anal. Biochem. 2010, 399, 284–292. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takeda, Y.; Kurota, Y.; Kato, T.; Ito, H.; Araki, A.; Nara, H.; Saitoh, S.; Tanaka, N.; Tsuchiya, N.; Asao, H. GPI-80 Augments NF-κB Activation in Tumor Cells. Int. J. Mol. Sci. 2021, 22, 12027. https://doi.org/10.3390/ijms222112027
Takeda Y, Kurota Y, Kato T, Ito H, Araki A, Nara H, Saitoh S, Tanaka N, Tsuchiya N, Asao H. GPI-80 Augments NF-κB Activation in Tumor Cells. International Journal of Molecular Sciences. 2021; 22(21):12027. https://doi.org/10.3390/ijms222112027
Chicago/Turabian StyleTakeda, Yuji, Yuta Kurota, Tomoyuki Kato, Hiromi Ito, Akemi Araki, Hidetoshi Nara, Shinichi Saitoh, Nobuyuki Tanaka, Norihiko Tsuchiya, and Hironobu Asao. 2021. "GPI-80 Augments NF-κB Activation in Tumor Cells" International Journal of Molecular Sciences 22, no. 21: 12027. https://doi.org/10.3390/ijms222112027
APA StyleTakeda, Y., Kurota, Y., Kato, T., Ito, H., Araki, A., Nara, H., Saitoh, S., Tanaka, N., Tsuchiya, N., & Asao, H. (2021). GPI-80 Augments NF-κB Activation in Tumor Cells. International Journal of Molecular Sciences, 22(21), 12027. https://doi.org/10.3390/ijms222112027





