Adaptive Responses of Citrus grandis Leaves to Copper Toxicity Revealed by RNA-Seq and Physiology
Abstract
:1. Introduction
2. Results
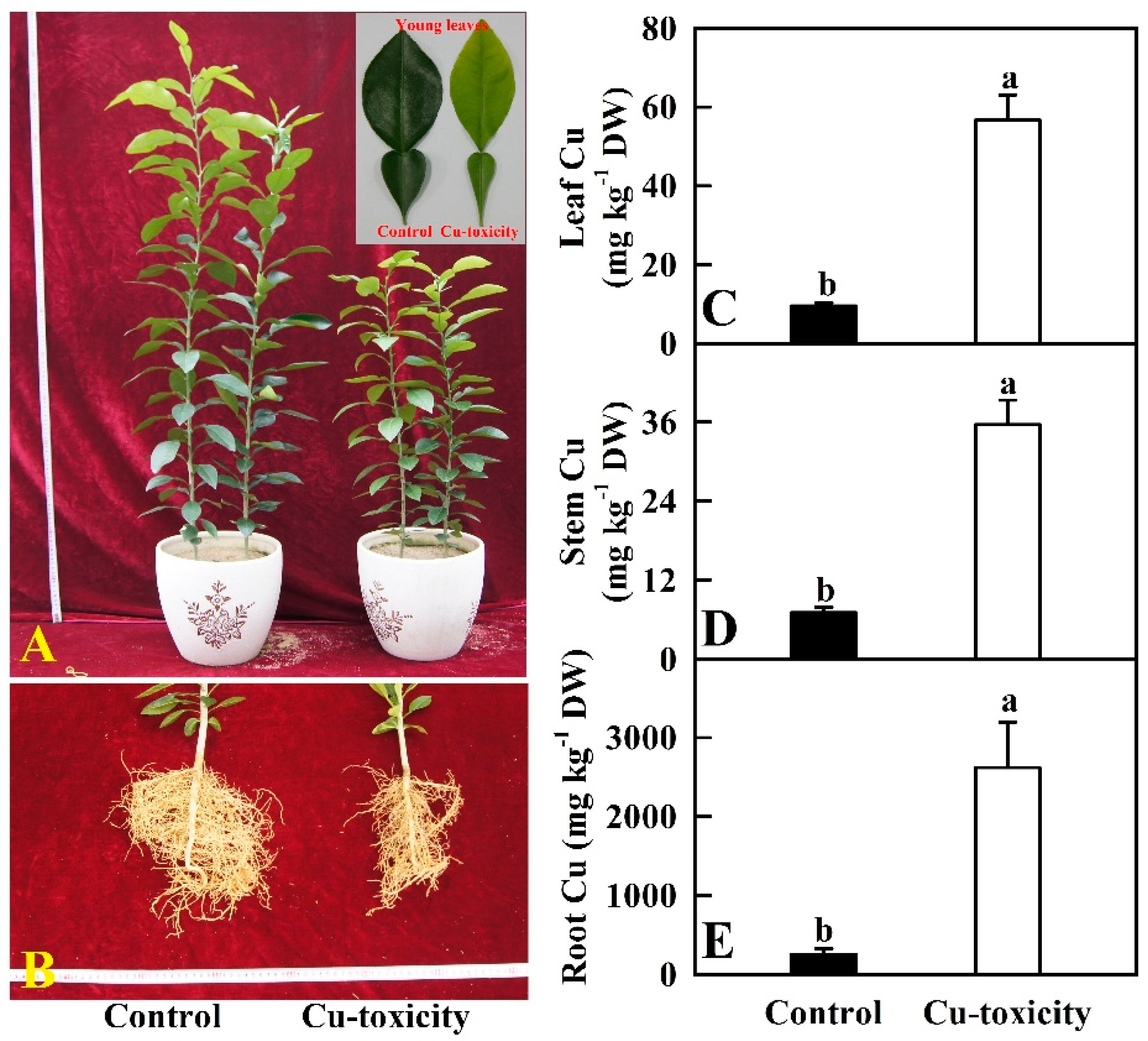
2.1. Seedling Growth and Cu Level in Roots, Stems and Leaves
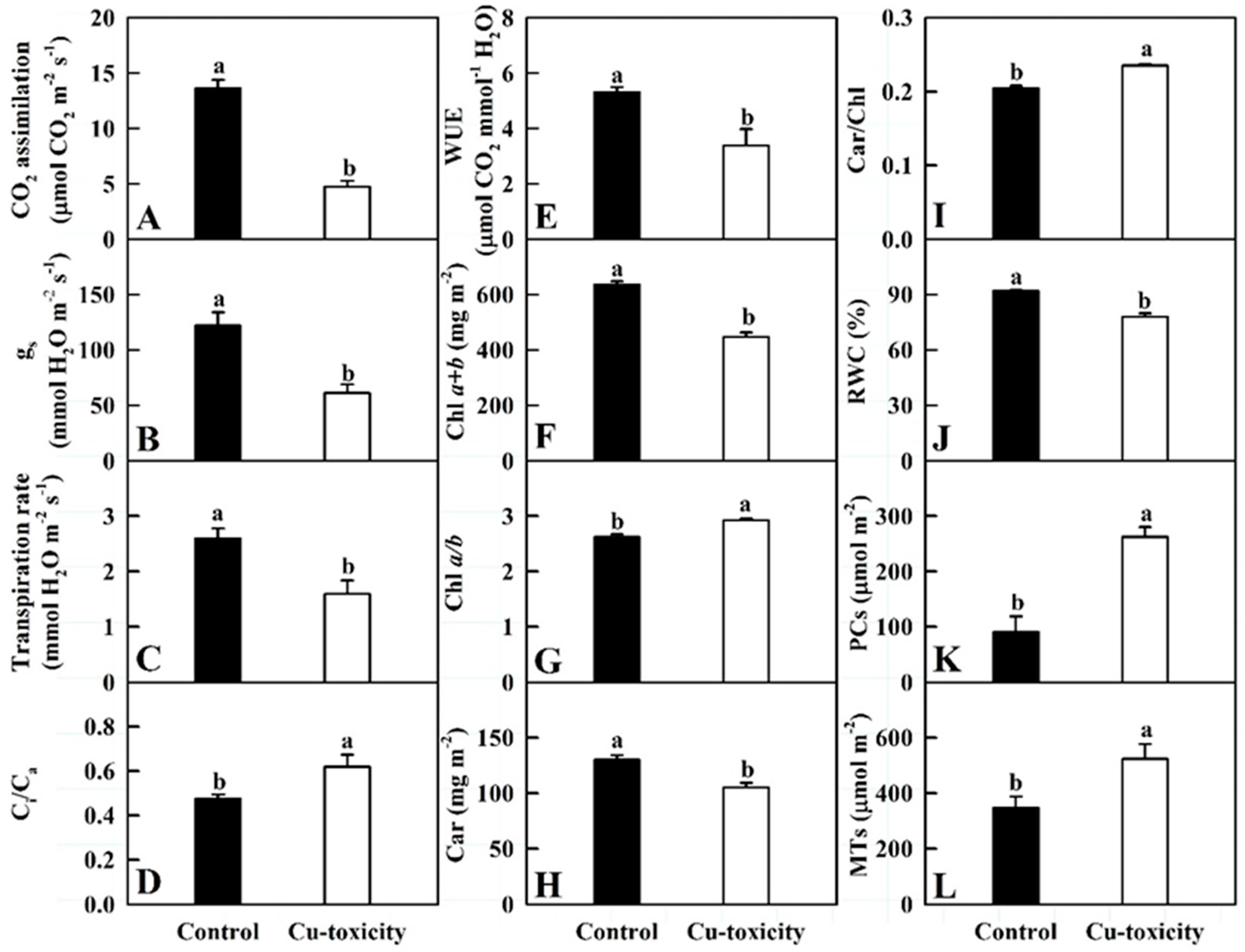
2.2. Gas Exchange, Pigments, RWC, PCs and MTs in Leaves
2.3. RNA-Seq and De Novo Assembly
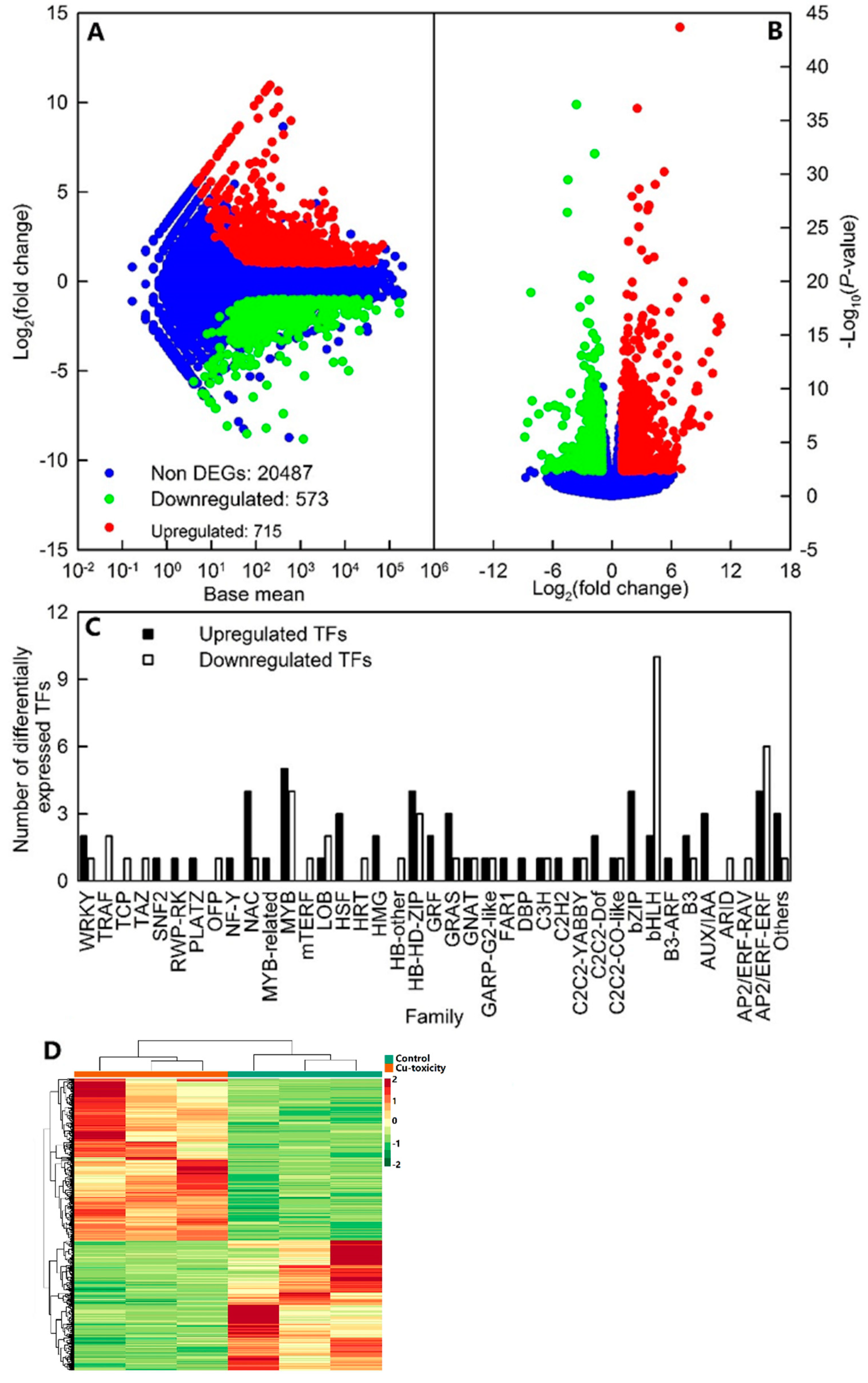
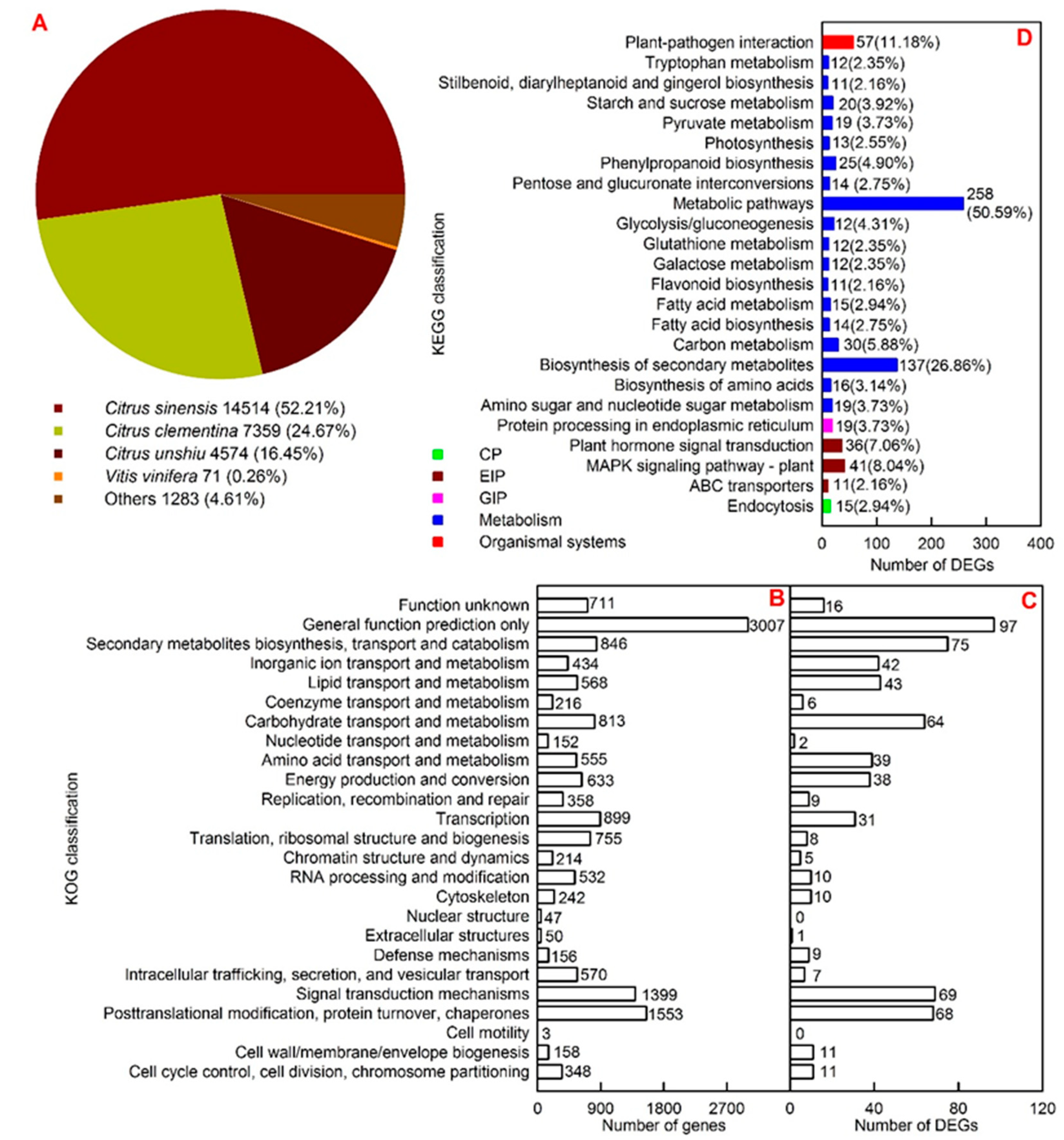
2.4. Functional Annotation and Cu-Toxicity-Responsive Genes
2.5. qRT-PCR Analysis
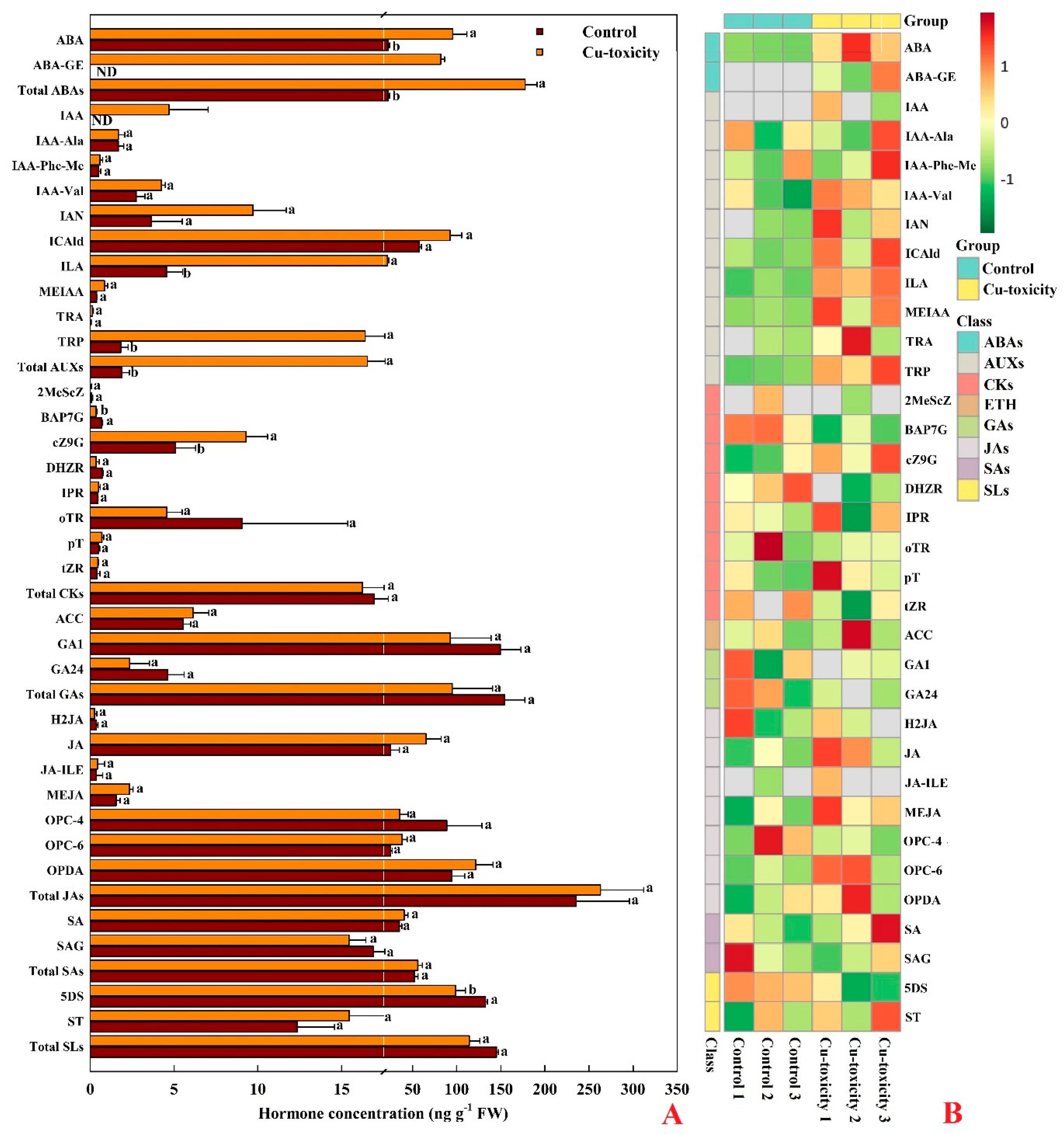
2.6. Hormones in Leaves
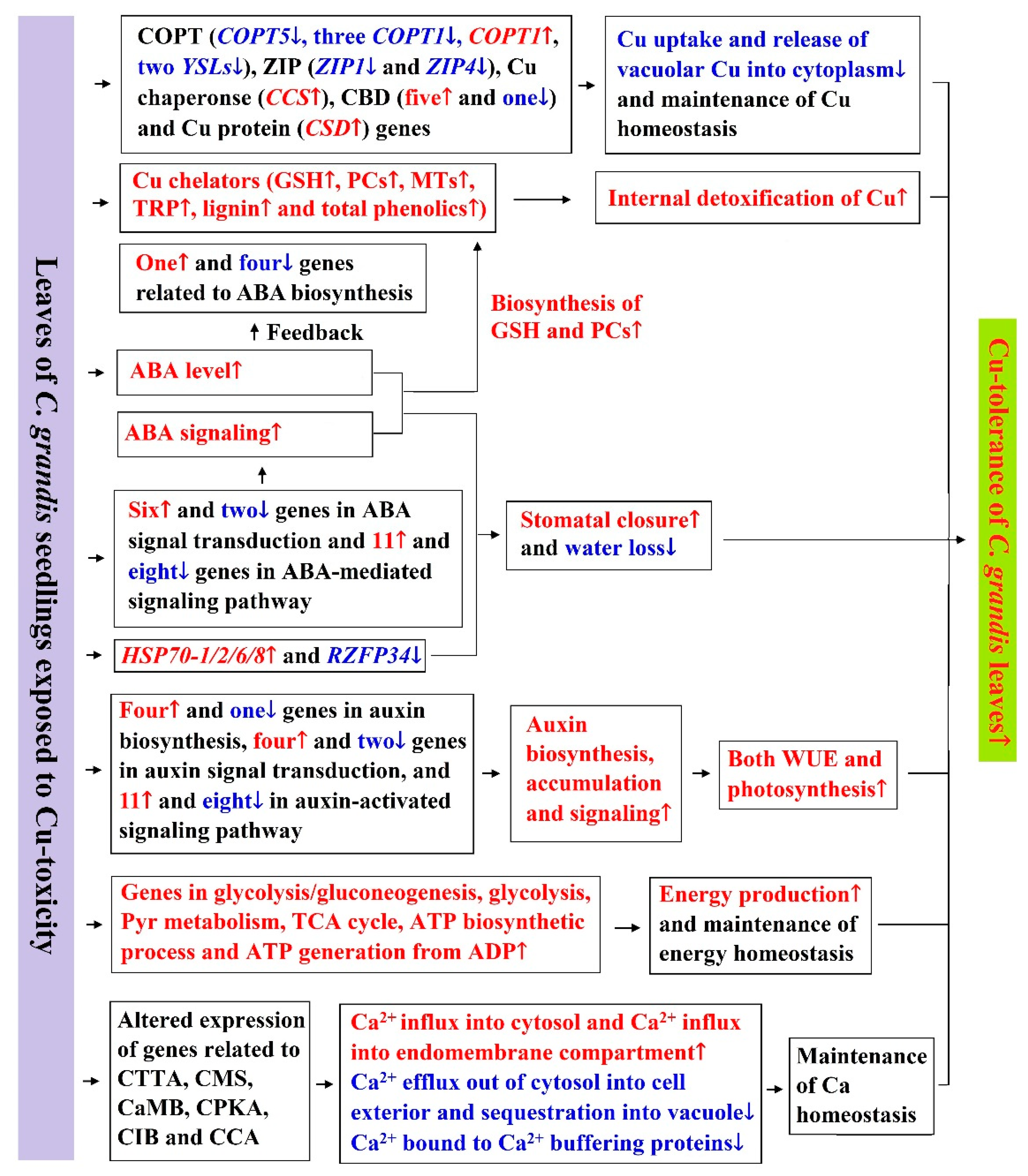
3. Discussion
3.1. Increased Immobilization of Cu in Roots, and Cu Homeostasis and Detoxification in Leaves
3.2. Cu-Toxic Effects on Cell Wall Metabolism in Leaves
3.3. Cu-Toxic Effects on Pigment Metabolism, Photosynthesis, and Carbon, Carbohydrate and Energy Metabolisms in Leaves
3.4. Cu-Toxic Effects on Thermal Dissipation, ROS Scavenging and Cell Redox Homeostasis in Leaves
3.5. Cu-Toxic Effects on Calcium Signaling and MAPK Signaling in Leaves
3.6. Cu-Toxic Effects on Biosynthesis and Signaling of Phytohormones in Leaves
4. Materials and Methods
4.1. Plant Culture and Cu Treatments
4.2. Cu Concentration in Leaves, Stems and Roots
4.3. Gas Exchange, Pigments, RWC, PCs and MTs in Leaves
4.4. Leaf RNA Extraction and RNA-Seq
4.5. RNA-Seq Analysis
4.6. qRT-PCR Validation
4.7. Extraction and Measurements of Hormones in Leaves
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Adrees, M.; Ali, S.; Rizwan, M.; Ibrahim, M.; Abbas, F.; Farid, M.; Ziaurrehman, M.; Irshad, M.K.; Bharwana, S.A. The effect of excess copper on growth and physiology of important food crops: A review. Environ. Sci. Pollut. Res. 2015, 22, 8148–8162. [Google Scholar] [CrossRef]
- Li, Q.; Chen, H.-H.; Qi, Y.-P.; Ye, X.; Yang, L.-T.; Huang, Z.-R.; Chen, L.-S. Excess copper effects on growth, uptake of water and nutrients, carbohydrates, and PSII photochemistry revealed by OJIP transients in Citrus seedlings. Environ. Sci. Pollut. Res. 2019, 26, 30188–30205. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Zhang, S.-Q.; Lin, W.-J.; Chen, H.-H.; Lin, F.; Zhu, D.-H.; Chen, L.-S.; Li, Y.; Guo, J.-X. Soil copper (Cu) nutrient status and its influencing factors in pomelo orchards in Pinghe county, Fujian Province. J. Fruit Sci. 2018, 35, 301–310. [Google Scholar]
- Li, Y.; Han, M.-Q.; Lin, F.; Ten, Y.; Lin, J.; Zhu, D.-H.; Guo, P.; Weng, Y.-B.; Chen, L.-S. Soil chemical properties, ‘Guanximiyou’ pummelo leaf mineral nutrient status and fruit quality in the southern region of Fujian province, China. J. Soil Sci. Plant Nutr. 2015, 15, 615–628. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.Y.; Liu, B.; Chen, G.F.; Wang, Y. Content of copper in soil, Citrus leaves, and branch roots of Citrus orchards in Guangxi. Southwest China J. Agric. Sci. 2007, 20, 1060–1063. [Google Scholar]
- Alva, A.K.; Huang, B.; Prakash, O.; Paramasivam, S. Effects of copper rates and soil pH on growth and nutrient uptake by Citrus seedlings. J. Plant Nutr. 1999, 22, 1687–1699. [Google Scholar] [CrossRef]
- Kumar, V.; Pandita, S.; Sidhu, G.P.S.; Sharma, A.; Khanna, K.; Kaur, P.; Bali, A.S.; Setia, R. Copper bioavailability, uptake, toxicity and tolerance in plants: A comprehensive review. Chemosphere 2021, 262, 127810. [Google Scholar] [CrossRef]
- Wan, H.; Du, J.; He, J.; Lyu, D.; Li, H. Copper accumulation, subcellular partitioning and physiological and molecular responses in relation to different copper tolerance in apple rootstocks. Tree Physiol. 2019, 39, 1215–1234. [Google Scholar] [CrossRef]
- Shabbir, Z.; Sardar, A.; Shabbir, A.; Abbas, G.; Shamshad, S.; Khalid, S.; Natasha; Murtaza, G.; Dumat, C.; Shahid, M. Copper uptake, essentiality, toxicity, detoxification and risk assessment in soil-plant environment. Chemosphere 2020, 259, 127436. [Google Scholar] [CrossRef]
- Cai, L.-Y.; Zhang, J.; Ren, Q.-Q.; Lai, Y.-H.; Peng, M.-Y.; Deng, C.-L.; Ye, X.; Yang, L.-T.; Yang, Z.-R.; Chen, L.-S. Increased pH-mediated alleviation of copper-toxicity and growth response function in Citrus sinensis seedlings. Sci. Hortic. 2021, 288, 110310. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, F.; Xia, Y.; Wang, G.; Shen, Z. Excess copper induces production of hydrogen peroxide in the leaf of Elsholtzia haichowensis through apoplastic and symplastic CuZn-superoxide dismutase. J. Hazard. Mater. 2018, 178, 834–843. [Google Scholar] [CrossRef] [PubMed]
- Hippler, F.W.R.; Boaretto, R.M.; Dovis, V.L.; Quaggio, J.A.; Azevedo, R.A.; Mattos, D., Jr. Oxidative stress induced by Cu nutritional disorders in Citrus depends on nitrogen and calcium availability. Sci. Rep. 2018, 8, 1641. [Google Scholar] [CrossRef]
- Kováčik, J.; Klejdus, B.; Hedbavny, J.; Štork, F.; Bačkor, M. Comparison of cadmium and copper effect on phenolic metabolism, mineral nutrients and stress-related parameters in Matricaria chamomilla plants. Plant Soil 2009, 320, 231–242. [Google Scholar] [CrossRef]
- Lequeux, H.; Hermans, C.; Lutts, S.; Verbruggen, N. Response to copper excess in Arabidopsis thaliana: Impact on the root system architecture, hormone distribution, lignin accumulation and mineral profile. Plant Physiol. Biochem. 2010, 48, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.L.; He, X.J.; Chen, M.; An, R.D.; An, X.L.; Li, J. Responses of nitrogen metabolism to copper stress in Luffa cylindrica roots. J. Soil Sci. Plant Nutr. 2014, 14, 616–624. [Google Scholar] [CrossRef]
- Xia, Y.; Yin, S.; Zhang, K.; Shi, X.; Shen, Z. Oswak11, a rice wall-associated kinase, regulates Cu detoxification by alteration the immobilization of cu in cell walls. Environ. Exp. Bot. 2018, 150, 99–105. [Google Scholar] [CrossRef]
- Shi, K.; Liu, X.; Zhu, Y.; Bai, Y.; Shan, D.; Zheng, X.; Wang, L.; Zhang, H.; Wang, C.; Yan, T.; et al. MdWRKY11 improves copper tolerance by directly promoting the expression of the copper transporter gene MdHMA5. Hortic. Res. 2020, 7, 105. [Google Scholar] [CrossRef]
- Wan, H.; Yang, F.; Zhuang, X.; Cao, Y.; He, J.; Li, H.; Qin, S.; Lyu, D. Malus rootstocks affect copper accumulation and tolerance in trees by regulating copper mobility, physiological responses, and gene expression patterns. Environ. Pollut. 2021, 287, 117610. [Google Scholar] [CrossRef]
- Wang, Q.Y.; Liu, J.S.; Hu, B. Integration of copper subcellular distribution and chemical forms to understand copper toxicity in apple trees. Environ. Exp. Bot. 2016, 123, 125–131. [Google Scholar] [CrossRef]
- Huang, W.-L.; Wu, F.-L.; Huang, H.-Y.; Huang, W.-T.; Deng, C.-L.; Yang, L.-T.; Huang, Z.-R.; Chen, L.-S. Excess copper-induced alterations of protein profiles and related physiological parameters in Citrus leaves. Plants 2020, 9, 291. [Google Scholar] [CrossRef] [Green Version]
- Printz, B.; Lutts, S.; Hausman, J.F.; Sergeant, K. Copper trafficking in plants and its implication on cell wall dynamics. Front. Plant Sci. 2016, 7, 601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouazizi, H.; Jouili, H.; Geitmann, A.; El Ferjani, E. Cell wall accumulation of Cu ions and modulation of lignifying enzymes in primary leaves of bean seedlings exposed to excess copper. Biol. Trace Elem. Res. 2011, 139, 97–107. [Google Scholar] [CrossRef]
- Vinit-Dunand, F.; Epron, D.; Alaoui-Sossé, B.; Badot, P.M. Effects of copper on growth and on photosynthesis of mature and expanding leaves in cucumber plants. Plant Sci. 2002, 163, 53–58. [Google Scholar] [CrossRef]
- Jalmi, S.K.; Bhagat, P.K.; Verma, D.; Noryang, S.; Tayyeba, S.; Singh, K.; Sharma, D.; Sinha, A.K. Traversing the links between heavy metal stress and plant signaling. Front. Plant Sci. 2018, 9, 12. [Google Scholar] [CrossRef]
- Thao, N.P.; Khan, M.I.; Thu, N.B.; Hoang, X.L.; Asgher, M.; Khan, N.A.; Tran, L.S. Role of ethylene and its cross talk with other signaling molecules in plant responses to heavy metal stress. Plant Physiol. 2015, 169, 73–84. [Google Scholar] [CrossRef] [Green Version]
- Zehra, A.; Choudhary, S.; Wani, K.I.; Naeem, M.; Khan, M.M.A.; Aftab, T. Exogenous abscisic acid mediates ROS homeostasis and maintains glandular trichome to enhance artemisinin biosynthesis in Artemisia annua under copper toxicity. Plant Physiol. Biochem. 2020, 156, 125–134. [Google Scholar] [CrossRef]
- Fariduddin, Q.; Yusuf, M.; Hayat, S.; Ahmad, A. Effect of 28-homobrassinolide on antioxidant capacity and photosynthesis in Brassica juncea plants exposed to different levels of copper. Environ. Exp. Bot. 2009, 66, 418–424. [Google Scholar] [CrossRef]
- Hu, Z.; Fu, Q.; Zheng, J.; Zhang, A.; Wang, H. Transcriptomic and metabolomic analyses reveal that melatonin promotes melon root development under copper stress by inhibiting jasmonic acid biosynthesis. Horti. Res. 2020, 7, 1–15. [Google Scholar] [CrossRef]
- Landa, P.; Dytrych, P.; Prerostova, S.; Petrova, S.; Vankova, R.; Vanek, T. Transcriptomic response of Arabidopsis thaliana exposed to CuO nanoparticles, bulk material, and ionic copper. Environ. Sci. Technol. 2017, 51, 10814–10824. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Trinh, N.N.; Fu, S.F.; Hsiung, Y.C.; Chia, L.C.; Lin, C.W.; Huang, H.J. Comparison of early transcriptome responses to copper and cadmium in rice roots. Plant Mol. Biol. 2013, 81, 507–522. [Google Scholar] [CrossRef]
- Zhang, Z.; Ke, M.; Qu, Q.; Peijnenburg, W.J.G.M.; Lu, T.; Zhang, Q.; Ye, Y.; Xu, P.; Du, B.; Sun, L.; et al. Impact of copper nanoparticles and ionic copper exposure on wheat (Triticum aestivum L.) root morphology and antioxidant response. Environ. Pollut. 2018, 239, 689–697. [Google Scholar] [CrossRef]
- Cao, Y.Y.; Qi, C.D.; Li, S.; Wang, Z.; Wang, X.; Wang, J.; Ren, S.; Li, X.; Zhang, N.; Guo, Y.D. Melatonin alleviates copper toxicity via improving copper sequestration and ROS scavenging in cucumber. Plant Cell Physiol. 2019, 60, 562–574. [Google Scholar] [CrossRef]
- Fu, X.Z.; Zhang, X.Y.; Qiu, J.Y.; Zhou, X.; Yuan, M.; He, Y.Z.; Chun, C.P.; Cao, L.; Ling, L.L.; Peng, L.Z. Whole-transcriptome RNA sequencing reveals the global molecular responses and ceRNA regulatory network of mRNAs, lncRNAs, miRNAs and circRNAs in response to copper toxicity in Ziyang Xiangcheng (Citrus junos Sieb. Ex Tanaka). BMC Plant Biol. 2019, 19, 509. [Google Scholar] [CrossRef] [Green Version]
- Leng, X.; Jia, H.; Sun, X.; Shangguan, L.; Wu, Q.; Wang, B.; Fang, J. Comparative transcriptome analysis of grapevine in response to copper stress. Sci. Rep. 2015, 5, 17749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Fang, X.; Wang, Z.; Shangguan, L.; Liu, T.; Chen, C.; Liu, Z.; Ge, M.; Zhang, C.; Zheng, T.; et al. Multi-omics analyses on the response mechanisms of ‘Shine Muscat’ grapevine to low degree of excess copper stress (Low-ECS). Environ. Pollut. 2021, 286, 117278. [Google Scholar] [CrossRef]
- Sudo, E.; Itouga, M.; Yoshida-Hatanaka, K.; Ono, Y.; Sakakibara, H. Gene expression and sensitivity in response to copper stress in rice leaves. J. Exp. Bot. 2008, 59, 3465–3474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, P.; Qi, Y.-P.; Huang, W.-L.; Yang, L.-T.; Huang, Z.-R.; Lai, N.-W.; Chen, L.-S. Aluminum-responsive genes revealed by RNA-Seq and related physiological responses in leaves of two Citrus species with contrasting aluminum-tolerance. Ecotoxicol. Environ. Saf. 2018, 158, 213–222. [Google Scholar] [CrossRef]
- Guo, P.; Qi, Y.-P.; Yang, L.-T.; Lai, N.-W.; Ye, X.; Yang, Y.; Chen, L.-S. Root adaptive responses to aluminum-treatment revealed by RNA-Seq in two Citrus species with different aluminum-tolerance. Front. Plant Sci. 2017, 8, 330. [Google Scholar] [CrossRef] [Green Version]
- Guo, F.; Yu, H.; Xu, Q.; Deng, X. Transcriptomic analysis of differentially expressed genes in an orange-pericarp mutant and wild type in pummelo (Citrus grandis). BMC Plant Biol. 2015, 15, 44. [Google Scholar] [CrossRef] [Green Version]
- Burkhead, J.L.; Reynolds, K.A.G.; Abdel-Ghany, S.E.; Cohu, C.M.; Pilon, M. Copper homeostasis. New Phytol. 2009, 182, 799–816. [Google Scholar] [CrossRef] [PubMed]
- Sancenón, V.; Puig, S.; Mira, H.; Thiele, D.J.; Peñarrubia, L. Identification of a copper transporter family in Arabidopsis thaliana. Plant Mol. Biol. 2003, 51, 577–587. [Google Scholar] [CrossRef] [Green Version]
- Klaumann, S.; Nickolaus, S.D.; Fürst, S.H.; Starck, S.; Schneider, S.; Neuhaus, E.H.; Trentmann, O. The tonoplast copper transporter COPT5 acts as an exporter and is required for interorgan allocation of copper in Arabidopsis thaliana. New Phytol. 2011, 192, 393–404. [Google Scholar] [CrossRef]
- Robinson, N.J.; Procter, C.M.; Connolly, E.L.; Guerinot, M.L. A ferric-chelate reductase for iron uptake from soils. Nature 1999, 397, 694–697. [Google Scholar] [CrossRef]
- Welch, R.M.; Norvell, W.A.; Schaefer, S.C.; Shaff, J.E.; Kochian, L.V. Induction of iron(III) and copper(II) reduction in pea roots by Fe and Cu status: Does the root-cell plasmalemma Fe(III)-chelate reductase perform a general role in regulation of cation uptake. Planta 1993, 190, 555–561. [Google Scholar] [CrossRef]
- Jeong, J.; Cohu, C.; Kerkeb, L.; Pilon, M.; Connolly, E.L.; Guerinot, M.L. Chloroplast Fe(III) chelate reductase activity is essential for seedling viability under iron limiting conditions. Proc. Natl. Acad. Sci. USA 2008, 105, 10619–10624. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Yamaji, N.; Yokosho, K.; Ma, J.F. YSL16 is a phloem-localized transporter of the copper-nicotianamine complex that is responsible for copper distribution in rice. Plant Cell 2012, 24, 3767–3782. [Google Scholar] [CrossRef] [Green Version]
- Blaby-Haas, C.E.; Padilla-Benavides, T.; Stübe, R.; Argüello, J.M.; Merchant, S.S. Evolution of a plant-specific copper chaperone family for chloroplast copper homeostasis. Proc. Natl. Acad. Sci. USA 2014, 111, E5480–E5487. [Google Scholar] [CrossRef] [Green Version]
- Del Pozo, T.; Cambiazo, V.; González, M. Gene expression profiling analysis of copper homeostasis in Arabidopsis thaliana. Biochem. Bioph. Res. Commun. 2010, 393, 248–252. [Google Scholar] [CrossRef]
- Schiavon, M.; Zhang, L.; Abdel-Ghany, S.E.; Pilon, M.; Malagoli, M.; Pilon-Smits, E.A.H. Variation in copper tolerance in Arabidopsis thaliana accessions Columbia, Landsberg erecta and Wassilewskija. Physiol. Plant. 2007, 129, 342–350. [Google Scholar] [CrossRef]
- Huang, H.-Y.; Ren, Q.-Q.; Lai, Y.-H.; Peng, M.-Y.; Zhang, J.; Yang, L.-T.; Huang, Z.-R.; Chen, L.-S. Metabolomics combined with physiology and transcriptomics reveals how Citrus grandis leaves cope with copper-toxicity. Ecotoxicol. Environ. Saf. 2021, 223, 112579. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Tong, H.; Selby, J.; Dewitt, J.; Peng, X.; He, Z.H. Involvement of a cell wall-associated kinase, WAKL4, in Arabidopsis mineral responses. Plant Physiol. 2005, 139, 1704–1716. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, T. Xyloglucans in the primary cell wall. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1989, 40, 139–168. [Google Scholar] [CrossRef]
- Zhu, X.F.; Shi, Y.Z.; Lei, G.J.; Fry, S.C.; Zhang, B.C.; Zhou, Y.H.; Braam, J.; Jiang, T.; Xu, X.Y.; Mao, C.Z.; et al. XTH31, encoding an in vitro XEH/XET-active enzyme, controls Al sensitivity by modulating in vivo XET action, cell wall xyloglucan content and Al binding capacity in Arabidopsis. Plant Cell 2012, 24, 4731–4747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.F.; Wan, J.X.; Sun, Y.; Shi, Y.Z.; Braam, J.; Li, G.X.; Zheng, S.J. Xyloglucan endotransglucosylase-hydrolase17 interacts with xyloglucan endotransglucosylase-hydrolase31 to confer xyloglucan endotransglucosylase action and affect aluminum sensitivity in Arabidopsis. Plant Physiol. 2014, 165, 1566–1574. [Google Scholar] [CrossRef]
- Jiang, H.-X.; Yang, L.-T.; Qi, Y.-P.; Lu, Y.B.; Huang, Z.-R.; Chen, L.-S. Root iTRAQ protein profile analysis of two Citrus species differing in aluminum-tolerance in response to long-term aluminum-toxicity. BMC Genomics 2015, 16, 949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.Q.; Xu, X.Y.; Gong, Q.Q.; Xie, C.; Fan, W.; Yang, J.L.; Lin, Q.S.; Zheng, S.J. Root proteome of rice studied by iTRAQ provides integrated insight into aluminum stress tolerance mechanisms in plants. J. Proteomics 2014, 98, 189–205. [Google Scholar] [CrossRef]
- Asada, K. 2000. The water-water cycle as alternative photon and electron sinks. Phil. Trans. R. Soc. Lond. B 2000, 355, 1419–1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohu, C.M.; Pilon, M. Regulation of superoxide dismutase expression by copper availability. Physiol. Plant. 2007, 129, 747–755. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Burkhead, J.L.; Gogolin, K.A.; Andrés-Colás, N.; Bodecker, J.R.; Puig, S.; Peñarrubia, L.; Pilon, M. AtCCS is a functional homolog of the yeast copper chaperone Ccs1/Lys7. FEBS Let. 2005, 579, 2307–2312. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.-H.; Kuo, W.-Y.; Weiss, C.; Jinn, T.L. Copper chaperone-dependent and -independent activation of three copper-zinc superoxide dismutase homologs localized in different cellular compartments in Arabidopsis. Plant Physiol. 2012, 158, 737–746. [Google Scholar] [CrossRef] [Green Version]
- Davletova, S.; Rizhsky, L.; Liang, H.; Zhong, S.; Oliver, D.J.; Coutu, J.; Shulaev, V.; Schlauch, K.; Mittleret, R. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 2005, 17, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Lim, J.D.; Hahn, S.J.; Yu, C.Y.; Chung, I.M. Expression of the glutathione S-transferase gene (NT107) in transgenic Dianthus superbus. Plant Cell Tissue Organ Cult. 2015, 80, 277–286. [Google Scholar] [CrossRef]
- Zhang, L.; Du, L.; Poovaiah, B.W. Calcium signaling and biotic defense responses in plants. Plant Signal. Behav. 2014, 9, e973818. [Google Scholar] [CrossRef] [Green Version]
- Krebs, J.; Agellon, L.B.; Michalak, M. Ca2+ homeostasis and endoplasmic reticulum (ER) stress: An integrated view of calcium signaling. Biochem. Bioph. Res. Co. 2015, 460, 114–121. [Google Scholar] [CrossRef]
- Teardo, E.; Carraretto, L.; De Bortoli, S.; Costa, A.; Behera, S.; Wagner, R.; Lo Schiavo, F.; Formentin, E.; Szabo, I. Alternative splicing-mediated targeting of the Arabidopsis glutamate receptor 3.5 to mitochondria affects organelle morphology. Plant Physiol. 2015, 167, 216–227. [Google Scholar] [CrossRef] [Green Version]
- Teardo, E.; Formentin, E.; Segalla, A.; Giacometti, G.M.; Marin, O.; Zanetti, M.; Lo Schiavo, F.; Zoratti, M.; Szabò, I. Dual localization of plant glutamate receptor AtGLR3.4 to plastids and plasmamembrane. Biochim. Biophys. Acta-Bioenergetics 2011, 1807, 359–367. [Google Scholar] [CrossRef]
- Yan, J.; Guan, L.; Sun, Y.; Zhu, Y.; Liu, L.; Lu, R.; Jiang, M.; Tan, M.; Zhang, A. Calcium and ZmCCaMK are involved in brassinosteroid-induced antioxidant defense in maize leaves. Plant Cell Physiol. 2015, 56, 883–896. [Google Scholar] [CrossRef] [Green Version]
- Ma, F.; Lu, R.; Liu, H.; Shi, B.; Zhang, J.; Tan, M.; Zhang, A.; Jiang, M. Nitric oxide-activated calcium/calmodulin-dependent protein kinase regulates the abscisic acid-induced antioxidant defence in maize. J. Exp. Bot. 2012, 63, 4835–4847. [Google Scholar] [CrossRef] [PubMed]
- Pearce, G.; Moura, D.S.; Stratmann, J.; Ryan, C.A. RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl. Acad. Sci. USA 2001, 98, 12843–12847. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Zayed, O.; Yu, Z.; Jiang, W.; Zhu, P.; Hsu, C.C.; Zhang, L.; Tao, W.A.; Lozano-Durán, R.; Zhu, J.K. Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, 13123–13128. [Google Scholar] [CrossRef] [Green Version]
- Yeh, C.M.; Chine, P.S.; Huang, H.J. Distinct signalling pathways for induction of MAP kinase activities by cadmium and copper in rice roots. J. Exp. Bot. 2007, 58, 659–671. [Google Scholar] [CrossRef] [Green Version]
- Burnett, E.C.; Desikan, R.; Moser, R.C.; Neill, S.J. ABA activation of an MBP kinase in Pisum sativum epidermal peels correlates with stomatal responses to ABA. J. Exp. Bot. 2000, 51, 197–205. [Google Scholar] [CrossRef]
- Agrawal, G.K.; Agrawal, S.K.; Shibato, J.; Iwahashi, H.; Rakwal, R. Novel rice MAP kinases OsMSRMK3 and OsWJUMK1 involved in encountering diverse environmental stresses and developmental regulation. Biochem. Biophys. Res. Commun. 2003, 300, 775–783. [Google Scholar] [CrossRef]
- Schweighofer, A.; Hirt, H.; Meskiene, I. Plant PP2C phosphatases: Emerging functions in stress signaling. Trends Plant Sci. 2004, 9, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Johan, S.K.; Bagri, J.; Pandey, G.K. ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PLoS ONE 2015, 10, e0125168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Zhu, Y.; Zhai, H.; Cai, H.; Ji, W.; Luo, X.; Li, J.; Bai, X. AtPP2CG1, a protein phosphatase 2C, positively regulates salt tolerance of Arabidopsis in abscisic acid-dependent manner. Biochem. Biophys. Res. Commun. 2012, 422, 710–715. [Google Scholar] [CrossRef] [PubMed]
- Ouzounidou, G.; Ilias, I. Hormone-induced protection of sunflower photosynthetic apparatus against copper toxicity. Biol. Plant. 2005, 49, 223–228. [Google Scholar] [CrossRef]
- Xiong, L.; Lee, H.; Ishitani, M.; Zhu, J.K. Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J. Biol. Chem. 2002, 277, 8588–8596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wang, Y.; Kai, W.; Zhao, B.; Chen, P.; Sun, L.; Ji, K.; Li, Q.; Dai, S.; Sun, Y.; et al. Transcriptional regulation of abscisic acid signal core components during cucumber seed germination and under Cu2+, Zn2+, NaCl and simulated acid rain stresses. Plant Physiol. Biochem. 2014, 76, 67–76. [Google Scholar] [CrossRef]
- Zengin, F.K.; Kirbag, S. Effects of copper on chlorophyll, proline, protein and abscisic acid level of sunflower (Helianthus annuus L.) seedlings. J. Environ. Biol. 2007, 28, 561–566. [Google Scholar]
- Nguyen, T.Q.; Sesin, V.; Kisiala, A.; Emery, R.J.N. Phytohormonal roles in plant responses to heavy metal stress: Implications for using macrophytes in phytoremediation of aquatic ecosystems. Environ. Toxicol. Chem. 2021, 40, 7–22. [Google Scholar] [CrossRef] [PubMed]
- Kuroha, T.; Tokunaga, H.; Kojima, M.; Ueda, N.; Ishida, T.; Nagawa, S.; Fukuda, H.; Sugimoto, K.; Sakakibara, H. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 2009, 21, 3152–3169. [Google Scholar] [CrossRef] [Green Version]
- Thomas, J.C.; Perron, M.; LaRosa, P.C.; Smigocki, A.C. Cytokinin and the regulation of a tobacco metallothionein-like gene during copper stress. Physiol. Plant. 2005, 123, 262–271. [Google Scholar] [CrossRef]
- Yang, T.-Y.; Cai, L.-Y.; Qi, Y.-P.; Yang, L.-T.; Lai, N.-W.; Chen, L.-S. Increasing nutrient solution pH alleviated aluminum-induced inhibition of growth and impairment of photosynthetic electron transport chain in Citrus sinensis seedlings. BioMed. Res. Int. 2019, 2019, 9058715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, P.; Qi, Y.-P.; Cai, Y.-T.; Yang, T.-Y.; Yang, L.-T.; Huang, Z.-R.; Chen, L.-S. Aluminum effects on photosynthesis, reactive oxygen species and methylglyoxal detoxification in two Citrus species differing in aluminum tolerance. Tree Physiol. 2018, 38, 1548–1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar]
- Long, A.; Zhang, J.; Yang, L.-T.; Ye, X.; Lai, N.-W.; Tan, L.-L.; Lin, D.; Chen, L.-S. Effects of low pH on photosynthesis, related physiological parameters and nutrient profile of Citrus. Front. Plant Sci. 2017, 8, 185. [Google Scholar] [CrossRef] [Green Version]
- Garg, N.; Kaur, H. Response of antioxidant enzymes, phytochelatins and glutathione production towards Cd and Zn stresses in Cajanus cajan (L.) Millsp. genotypes colonized by arbuscular mycorrhizal fungi. J. Agron. Crop Sci. 2013, 199, 118–133. [Google Scholar] [CrossRef]
- Malik, J.A.; Goel, S.; Kaur, N.; Sharma, S.; Singh, I.; Nayyar, H. Selenium antagonises the toxic effects of arsenic on mungbean (Phaseolus aureus Roxb.) plants by restricting its uptake and enhancing the antioxidative and detoxification mechanisms. Environ. Exp. Bot. 2012, 77, 242–248. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnol. 2015, 33, 290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Wu, Q.; He, W.; He, T.; Wu, Q.; Miao, Y. Combined de novo transcriptome and metabolome analysis of common bean response to Fusarium oxysporum f. sp. phaseoli infection. Int. J. Mol. Sci. 2019, 20, 6278. [Google Scholar] [CrossRef] [Green Version]
- Mostafa, H.H.A.; Wang, H.; Song, J.; Li, X. Effects of genotypes and explants on garlic callus production and endogenous hormones. Sci. Rep. 2020, 10, 4867. [Google Scholar] [CrossRef] [Green Version]
| Accession No. | KEGG | Swiss-Prot | Log2(FC) |
|---|---|---|---|
| Cu ion transmembrane transporter activity (GO:0005375) | |||
| Cg4g018610 | Solute carrier family 31 (copper transporter), member 1 | Copper transporter 5; AtCOPT5 | −1.317 |
| Cg8g023350 | Solute carrier family 31 (copper transporter), member 1 | Copper transporter 1; AtCOPT1 | −3.606 |
| Cg8g023360 | Solute carrier family 31 (copper transporter), member 1 | Copper transporter 1; AtCOPT1 | −1.076 |
| Cg8g023380 | Solute carrier family 31 (copper transporter), member 1 | Copper transporter 1; AtCOPT1 | −8.212 |
| Cg6g005770 | Solute carrier family 31 (copper transporter), member 1 | Copper transporter 1; AtCOPT1 | 3.407 |
| Yellow Stripe-Like (YSL) family | |||
| Cg5g018670 | Fanconi-associated nuclease 1 [EC:3.1.21.- 3.1.4.1] | Metal-nicotianamine transporter YSL3; Protein YELLOW STRIPE LIKE 3; AtYSL3 | −1.609 |
| Cg5g020560 | Fanconi-associated nuclease 1 [EC:3.1.21.- 3.1.4.1] | Probable metal-nicotianamine transporter YSL5; Protein YELLOW STRIPE LIKE 5; AtYSL5 | −1.066 |
| Cu ion binding (GO:0005507) and/or Cu proteins | |||
| Cg1g028930 | l-ascorbate oxidase [EC:1.10.3.3] | l-ascorbate oxidase | −3.067 |
| Cg2g001710 | Enoyl-[acyl-carrier protein] reductase I [EC:1.3.1.9 1.3.1.10] | Enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic | 1.679 |
| Cg2g018560 | Iron transport multicopper oxidase | l-ascorbate oxidase homolog | 1.069 |
| Cg3g024840 | Iron transport multicopper oxidase | l-ascorbate oxidase homolog | 1.455 |
| Cg3g024680 | Plastocyanin | Plastocyanin, chloroplastic | −1.448 |
| Cg5g007370 | Glutamate dehydrogenase (NAD(P)+) [EC:1.4.1.3] | Glutamate dehydrogenase 2 | 1.745 |
| Cg5g009340 | Copper chaperone for superoxide dismutase | Copper chaperone for superoxide dismutase, chloroplastic/cytosolic; AtCCS | 1.588 |
| Cg7g012360 | Glutathione S-transferase [EC:2.5.1.18] | Glutathione S-transferase F9 | −1.318 |
| Cg8g018870 | Superoxide dismutase, Cu-Zn family [EC:1.15.1.1] | Superoxide dismutase [Cu-Zn], chloroplastic | 1.344 |
| Cg9g013180 | Cytochrome c oxidase subunit 3 | Cytochrome c oxidase subunit 3 | −1.324 |
| CgUng010240 | Cytochrome c oxidase subunit 2 | Uncharacterized mitochondrial protein AtMg00530 | −1.581 |
| Cu chaperones | |||
| Cg5g009340 | Copper chaperone for superoxide dismutase | Copper chaperone for superoxide dismutase, chloroplastic/cytosolic; AtCCS | 1.588 |
| Others | |||
| Cg3g000750 | Cd2+/Zn2+-exporting ATPase [EC:3.6.3.3 3.6.3.5] | Cadmium/zinc-transporting ATPase HMA2 | −1.403 |
| Cg4g006740 | Solute carrier family 39 (zinc transporter), member 1/2/3 | Zinc transporter 4, chloroplastic; ZRT/IRT-like protein 4 | −1.718 |
| Cg8g022750 | Solute carrier family 39 (zinc transporter), member 1/2/3 | Zinc transporter 1; ZRT/IRT-like protein 1; OsZIP1 | −2.937 |
| Cg5g041700 | Ferric-chelate reductase [EC:1.16.1.7] | Ferric reduction oxidase 2; AtFRO2; EC = 1.16.1.7; Ferric-chelate reductase 2 | 3.591 |
| Cg1g023140 | Ferric-chelate reductase [EC:1.16.1.7] | (RefSeq) ferric reduction oxidase 7, chloroplastic-like (A) | NAC domain-containing protein 104 {ECO:0000305} | 3.271 |
| Cg2g021360 | Ferric-chelate reductase [EC:1.16.1.7] | (RefSeq) ferric reduction oxidase 7, chloroplastic-like (A) | NAC domain-containing protein 72 | 2.012 |
| Cg5g010410 | Ferric-chelate reductase [EC:1.16.1.7] | (RefSeq) ferric reduction oxidase 7, chloroplastic-like (A) | NAC domain-containing protein 100 {ECO:0000303|PubMed:15029955} | 1.441 |
| Cg6g025130 | Ferric-chelate reductase [EC:1.16.1.7] | (RefSeq) ferric reduction oxidase 7, chloroplastic-like (A) | NAC domain-containing protein 100 {ECO:0000303|PubMed:15029955} | 1.777 |
| Cg8g013730 | Ferric-chelate reductase [EC:1.16.1.7] | (RefSeq) ferric reduction oxidase 7, chloroplastic-like (A) | NAC domain-containing protein 90 | −1.805 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, F.; Huang, H.; Peng, M.; Lai, Y.; Ren, Q.; Zhang, J.; Huang, Z.; Yang, L.; Rensing, C.; Chen, L. Adaptive Responses of Citrus grandis Leaves to Copper Toxicity Revealed by RNA-Seq and Physiology. Int. J. Mol. Sci. 2021, 22, 12023. https://doi.org/10.3390/ijms222112023
Wu F, Huang H, Peng M, Lai Y, Ren Q, Zhang J, Huang Z, Yang L, Rensing C, Chen L. Adaptive Responses of Citrus grandis Leaves to Copper Toxicity Revealed by RNA-Seq and Physiology. International Journal of Molecular Sciences. 2021; 22(21):12023. https://doi.org/10.3390/ijms222112023
Chicago/Turabian StyleWu, Fenglin, Huiyu Huang, Mingyi Peng, Yinhua Lai, Qianqian Ren, Jiang Zhang, Zengrong Huang, Lintong Yang, Christopher Rensing, and Lisong Chen. 2021. "Adaptive Responses of Citrus grandis Leaves to Copper Toxicity Revealed by RNA-Seq and Physiology" International Journal of Molecular Sciences 22, no. 21: 12023. https://doi.org/10.3390/ijms222112023
APA StyleWu, F., Huang, H., Peng, M., Lai, Y., Ren, Q., Zhang, J., Huang, Z., Yang, L., Rensing, C., & Chen, L. (2021). Adaptive Responses of Citrus grandis Leaves to Copper Toxicity Revealed by RNA-Seq and Physiology. International Journal of Molecular Sciences, 22(21), 12023. https://doi.org/10.3390/ijms222112023








