The Great Escape: mRNA Export through the Nuclear Pore Complex
Abstract
1. Introduction
2. Nuclear Processing of RNA
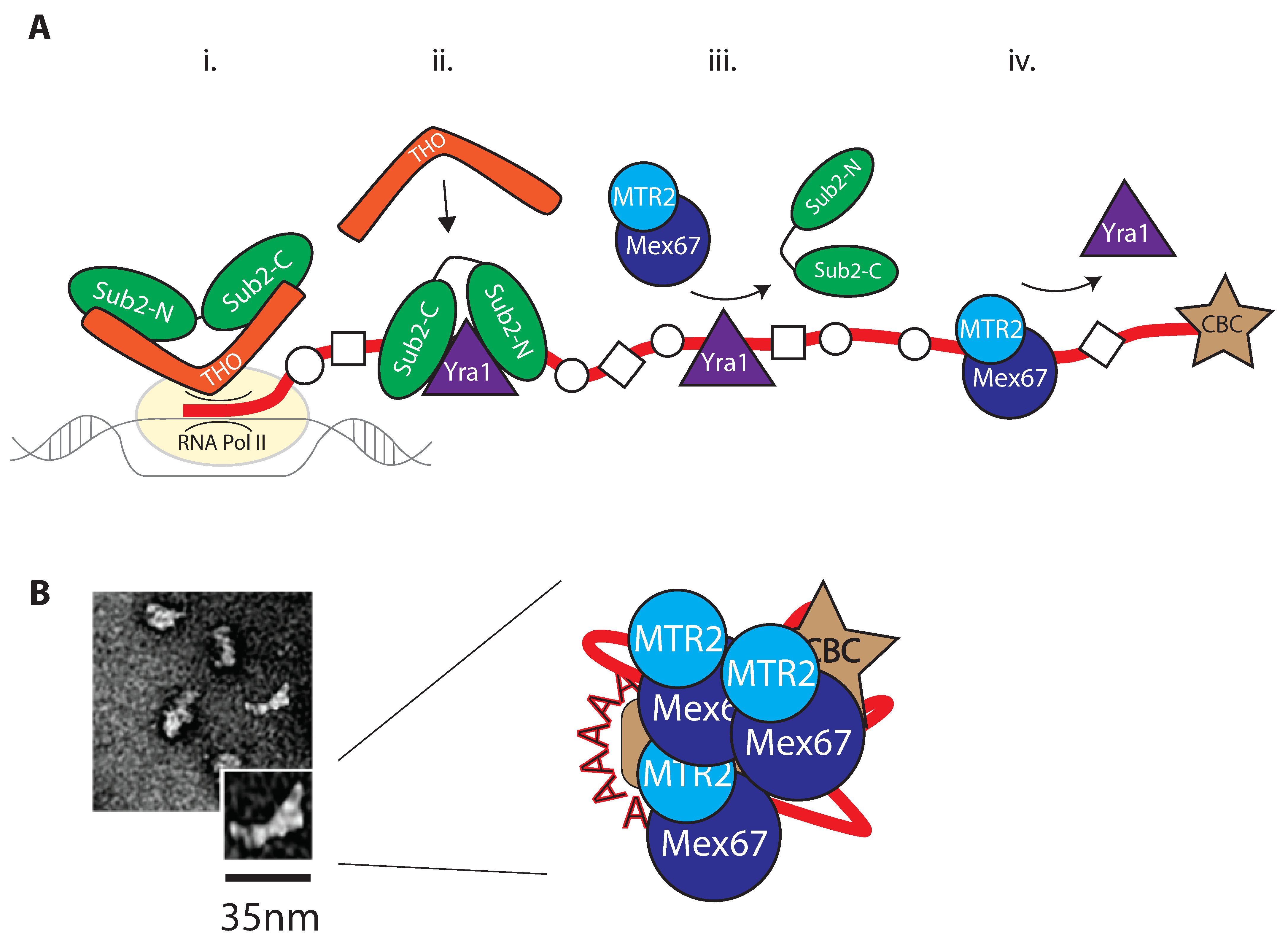
2.1. Remodeling by RNA Binding Proteins (RBPs)
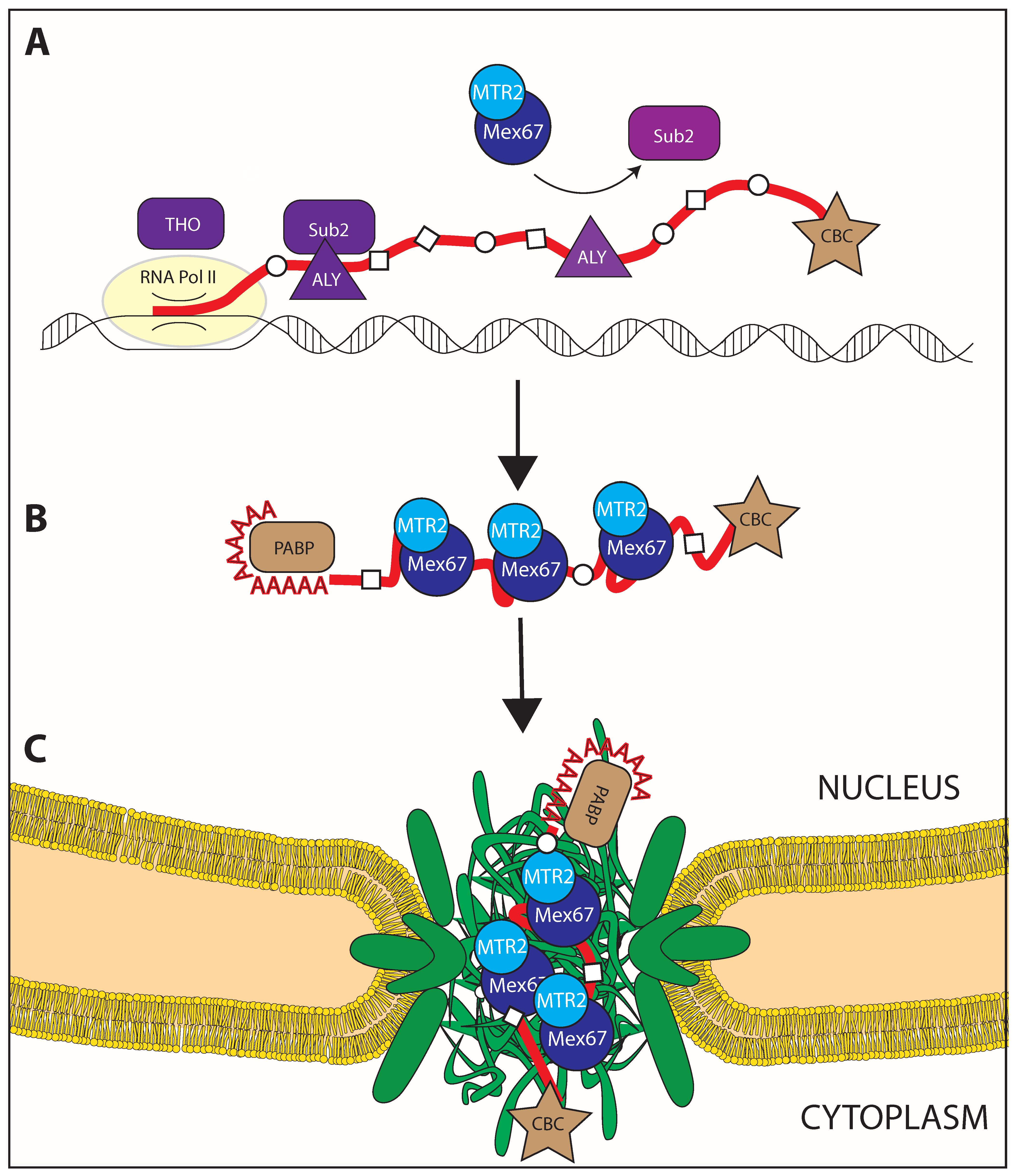
2.2. Mex67/MTR2 Is the Key Driver of mRNA Export
3. mRNP Export through the Nuclear Pore Complex
3.1. Remodeling upon Entering the Nuclear Pore Complex
3.2. Loading of Mex67/MTR2 on mRNP
3.3. Crosstalk with Positioning of Factors on the mRNA
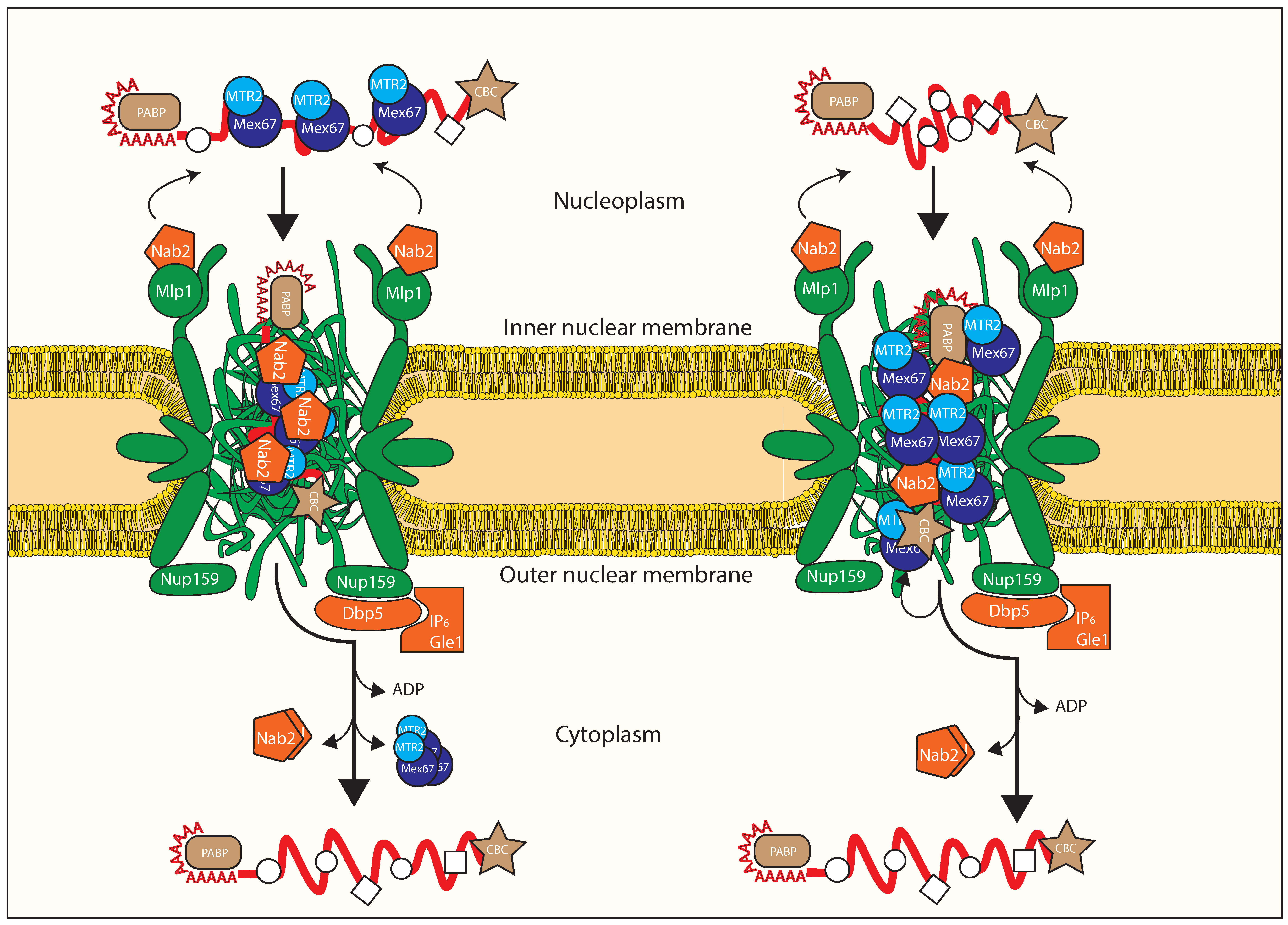
3.4. Remodeling at the Nuclear Basket
3.5. Translocation through the Central Channel of the NPC
3.6. Remodeling upon Exiting the Nuclear Pore Complex
4. Conclusions and Future Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Sloan, K.E. The mitochondrial epitranscriptome: The roles of RNA modifications in mitochondrial translation and human disease. Cell. Mol. Life Sci. 2018, 75, 241–260. [Google Scholar] [CrossRef] [PubMed]
- Henras, A.K.; Plisson-Chastang, C.; O’Donohue, M.F.; Chakraborty, A.; Gleizes, P.E. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 2015, 6, 225–242. [Google Scholar] [CrossRef]
- Hentze, M.W.; Castello, A.; Schwarzl, T.; Preiss, T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018, 19, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.Y.; Dunham, W.H.; Hong, S.J.; Knight, J.D.; Bashkurov, M.; Chen, G.I.; Bagci, H.; Rathod, B.; MacLeod, G.; Eng, S.W.M.; et al. High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Mol. Cell 2018, 69, 517–532.e11. [Google Scholar] [CrossRef]
- Adivarahan, S.; Livingston, N.; Nicholson, B.; Rahman, S.; Wu, B.; Rissland, O.S.; Zenklusen, D. Spatial Organization of Single mRNPs at Different Stages of the Gene Expression Pathway. Mol. Cell 2018, 72, 727–738.e5. [Google Scholar] [CrossRef]
- Metkar, M.; Ozadam, H.; Lajoie, B.R.; Imakaev, M.; Mirny, L.A.; Dekker, J.; Moore, M.J. Higher-Order Organization Principles of Pre-translational mRNPs. Mol. Cell 2018, 72, 715–726.e3. [Google Scholar] [CrossRef] [PubMed]
- Saguez, C.; Olesen, J.R.; Jensen, T.H. Formation of export-competent mRNP: Escaping nuclear destruction. Curr. Opin. Cell Biol. 2005, 17, 287–293. [Google Scholar] [CrossRef]
- Hershey, J.W.B.; Sonenberg, N.; Mathews, M.B. Principles of translational control: An overview. Cold Spring Harb. Perspect. Biol. 2012, 4, a011528. [Google Scholar] [CrossRef] [PubMed]
- Gerovac, M.; Tampé, R. Control of mRNA Translation by Versatile ATP-Driven Machines. Trends Biochem. Sci. 2019, 44, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.S.; von Lindern, M. RNA binding proteins and regulation of mRNA translation in erythropoiesis. Front. Physiol. 2018, 9, 910. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.R.; Pratt, G.A.; Martinez, F.J.; Yeo, G.W.; Lykke-Andersen, J. Target Discrimination in Nonsense-Mediated mRNA Decay Requires Upf1 ATPase Activity. Mol. Cell 2015, 59, 413–425. [Google Scholar] [CrossRef]
- Baltz, A.G.; Munschauer, M.; Schwanhäusser, B.; Vasile, A.; Murakawa, Y.; Schueler, M.; Youngs, N.; Penfold-Brown, D.; Drew, K.; Milek, M.; et al. The mRNA-Bound Proteome and Its Global Occupancy Profile on Protein-Coding Transcripts. Mol. Cell 2012, 46, 674–690. [Google Scholar] [CrossRef]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Preiss, T.; Steinmetz, L.M.; et al. Insights into RNA Biology from an Atlas of Mammalian mRNA-Binding Proteins. Cell 2012, 149, 1393–1406. [Google Scholar] [CrossRef]
- Kwon, S.C.; Yi, H.; Eichelbaum, K.; Foehr, S.; Fischer, B.; You, K.T.; Castello, A.; Krijgsveld, J.; Hentze, M.; Kim, V.N. The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 2013, 20, 1122–1130. [Google Scholar] [CrossRef]
- Ainger, K.; Avossa, D.; Diana, A.S.; Barry, C.; Barbarese, E.; Carson, J.H. Transport and localization elements in myelin basic protein mRNA. J. Cell Biol. 1997, 138, 1077–1087. [Google Scholar] [CrossRef]
- Sharma, D.; Zagore, L.L.; Brister, M.M.; Ye, X.; Crespo-Hernández, C.E.; Licatalosi, D.D.; Jankowsky, E. The kinetic landscape of an RNA-binding protein in cells. Nature 2021, 591, 152–156. [Google Scholar] [CrossRef]
- Kislauskis, E.H.; Zhu, X.; Singer, R.H. Sequences Responsible for Intracellular Localization of -Actin Messenger RNA Also Affect Cell Phenotype. J. Cell Biol. 1994, 127, 441–451. [Google Scholar] [CrossRef]
- Blichenberg, A.; Schwanke, B.; Rehbein, M.; Garner, C.C.; Richter, D.; Kindler, S. Identification of a cis -Acting Dendritic Targeting Element in MAP2 mRNAs Identification of a cis -Acting Dendritic Targeting Element in MAP2 mRNAs. J. Neurosci. 1999, 19, 8818–8829. [Google Scholar] [CrossRef]
- Kiebler, M.A.; Desgroseillers, L. Molecular Insights into mRNA Transport and Local Translation in the Mammalian Nervous System. Neuron 2000, 25, 19–28. [Google Scholar] [CrossRef]
- Raju, C.S.; Göritz, C.; Nord, Y.; Hermanson, O.; López-Iglesias, C.; Visa, N.; Castelo-Branco, G.; Percipalle, P. In Cultured Oligodendrocytes the A/B-type hnRNP CBF-A Accompanies MBP mRNA Bound to mRNA Trafficking Sequences. Mol. Biol. Cell 2008, 19, 3008–3019. [Google Scholar] [CrossRef] [PubMed]
- Lange, A.; Mills, R.E.; Lange, C.J.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical Nuclear Localization Signals: Definition, Function, and Interaction with Importin α. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef]
- Eliscovich, C.; Buxbaum, A.R.; Katz, Z.B.; Singer, R.H. mRNA on the move: The road to its biological destiny. J. Biol. Chem. 2013, 288, 20361–20368. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Gkogkas, C.G.; Sonenberg, N.; Holt, C.E. Remote control of gene function by local translation. Cell 2014, 157, 26–40. [Google Scholar] [CrossRef]
- van Nostrand, E.L.; Freese, P.; Pratt, G.A.; Wang, X.; Wei, X.; Xiao, R.; Blue, S.M.; Chen, J.-Y.; Cody, N.A.L.; Dominguez, D.; et al. A large-scale binding and functional map of human RNA-binding proteins. Nature 2020, 583, 711–719. [Google Scholar] [CrossRef]
- Khong, A.; Parker, R. MRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction. bioRxiv 2018, 217, 4124–4140. [Google Scholar] [CrossRef]
- Kapeli, K.; Martinez, F.J.; Yeo, G.W. Genetic mutations in RNA-binding proteins and their roles in ALS. Hum. Genet. 2017, 136, 1193–1214. [Google Scholar] [CrossRef]
- Valkov, E.; Dean, J.C.; Jani, D.; Kuhlmann, S.I.; Stewart, M. Structural basis for the assembly and disassembly of mRNA nuclear export complexes. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 578–592. [Google Scholar] [CrossRef]
- Niño, C.A.; Hérissant, L.; Babour, A.; Dargemont, C. MRNA nuclear export in yeast. Chem. Rev. 2013, 113, 8523–8545. [Google Scholar] [CrossRef]
- Natalizio, B.J.; Wente, S.R. Postage for the messenger: Designating routes for nuclear mRNA export. Trends Cell Biol. 2013, 23, 365–373. [Google Scholar] [CrossRef]
- Martinez-Rucobo, F.W.; Kohler, R.; van de Waterbeemd, M.; Heck, A.J.; Hemann, M.; Herzog, F.; Stark, H.; Cramer, P. Molecular Basis of Transcription-Coupled Pre-mRNA Capping. Mol. Cell 2015, 58, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Dufu, K.; Lee, C.-S.; Hsu, J.L.; Dias, A.; Reed, R. Human mRNA Export Machinery Recruited to the 5′ End of mRNA. Cell 2006, 127, 1389–1400. [Google Scholar] [CrossRef] [PubMed]
- Zahler, A.M.; Lane, W.S.; Stolk, J.A.; Roth, M.B. SR proteins: A conserved family of pre-mRNA splicing factors. Genes Dev. 1992, 6, 837–847. [Google Scholar] [CrossRef]
- Müller-McNicoll, M.; Botti, V.; Domingues, A.M.D.J.; Brandl, H.; Schwich, O.D.; Steiner, M.C.; Curk, T.; Poser, I.; Zarnack, K.; Neugebauer, K.M. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 2016, 30, 553–566. [Google Scholar] [CrossRef]
- Jeong, S. SR proteins: Binders, regulators, and connectors of RNA. Mol. Cells 2017, 40, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Windgassen, M.; Krebber, H. Identification of Gbp2 as a novel poly(A)+ RNA-binding protein involved in the cytoplasmic delivery of messenger RNAs in yeast. EMBO Rep. 2003, 4, 278–283. [Google Scholar] [CrossRef]
- Häcker, S.; Krebber, P. Differential Export Requirements for Shuttling Serine/Arginine-type mRNA-binding Proteins. J. Biol. Chem. 2004, 279, 5049–5052. [Google Scholar] [CrossRef]
- Lei, E.P.; Krebber, H.; Silver, P.A. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 2001, 15, 1771–1782. [Google Scholar] [CrossRef]
- Krebber, H.; Sturm, D.; Windgassen, M.; Cajigas, I.; González, C.; Seedorf, M.; Bastians, H. Yeast shuttling SR-proteins Npl3p, Gbp2p and Hrb1p are part of the translating mRNPs and Npl3p can function as a translational repressor. GBM Annu. Fall Meet. Berl. Potsdam. 2005, 2005, 10479–10491. [Google Scholar] [CrossRef]
- Le Hir, H.; Gatfield, D.; Izaurralde, E.; Moore, M.J. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001, 20, 4987–4997. [Google Scholar] [CrossRef]
- Singh, G.; Kucukural, A.; Cenik, C.; Leszyk, J.D.; Shaffer, S.A.; Weng, Z.; Moore, M.J. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 2012, 151, 750–764. [Google Scholar] [CrossRef] [PubMed]
- Viphakone, N.; Sudbery, I.; Griffith, L.; Heath, C.G.; Sims, D.; Wilson, S.A. Co-transcriptional Loading of RNA Export Factors Shapes the Human Transcriptome. Mol. Cell 2019, 75, 310–323.e8. [Google Scholar] [CrossRef]
- Boehm, V.; Gehring, N.H. Exon Junction Complexes: Supervising the Gene Expression Assembly Line. Trends Genet. 2016, 32, 724–735. [Google Scholar] [CrossRef]
- Le Hir, H.; Sauliere, J.; Wang, Z. The exon junction complex as a node of post-transcriptional networks. Nat. Rev. Mol. Cell Biol. 2016, 17, 41–54. [Google Scholar] [CrossRef]
- Ares, M.; Grate, L.; Pauling, M.H. A handful of intron-containing genes produces the lion’s share of yeast mRNA. RNA 1999, 5, 1138–1139. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.T.; Hautbergue, G.M.; Walsh, M.J.; Viphakone, N.; van Dijk, T.B.; Philipsen, S.; Wilson, S.A. Chtop is a component of the dynamic TREX mRNA export complex. EMBO J. 2013, 32, 473–486. [Google Scholar] [CrossRef]
- Baejen, C.; Torkler, P.; Gressel, S.; Essig, K.; Söding, J.; Cramer, P. Transcriptome Maps of mRNP Biogenesis Factors Define Pre-mRNA Recognition. Mol. Cell 2014, 55, 745–757. [Google Scholar] [CrossRef]
- Pandya-Jones, A.; Bhatt, D.M.; Lin, C.-H.; Tong, A.-J.; Smale, S.T.; Black, D.L. Splicing kinetics and transcript release from the chromatin compartment limit the rate of Lipid A-induced gene expression. RNA 2013, 19, 811–827. [Google Scholar] [CrossRef]
- Mauger, O.; Lemoine, F.; Scheiffele, P. Targeted Intron Retention and Excision for Rapid Gene Regulation in Response to Neuronal Activity. Neuron 2016, 92, 1266–1278. [Google Scholar] [CrossRef]
- Brody, Y.; Neufeld, N.; Bieberstein, N.; Causse, S.; Böhnlein, E.-M.; Neugebauer, K.M.; Darzacq, X.; Shav-Tal, Y. The In Vivo Kinetics of RNA Polymerase II Elongation during Co-Transcriptional Splicing. PLoS Biol. 2011, 9, e1000573. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.T.; Holloway, M.P.; Altura, R.A. The CRM1 nuclear export protein in normal development and disease. Int. J. Biochem. Mol. Biol. 2012, 3, 137–151. [Google Scholar]
- Carmody, S.R.; Wente, S.R. mRNA nuclear export at a glance. J. Cell Sci. 2009, 122, 1933–1937. [Google Scholar] [CrossRef]
- Segref, A.; Sharma, K.; Doye, V.; Hellwig, A.; Huber, J.; Lührmann, R.; Hurt, E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997, 16, 3256–3271. [Google Scholar] [CrossRef]
- Katahira, J.; Sträßer, K.; Podtelejnikov, A.; Mann, M.; Jung, J.U.; Hurt, E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999, 18, 2593–2609. [Google Scholar] [CrossRef]
- Grüter, P.; Tabernero, C.; von Kobbe, C.; Schmitt, C.; Saavedra, C.; Bachi, A.; Wilm, M.; Felber, B.K.; Izaurralde, E. TAP, the Human Homolog of Mex67p, Mediates CTE-Dependent RNA Export from the Nucleus. Mol. Cell 1998, 1, 649–659. [Google Scholar] [CrossRef]
- Braun, I.C.; Rohrbach, E.; Schmitt, C.; Izaurralde, E. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J. 1999, 18, 1953–1965. [Google Scholar] [CrossRef]
- Aibara, S.; Katahira, J.; Valkov, E.; Stewart, M. The principal mRNA nuclear export factor NXF1:NXT1 forms a symmetric binding platform that facilitates export of retroviral CTE-RNA. Nucleic Acids Res. 2015, 43, 1883–1893. [Google Scholar] [CrossRef]
- Xie, Y.; Ren, Y. Mechanisms of nuclear mRNA export: A structural perspective. Traffic 2019, 20, 829–840. [Google Scholar] [CrossRef]
- Bachi, A.; Braun, I.C.; Rodrigues, J.P.; Panté, N.; Ribbeck, K.; VON Kobbe, C.; Kutay, U.; Wilm, M.; Görlich, D.; Carmo-Fonseca, M.; et al. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 2000, 6, 136–158. [Google Scholar] [CrossRef]
- Sträßer, K.; Baßler, J.; Hurt, E. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J. Cell Biol. 2000, 150, 695–706. [Google Scholar] [CrossRef]
- Strawn, L.A.; Shen, T.; Wente, S.R. The GLFG Regions of Nup116p and Nup100p Serve as Binding Sites for Both Kap95p and Mex67p at the Nuclear Pore Complex. J. Biol. Chem. 2001, 276, 6445–6452. [Google Scholar] [CrossRef]
- Adams, R.L.; Terry, L.J.; Wente, S.R. Nucleoporin FG domains facilitate mRNP remodeling at the cytoplasmic face of the nuclear pore complex. Genetics 2014, 197, 1213–1224. [Google Scholar] [CrossRef] [PubMed]
- Fribourg, S.; Braun, I.C.; Izaurralde, E.; Conti, E. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol. Cell 2001, 8, 645–656. [Google Scholar] [CrossRef]
- Grant, R.P.; Hurt, E.; Neuhaus, D.; Stewart, M. Structure of the C-terminal FG-nucleoporin binding domain of Tap/NXF1. Nat. Struct. Biol. 2002, 9, 247–251. [Google Scholar] [CrossRef]
- Grant, R.P.; Neuhaus, D.; Stewart, M. Structural basis for the interaction between the Tap/NXF1 UBA domain and FG nucleoporins at 1 Å resolution. J. Mol. Biol. 2003, 326, 849–858. [Google Scholar] [CrossRef]
- Terry, L.J.; Wente, S.R. Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J. Cell Biol. 2007, 178, 1121–1132. [Google Scholar] [CrossRef]
- Tetenbaum-Novatt, J.; Hough, L.E.; Mironska, R.; McKenney, A.S.; Rout, M.P. Nucleocytoplasmic transport: A role for nonspecific competition in karyopherin-nucleoporin interactions. Mol. Cell. Proteom. 2012, 11, 31–46. [Google Scholar] [CrossRef]
- Smith, C.; Lari, A.; Derrer, C.P.; Ouwehand, A.; Rossouw, A.; Huisman, M.; Dange, T.; Hopman, M.; Joseph, A.; Zenklusen, D.; et al. In vivo single-particle imaging of nuclear mRNA export in budding yeast demonstrates an essential role for Mex67p. J. Cell Biol. 2015, 211, 1121–1130. [Google Scholar] [CrossRef] [PubMed]
- Aibara, S.; Valkov, E.; Lamers, M.; Stewart, M. Domain organization within the nuclear export factor Mex67:Mtr2 generates an extended mRNA binding surface. Nucleic Acids Res. 2015, 43, 1927–1936. [Google Scholar] [CrossRef]
- Beck, M.; Schmidt, A.; Malmström, J.; Claassen, M.; Ori, A.; Szymborska-Mell, A.; Herzog, F.; Rinner, O.; Ellenberg, J.; Aebersold, R. The quantitative proteome of a human cell line. Mol. Syst. Biol. 2011, 7, 549. [Google Scholar] [CrossRef]
- Maeshima, K.; Iino, H.; Hihara, S.; Funakoshi, T.; Watanabe, A.; Nishimura, M.; Nakatomi, R.; Yahata, K.; Imamoto, F.; Hashikawa, T.; et al. Nuclear pore formation but not nuclear growth is governed by cyclin-dependent kinases (Cdks) during interphase. Nat. Struct. Mol. Biol. 2010, 17, 1065–1071. [Google Scholar] [CrossRef]
- Winey, M.; Yarar, D.; Giddings, T.H.; Mastronarde, D.N. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol. Biol. Cell 1997, 8, 2119–2132. [Google Scholar] [CrossRef] [PubMed]
- Sankhala, R.S.; Lokareddy, R.K.; Begum, S.; Pumroy, R.; Gillilan, R.E.; Cingolani, G. Three-dimensional context rather than NLS amino acid sequence determines importin α subtype specificity for RCC1. Nat. Commun. 2017, 8, 979. [Google Scholar] [CrossRef] [PubMed]
- Niepel, M.; Molloy, K.R.; Williams, R.; Farr, J.C.; Meinema, A.C.; Vecchietti, N.; Cristea, I.M.; Chait, B.T.; Rout, M.P.; Strambio-De-Castillia, C. The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol. Biol. Cell 2013, 24, 3920–3938. [Google Scholar] [CrossRef] [PubMed]
- Rabut, G.; Doye, V.; Ellenberg, J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 2004, 6, 1114–1121. [Google Scholar] [CrossRef] [PubMed]
- Schuller, S.K.; Schuller, J.M.; Prabu, J.R.; Baumgärtner, M.; Bonneau, F.; Basquin, J.; Conti, E. Structural insights into the nucleic acid remodeling mechanisms of the yeast THO-Sub2 complex. eLife 2020, 9, e61467. [Google Scholar] [CrossRef]
- Batisse, J.; Batisse, C.; Budd, A.; Böttcher, B.; Hurt, E. Purification of nuclear poly(A)-binding protein Nab2 reveals association with the yeast transcriptome and a messenger ribonucleoprotein core structure. J. Biol. Chem. 2009, 284, 34911–34917. [Google Scholar] [CrossRef] [PubMed]
- Meinel, D.; Burkert-Kautzsch, C.; Kieser, A.; O’Duibhir, E.; Siebert, M.; Mayer, A.; Cramer, P.; Söding, J.; Holstege, F.C.P.; Sträßer, K. Recruitment of TREX to the Transcription Machinery by Its Direct Binding to the Phospho-CTD of RNA Polymerase II. PLoS Genet. 2013, 9, e1003914. [Google Scholar] [CrossRef]
- Pühringer, T.; Hohmann, U.; Fin, L.; Pacheco-Fiallos, B.; Schellhaas, U.; Brennecke, J.; Plaschka, C. Structure of the human core transcription-export complex reveals a hub for multivalent interactions. eLife 2020, 9, e61503. [Google Scholar] [CrossRef]
- Ren, Y.; Schmiege, P.; Blobel, G. Structural and biochemical analyses of the DEAD-box ATPase Sub2 in association with THO or Yra1. eLife 2017, 6, e20070. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Luo, M.-J.; Sträßer, K.; Katahira, J.; Hurt, E.; Reed, R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nat. Cell Biol. 2000, 407, 401–405. [Google Scholar] [CrossRef]
- Reed, R.; Magni, K. A new view of mRNA export: Separating the wheat from the chaff. Nat. Cell Biol. 2001, 3, 201–204. [Google Scholar] [CrossRef]
- Straßer, K.; Hurt, E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000, 19, 410–420. [Google Scholar]
- Duncan, K.; Umen, J.G.; Guthrie, C. A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Curr. Biol. 2000, 10, 687–696. [Google Scholar] [CrossRef]
- Iglesias, N.; Tutucci, E.; Gwizdek, C.; Vinciguerra, P.; Von Dach, E.; Corbett, A.H.; Dargemont, C.; Stutz, F. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 2010, 24, 1927–1938. [Google Scholar] [CrossRef]
- Xie, Y.; Clarke, B.P.; Kim, Y.J.; Ivey, A.L.; Hill, P.S.; Shi, Y.; Ren, Y. Cryo-EM structure of the yeast TREX complex and coordination with the SR-like protein Gbp2. eLife 2021, 10, e65699. [Google Scholar] [CrossRef]
- Hackmann, A.; Wu, H.; Schneider, U.-M.; Meyer, K.; Jung, K.; Krebber, H. Quality control of spliced mRNAs requires the shuttling SR proteins Gbp2 and Hrb1. Nat. Commun. 2014, 5, 3123. [Google Scholar] [CrossRef] [PubMed]
- Khan, G.A.; Deforges, J.; Reis, R.S.; Hsieh, Y.-F.; Montpetit, J.; Antosz, W.; Santuari, L.; Hardtke, C.; Grasser, K.D.; Poirier, Y. The transcription and export complex THO/TREX contributes to transcription termination in plants. PLoS Genet. 2020, 16, e1008732. [Google Scholar] [CrossRef]
- Fischer, T.; Sträßer, K.; Racz, A.; Rodriguez-Navarro, S.; Oppizzi, M.; Ihrig, P.; Lechner, J.; Hurt, E. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002, 21, 5843–5852. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Hellerschmied, D.; Schubert, T.; Amlacher, S.; Vinayachandran, V.; Reja, R.; Pugh, B.F.; Clausen, T.; Köhler, A. The Nuclear Pore-Associated TREX-2 Complex Employs Mediator to Regulate Gene Expression. Cell 2015, 162, 1016–1028. [Google Scholar] [CrossRef]
- Jani, D.; Lutz, S.; Marshall, N.J.; Fischer, T.; Köhler, A.; Ellisdon, A.M.; Hurt, E.; Stewart, M. Sus1, Cdc31, and the Sac3 CID Region Form a Conserved Interaction Platform that Promotes Nuclear Pore Association and mRNA Export. Mol. Cell 2009, 33, 727–737. [Google Scholar] [CrossRef]
- Schubert, T.; Köhler, A. Mediator and TREX-2: Emerging links between transcription initiation and mRNA export. Nucleus 2016, 7, 126–131. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Umlauf, D.; Bonnet, J.; Waharte, F.; Fournier, M.; Stierle, M.; Fischer, B.; Brino, L.; Devys, D.; Tora, L. The human TREX-2 complex is stably associated with the nuclear pore basket. J. Cell Sci. 2013, 126, 2656–2667. [Google Scholar] [CrossRef] [PubMed]
- Jani, D.; Valkov, E.; Stewart, M. Structural basis for binding the TREX2 complex to nuclear pores, GAL1 localisation and mRNA export. Nucleic Acids Res. 2014, 42, 6686–6697. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M. Structure and Function of the TREX-2 Complex. Subcell. Biochem. 2019, 93, 461–470. [Google Scholar]
- Aibara, S.; Gordon, J.M.B.; Riesterer, A.S.; McLaughlin, S.H.; Stewart, M. Structural basis for the dimerization of Nab2 generated by RNA binding provides insight into its contribution to both poly(A) tail length determination and transcript compaction in Saccharomyces cerevisiae. Nucleic Acids Res. 2017, 45, 1529–1538. [Google Scholar] [CrossRef]
- Green, D.M.; Marfatia, K.A.; Crafton, E.B.; Zhang, X.; Cheng, X.; Corbett, A.H. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 2002, 277, 7752–7760. [Google Scholar] [CrossRef] [PubMed]
- Fasken, M.B.; Stewart, M.; Corbett, A.H. Functional significance of the interaction between the mRNA-binding protein, Nab2, and the nuclear pore-associated protein, mlp1, in mRNA export. J. Biol. Chem. 2008, 283, 27130–27143. [Google Scholar] [CrossRef]
- Galy, V.; Gadal, O.; Fromont-Racine, M.; Romano, A.; Jacquier, A.; Nehrbass, U. Nuclear Retention of Unspliced mRNAs in Yeast Is Mediated by Perinuclear Mlp1. Cell 2004, 116, 63–73. [Google Scholar] [CrossRef]
- Adams, R.L.; Wente, S.R. Dbp5 associates with RNA-bound Mex67 and Nab2 and its localization at the nuclear pore complex is sufficient for mRNP export and cell viability. PLoS Genet. 2020, 16, e1009033. [Google Scholar] [CrossRef]
- Lee, E.S.; Wolf, E.J.; Ihn, S.S.J.; Smith, H.W.; Emili, A.; Palazzo, A.F. TPR is required for the efficient nuclear export of mRNAs and lncRNAs from short and intron-poor genes. Nucleic Acids Res. 2020, 48, 11645–11663. [Google Scholar] [CrossRef] [PubMed]
- Soheilypour, M.; Mofrad, M.R.K. Quality control of mRNAs at the entry of the nuclear pore: Cooperation in a complex molecular system. Nucleus 2018, 9, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Dreyfuss, G.; Kim, V.N.; Kataoka, N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002, 3, 195–205. [Google Scholar] [CrossRef]
- Müller-Mcnicoll, M.; Neugebauer, K.M. How cells get the message: Dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet. 2013, 14, 275–287. [Google Scholar] [CrossRef]
- Mehlin, H.; Daneholt, B.; Skoglund, U. Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell 1992, 69, 605–613. [Google Scholar] [CrossRef]
- Grünwald, D.; Singer, R.H. In vivo imaging of labelled endogenous Β-actin mRNA during nucleocytoplasmic transport. Nature 2010, 467, 604–607. [Google Scholar] [CrossRef]
- Mor, A.; Suliman, S.; Ben-Yishay, R.; Yunger, S.; Brody, Y.; Shav-Tal, Y. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat. Cell Biol. 2010, 12, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Liu, Z.; Michelotti, N.; Pitchiaya, S.; Veerapaneni, R.; Androsavich, J.R.; Walter, N.G.; Yang, W. High-resolution three-dimensional mapping of mRNA export through the nuclear pore. Nat. Commun. 2013, 4, 2414. [Google Scholar] [CrossRef]
- Siebrasse, J.P.; Kaminski, T.; Kubitscheck, U. Nuclear export of single native mRNA molecules observed by light sheet fluorescence microscopy. Proc. Natl. Acad. Sci. USA 2012, 109, 9426–9431. [Google Scholar] [CrossRef]
- Rout, M.P.; Blobel, G. Isolation of the yeast nuclear pore complex. J. Cell Biol. 1993, 123, 771–783. [Google Scholar] [CrossRef]
- Hoelz, A.; Debler, E.W.; Blobel, G. The Structure of the nuclear pore complex. Annu. Rev. Biochem. 2011, 80, 613–643. [Google Scholar] [CrossRef]
- Derrer, C.P.; Mancini, R.; Vallotton, P.; Huet, S.; Weis, K.; Dultz, E. The RNA export factor Mex67 functions as a mobile nucleoporin. J. Cell Biol. 2019, 218, 3967–3976. [Google Scholar] [CrossRef]
- Ben-Yishay, R.; Mor, A.; Shraga, A.; Ashkenazy-Titelman, A.; Kinor, N.; Schwed-Gross, A.; Jacob, A.; Kozer, N.; Kumar, P.; Garini, Y.; et al. Imaging within single NPCs reveals NXF1’s role in mRNA export on the cytoplasmic side of the pore. J. Cell Biol. 2019, 218, 2962–2981. [Google Scholar] [CrossRef]
- Frey, S.; Görlich, D. A Saturated FG-Repeat Hydrogel Can Reproduce the Permeability Properties of Nuclear Pore Complexes. Cell 2007, 130, 512–523. [Google Scholar] [CrossRef]
- Li, C.; Goryaynov, A.; Yang, W. The selective permeability barrier in the nuclear pore complex. Nucleus 2016, 7, 430–446. [Google Scholar] [CrossRef]
- Aramburu, I.V.; Lemke, E.A. Floppy but not sloppy: Interaction mechanism of FG-nucleoporins and nuclear transport receptors. Semin. Cell Dev. Biol. 2017, 68, 34–41. [Google Scholar] [CrossRef]
- Celetti, G.; Paci, G.; Caria, J.; VanDelinder, V.; Bachand, G.; Lemke, E.A. The liquid state of FG-nucleoporins mimics permeability barrier properties of nuclear pore complexes. J. Cell Biol. 2019, 219, e201907157. [Google Scholar] [CrossRef]
- Paradise, A.; Levin, M.K.; Korza, G.; Carson, J.H. Significant Proportions of Nuclear Transport Proteins with Reduced Intracellular Mobilities Resolved by Fluorescence Correlation Spectroscopy. J. Mol. Biol. 2007, 365, 50–65. [Google Scholar] [CrossRef]
- Tokunaga, M.; Imamoto, N.; Sakata-Sogawa, K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 2008, 5, 159–161. [Google Scholar] [CrossRef]
- Kapinos, L.E.; Schoch, R.L.; Wagner, R.S.; Schleicher, K.D.; Lim, R.Y.H. Karyopherin-centric control of nuclear pores based on molecular occupancy and kinetic analysis of multivalent binding with FG nucleoporins. Biophys. J. 2014, 106, 1751–1762. [Google Scholar] [CrossRef]
- Ribbeck, K.; Görlich, D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001, 20, 1320–1330. [Google Scholar] [CrossRef]
- Jovanovic-Talisman, T.; Zilman, A. Protein Transport by the Nuclear Pore Complex: Simple Biophysics of a Complex Biomachine. Biophys. J. 2017, 113, 6–14. [Google Scholar] [CrossRef]
- Lim, R.Y.H.; Kapinos, L.E. How to operate a nuclear pore complex by kap-centric control. Nucleus 2015, 6, 366–372. [Google Scholar] [CrossRef]
- Snay-Hodge, C.A.; Colot, H.V.; Goldstein, A.L.; Cole, C.N. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998, 17, 2663–2676. [Google Scholar] [CrossRef] [PubMed]
- Liker, E.; Fernandez, E.; Izaurralde, E.; Conti, E. The structure of the mRNA export factor TAP reveals a cis arrangement of a non-canonical RNP domain and an LRR domain. EMBO J. 2000, 19, 5587–5598. [Google Scholar] [CrossRef]
- Lund, M.K.; Guthrie, C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol. Cell 2005, 20, 645–651. [Google Scholar] [CrossRef]
- Cole, C.N.; Scarcelli, J.J. Unravelling mRNA export. Nat. Cell Biol. 2006, 8, 645–647. [Google Scholar] [CrossRef]
- Weirich, C.S.; Erzberger, J.P.; Flick, J.S.; Berger, J.M.; Thorner, J.; Weis, K. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol. 2006, 8, 668–676. [Google Scholar] [CrossRef]
- Tran, E.J.; Zhou, Y.; Corbett, A.H.; Wente, S.R. The DEAD-Box Protein Dbp5 Controls mRNA Export by Triggering Specific RNA:Protein Remodeling Events. Mol. Cell 2007, 28, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Montpetit, B.; Thomsen, N.D.; Helmke, K.J.; Seeliger, M.; Berger, J.M.; Weis, K. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature 2011, 472, 238–244. [Google Scholar] [CrossRef]
- Folkmann, A.W.; Noble, K.N.; Cole, C.N.; Wente, S.R. Dbp5, Gle1-IP6 and Nup159: A working model for mRNP export. Nucleus 2011, 2, 540–548. [Google Scholar] [CrossRef]
- Schmitt, C.; von Kobbe, C.; Bachi, A.; Panté, N.; Rodrigues, J.P.; Boscheron, C.; Rigaut, G.; Wilm, M.; Séraphin, B.; Carmo-Fonseca, M.; et al. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 1999, 18, 4332–4347. [Google Scholar] [CrossRef]
- Weirich, C.S.; Erzberger, J.P.; Berger, J.M.; Weis, K. The N-terminal domain of Nup159 forms a β-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Mol. Cell 2004, 16, 749–760. [Google Scholar] [CrossRef]
- von Moeller, H.; Basquin, C.; Conti, E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat. Struct. Mol. Biol. 2009, 16, 247–254. [Google Scholar] [CrossRef]
- Alcázar-Román, A.R.; Tran, E.J.; Guo, S.; Wente, S.R. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 2006, 8, 711–716. [Google Scholar] [CrossRef]
- Dossani, Z.Y.; Weirich, C.S.; Erzberger, J.P.; Berger, J.M.; Weis, K. Structure of the C-terminus of the mRNA export factor Dbp5 reveals the interaction surface for the ATPase activator Gle1. Proc. Natl. Acad. Sci. USA 2009, 106, 16251–16256. [Google Scholar] [CrossRef]
- Hodge, C.A.; Tran, E.; Noble, K.N.; Alcazar-Roman, A.R.; Ben-Yishay, R.; Scarcelli, J.J.; Folkmann, A.W.; Shav-Tal, Y.; Wente, S.R.; Cole, C.N. The Dbp5 cycle at the nuclear pore complex during mRNA export I: Dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev. 2011, 25, 1052–1064. [Google Scholar] [CrossRef]
- Noble, K.N.; Tran, E.; Alcazar-Roman, A.R.; Hodge, C.A.; Cole, C.N.; Wente, S.R. The Dbp5 cycle at the nuclear pore complex during mRNA export II: Nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes Dev. 2011, 25, 1065–1077. [Google Scholar] [CrossRef]
- Collins, R.; Karlberg, T.; Lehtiö, L.; Schütz, P.; Berg, S.V.D.; Dahlgren, L.-G.; Hammarström, M.; Weigelt, J.; Schüler, H. The DEXD/H-box RNA helicase DDX19 is regulated by an α-helical switch. J. Biol. Chem. 2009, 284, 10296–10300. [Google Scholar] [CrossRef]
- de Magistris, P.; Tatarek-Nossol, M.; Dewor, M.; Antonin, W. A self-inhibitory interaction within Nup155 and membrane binding are required for nuclear pore complex formation. J. Cell Sci. 2018, 131, jcs208538. [Google Scholar] [CrossRef]
- Grandi, P.; Emig, S.; Weise, C.; Hucho, F.; Pohl, T.; Hurt, E.C. A novel nuclear pore protein Nup82p which specifically binds to a fraction of Nsp1p. J. Cell Biol. 1995, 130, 1263–1273. [Google Scholar] [CrossRef]
- Kraemer, D.M.; Strambio-de-Castillia, C.; Blobel, G.; Rout, M.P. The essential yeast nucleoporin NUP159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J. Biol. Chem. 1995, 270, 19017–19021. [Google Scholar] [CrossRef]
- Belgareh, N.; Snay-Hodge, C.; Pasteau, F.; Dagher, S.; Cole, C.N.; Doye, V. Functional characterization of a Nup159p-containing nuclear pore subcomplex. Mol. Biol. Cell 1998, 9, 3475–3492. [Google Scholar] [CrossRef]
- Gaik, M.; Flemming, D.; Von Appen, A.; Kastritis, P.; Mücke, N.; Fischer, J.; Stelter, P.; Ori, A.; Bui, K.H.; Baßler, J.; et al. Structural basis for assembly and function of the Nup82 complex in the nuclear pore scaffold. J. Cell Biol. 2015, 208, 283–297. [Google Scholar] [CrossRef]
- Trahan, C.; Oeffinger, M. Targeted cross-linking-mass spectrometry determines vicinal interactomes within heterogeneous RNP complexes. Nucleic Acids Res. 2016, 44, 1354–1369. [Google Scholar] [CrossRef]
- Fernandez-Martinez, J.; Kim, S.J.; Shi, Y.; Upla, P.; Pellarin, R.; Gagnon, M.; Chemmama, I.E.; Wang, J.; Nudelman, I.; Zhang, W.; et al. Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform. Cell 2016, 167, 1215–1228.e25. [Google Scholar] [CrossRef] [PubMed]
- Kosinski, J.; Mosalaganti, S.; von Appen, A.; Teimer, R.; DiGuilio, A.L.; Wan, W.; Bui, K.H.; Hagen, W.J.; Briggs, J.A.G.; Glavy, J.S.; et al. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science 2016, 352, 363–365. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.H.; Stuwe, T.; Schilbach, S.; Rundlet, E.J.; Perriches, T.; Mobbs, G.; Fan, Y.; Thierbach, K.; Huber, F.M.; Collins, L.N.; et al. Architecture of the symmetric core of the nuclear pore. Science 2016, 352, aaf1015. [Google Scholar] [CrossRef] [PubMed]
- Stuwe, T.; Bley, C.J.; Thierbach, K.; Petrovic, S.; Schilbach, S.; Mayo, D.J.; Perriches, T.; Rundlet, E.J.; Jeon, Y.E.; Collins, L.N.; et al. Architecture of the fungal nuclear pore inner ring complex. Science 2015, 350, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Nousiainen, H.O.; Kestilä, M.; Pakkasjärvi, N.; Honkala, H.; Kuure, S.; Tallila, J.; Vuopala, K.; Ignatius, J.; Herva, R.; Peltonen, L. Mutations in mRNA export mediator GLE1 result in a fetal motoneuron disease. Nat. Genet. 2008, 40, 155–157. [Google Scholar] [CrossRef]
- Kaneb, H.M.; Folkmann, A.W.; Belzil, V.V.; Jao, L.-E.; Leblond, C.S.; Girard, S.; Daoud, H.; Noreau, A.; Rochefort, D.; Hince, P.; et al. Deleterious mutations in the essential mRNA metabolism factor, hGle1, in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2015, 24, 1363–1373. [Google Scholar] [CrossRef] [PubMed]
- Folkmann, A.W.; Dawson, T.R.; Wente, S.R. Insights into mRNA export-linked molecular mechanisms of human disease through a Gle1 structure-function analysis. Adv. Biol. Regul. 2014, 54, 74–91. [Google Scholar] [CrossRef]
- Muhlrad, D.; Parker, R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992, 6, 2100–2111. [Google Scholar] [CrossRef] [PubMed]
- Shyu, A.B.; Belasco, J.G.; Greenberg, M.E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991, 5, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.Y.; Mugler, C.F.; Heinrich, S.; Vallotton, P.; Weis, K. Non-invasive measurement of mRNA decay reveals translation initiation as the major determinant of mRNA stability. eLife 2018, e32536. [Google Scholar] [CrossRef]
- Eisen, T.J.; Eichhorn, S.W.; Subtelny, A.O.; Lin, K.S.; McGeary, S.E.; Gupta, S.; Bartel, D.P. The Dynamics of Cytoplasmic mRNA Metabolism. Mol. Cell 2020, 77, 786–799.e10. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Kim, J.R.; Van Bruggen, R.; Park, J. RNA-Binding Proteins in Amyotrophic Lateral Sclerosis. Mol. Cells 2018, 41, 818–829. [Google Scholar]
- Fazal, F.M.; Han, S.; Parker, K.R.; Kaewsapsak, P.; Xu, J.; Boettiger, A.N.; Chang, H.Y.; Ting, A.Y. Atlas of Subcellular RNA Localization Revealed by APEX-Seq. Cell 2019, 178, 473–490.e26. [Google Scholar] [CrossRef] [PubMed]
- Caspi, Y.; Zbaida, D.; Cohen, H.; Elbaum, M. Synthetic mimic of selective transport through the nuclear pore complex. Nano Lett. 2008, 8, 3728–3734. [Google Scholar] [CrossRef]
- Lim, R.Y.H.; Huang, N.-P.; Köser, J.; Deng, J.; Lau, K.H.A.; Schwarz-Herion, K.; Fahrenkrog, B.; Aebi, U. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc. Natl. Acad. Sci. USA 2006, 103, 9512–9517. [Google Scholar] [CrossRef]
- Jovanovic-Talisman, T.; Novatt, J.; McKenney, A.S.; Zilman, A.; Peters, R.; Rout, M.; Chait, B.T. Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature 2009, 457, 1023–1027. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, S.W.; Kapinos, L.; Blosser, T.R.; Magalhães, T.; Van Nies, P.; Lim, R.; Dekker, C. Single-molecule transport across an individual biomimetic nuclear pore complex. Nat. Nanotechnol. 2011, 6, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Fisher, P.D.E.; Shen, Q.; Akpinar, B.; Davis, L.; Chung, K.; Baddeley, D.; Saric, A.; Melia, T.J.; Hoogenboom, B.; Lin, C.; et al. A Programmable DNA Origami Platform for Organizing Intrinsically Disordered Nucleoporins within Nanopore Confinement. ACS Nano 2018, 12, 1508–1518. [Google Scholar] [CrossRef]
- Ketterer, P.; Ananth, A.N.; Trip, D.L.; Mishra, A.; Bertosin, E.; Ganji, M.; Van Der Torre, J.; Onck, P.; Dietz, H.; Dekker, C. DNA origami scaffold for studying intrinsically disordered proteins of the nuclear pore complex. Nat. Commun. 2018, 9, 902. [Google Scholar] [CrossRef] [PubMed]

| Yeast Protein Name | Human Protein Name | Role |
|---|---|---|
| mRNP Protein | ||
| Mex67/MTR2 | NXF1/NXT1, Tap/p15 | Interacts with FG nups to export mRNA |
| RNA Binding Proteins—RBPs | ||
| Cbc1/Cbc2 | Cbp20/Cbp80 | Members of the Cap Binding Complex |
| - | SRSF1-12 | SR proteins, bind mRNP during splicing |
| Npl3 | - | SR protein, binds mRNP during splicing |
| Gbp2 | - | SR protein, binds mRNP and contributes to loading Mex67/MTR2 |
| Hrb1 | - | SR protein, binds mRNP and contributes to loading Mex67/MTR2 |
| NPC—Nucleoplasmic Face | ||
| Sub2 | UAP56 | DEAD-box ATPase, member of the TREX complex, remodels mRNPs |
| Yra1 | ALY (REF, THOC4) | Adaptor protein, member of the TREX complex |
| THO | THO | A pentameric complex, component of the TREX. Includes Tho2, Hpr1, Mft1, Thp2 and Tex1 |
| Nab2 | ZC3H14 | Poly-A Binding protein, signals mature mRNPs for export |
| TREX-2 | TREX-2 | A multisubunit complex, anchored to the NPC basket (via Nup153 and TPR in metazoans, via Nup1 in yeast) |
| NPC—Cytoplasmic Face | ||
| Dbp5 | DDX19 | DEAD-box ATPase, remodels mRNPs as they exit the NPC |
| Gle1 | GLE1 | Member of the mRNA export platform, stimulates ATPase activity of Dbp5 |
| Nup42 | NUP42 | FG-nucleoporin, anchors Dbp5 and Gle1 |
| Nup159 | NUP214 | FG-nucleoporin, anchors Dbp5 to the cytoplasmic side of NPC |
| Nup116 | Nup98 | FG-nucleoporin, binds Mex67/MTR2 |
| Nup82 | Nup88 | Member of the Nup82 complex, mRNP remodeling platform |
| NSP1 | Nup62 | Member of the Nup82 complex, mRNP remodeling platform |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Magistris, P. The Great Escape: mRNA Export through the Nuclear Pore Complex. Int. J. Mol. Sci. 2021, 22, 11767. https://doi.org/10.3390/ijms222111767
De Magistris P. The Great Escape: mRNA Export through the Nuclear Pore Complex. International Journal of Molecular Sciences. 2021; 22(21):11767. https://doi.org/10.3390/ijms222111767
Chicago/Turabian StyleDe Magistris, Paola. 2021. "The Great Escape: mRNA Export through the Nuclear Pore Complex" International Journal of Molecular Sciences 22, no. 21: 11767. https://doi.org/10.3390/ijms222111767
APA StyleDe Magistris, P. (2021). The Great Escape: mRNA Export through the Nuclear Pore Complex. International Journal of Molecular Sciences, 22(21), 11767. https://doi.org/10.3390/ijms222111767





