Proteasome Inhibitors and Their Pharmacokinetics, Pharmacodynamics, and Metabolism
Abstract
1. Introduction
2. Pharmacokinetic, Pharmacodynamics, and Metabolism of Boronic Acid-Based Dual Proteasome Inhibitors
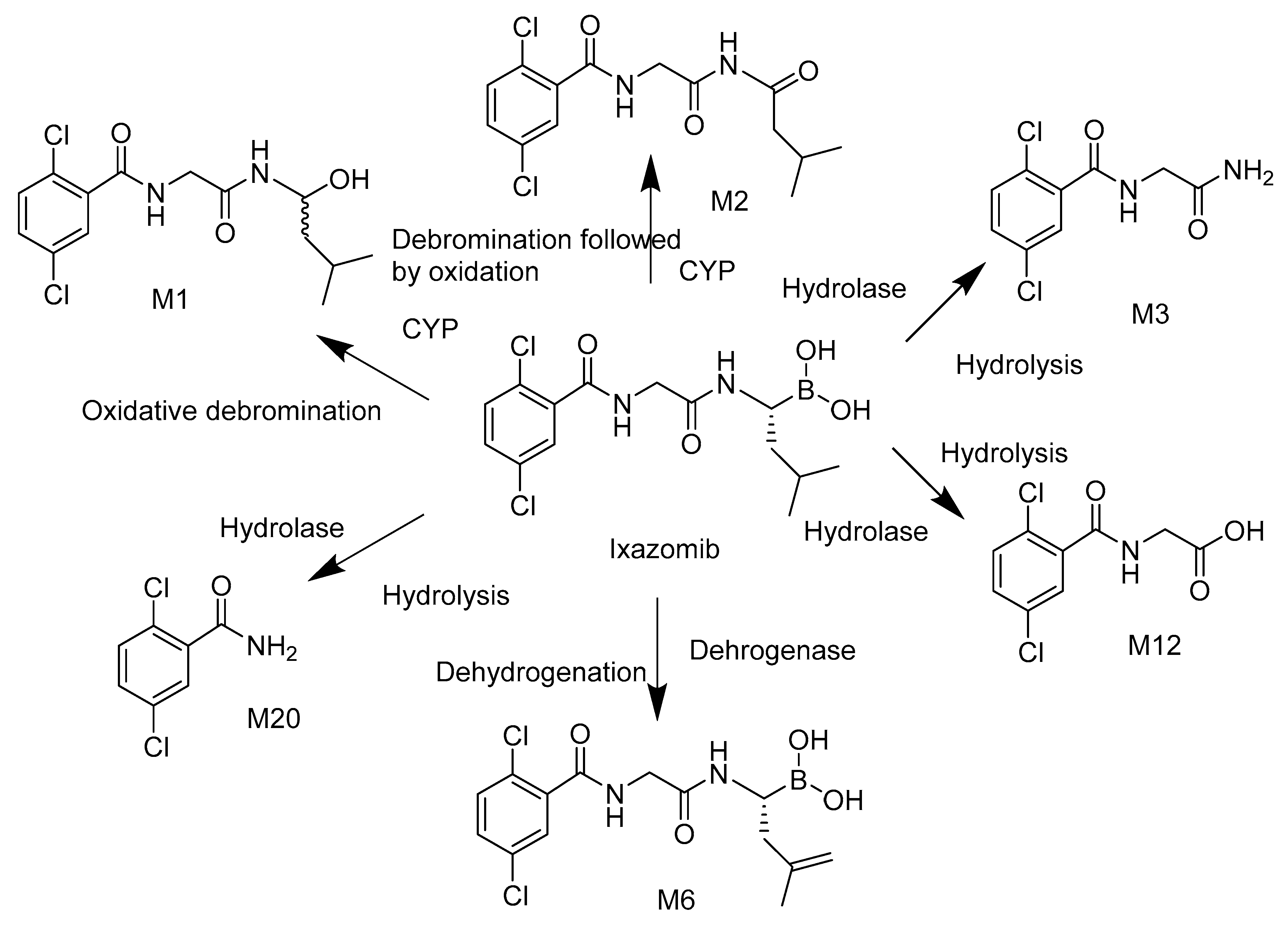
Ixazomib
3. Pharmacokinetics, Pharmacodynamics, and Metabolism of Epoxide-Based Proteasome Inhibitors
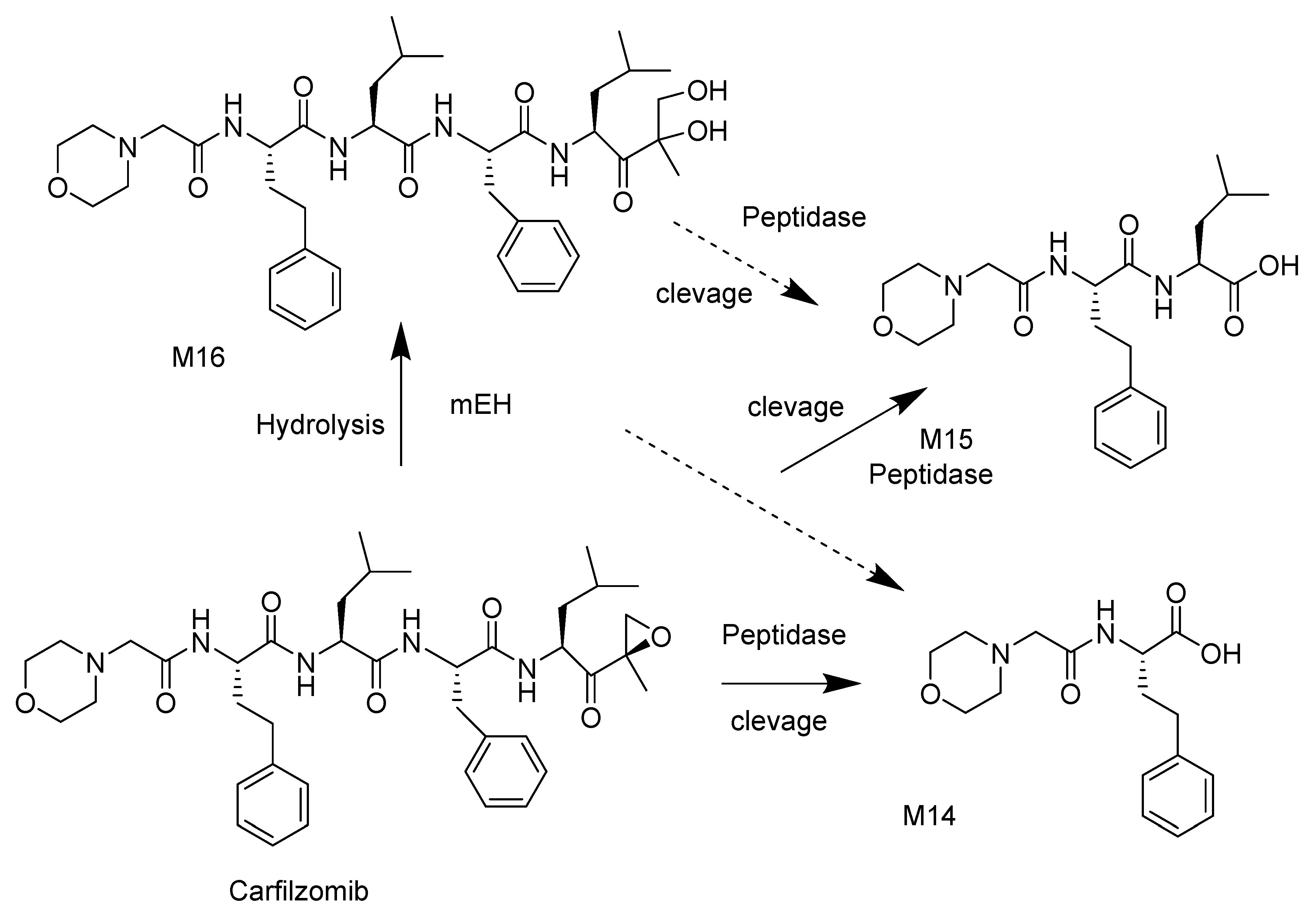
3.1. Carfilzomib
3.2. Oprozomib
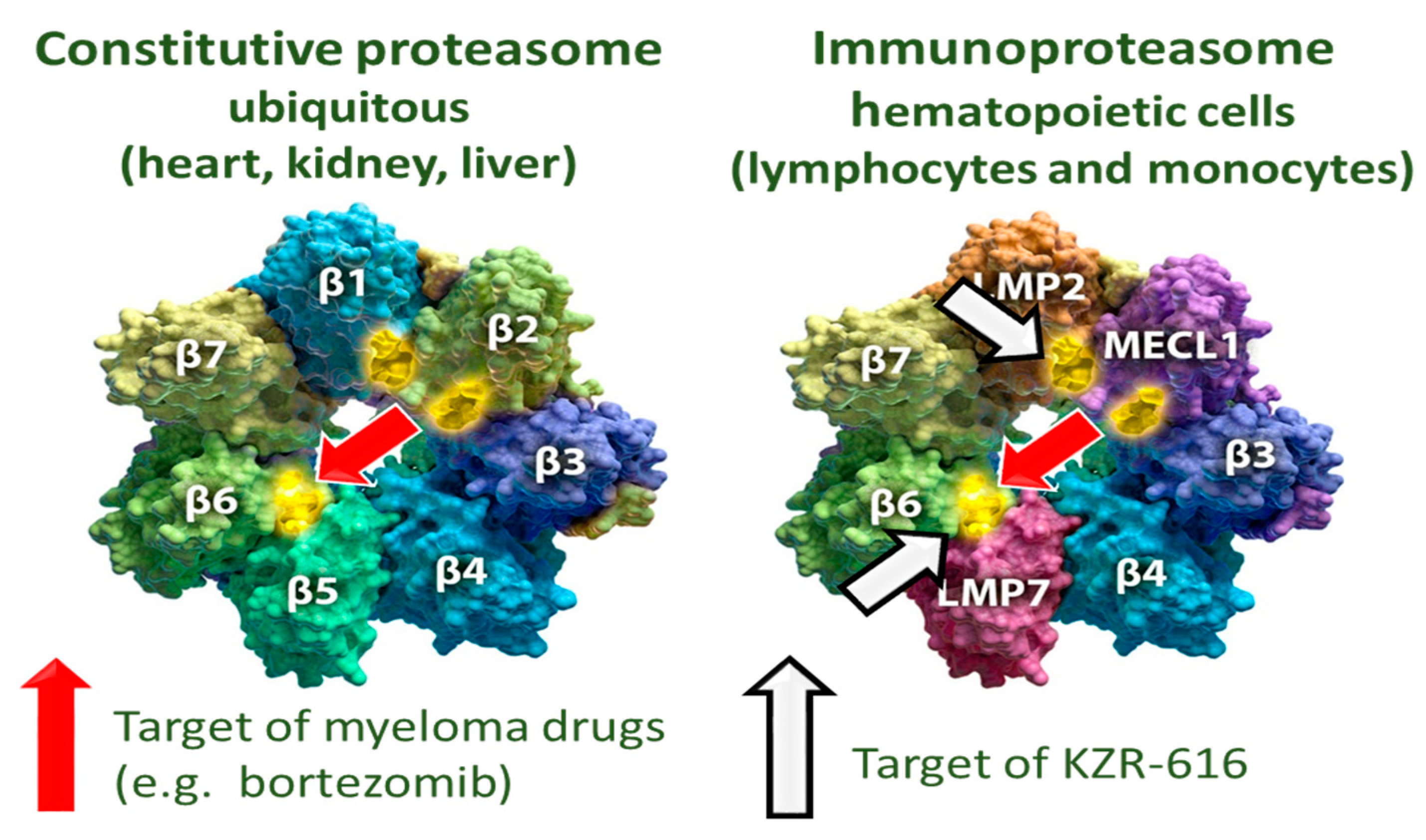
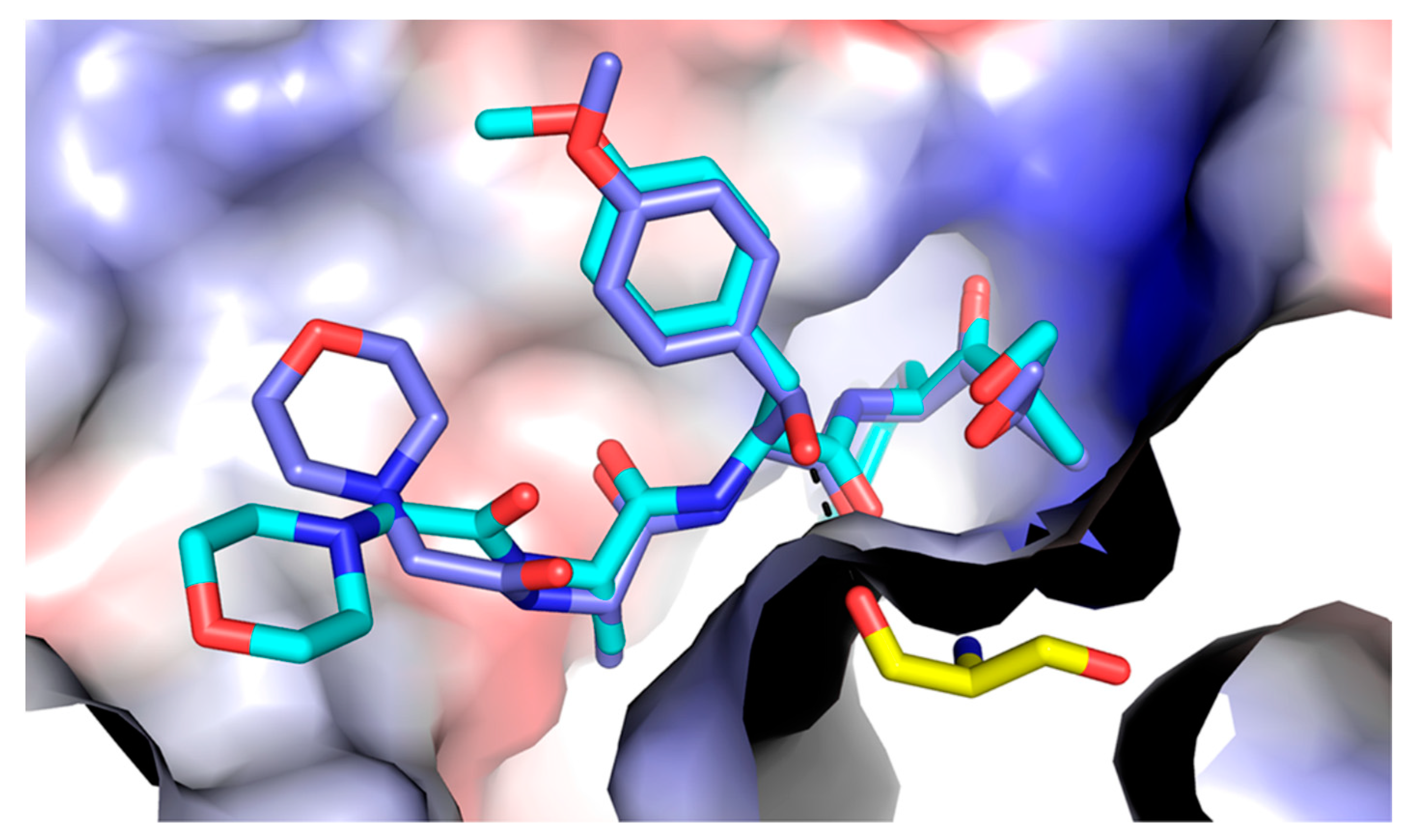
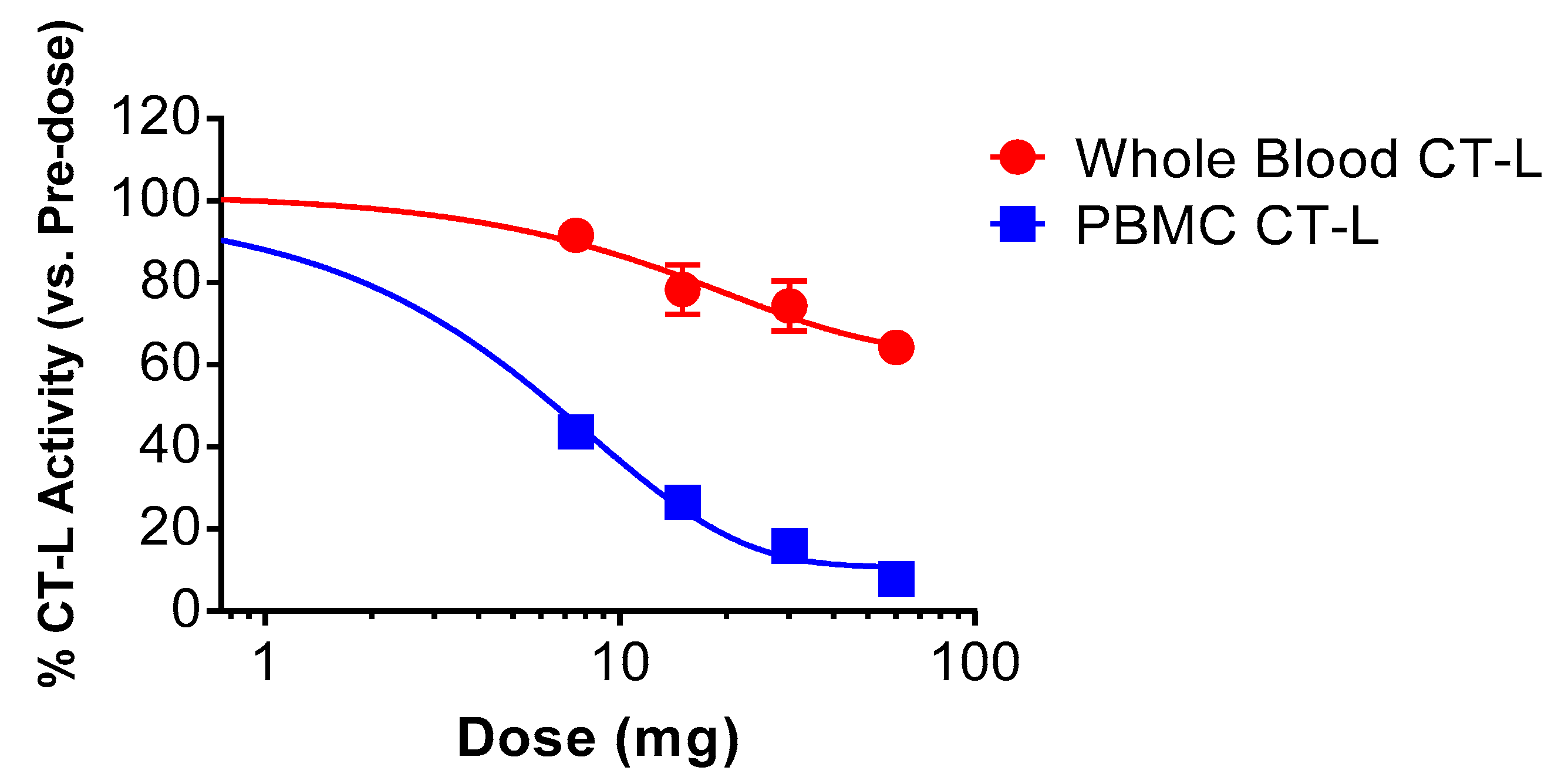
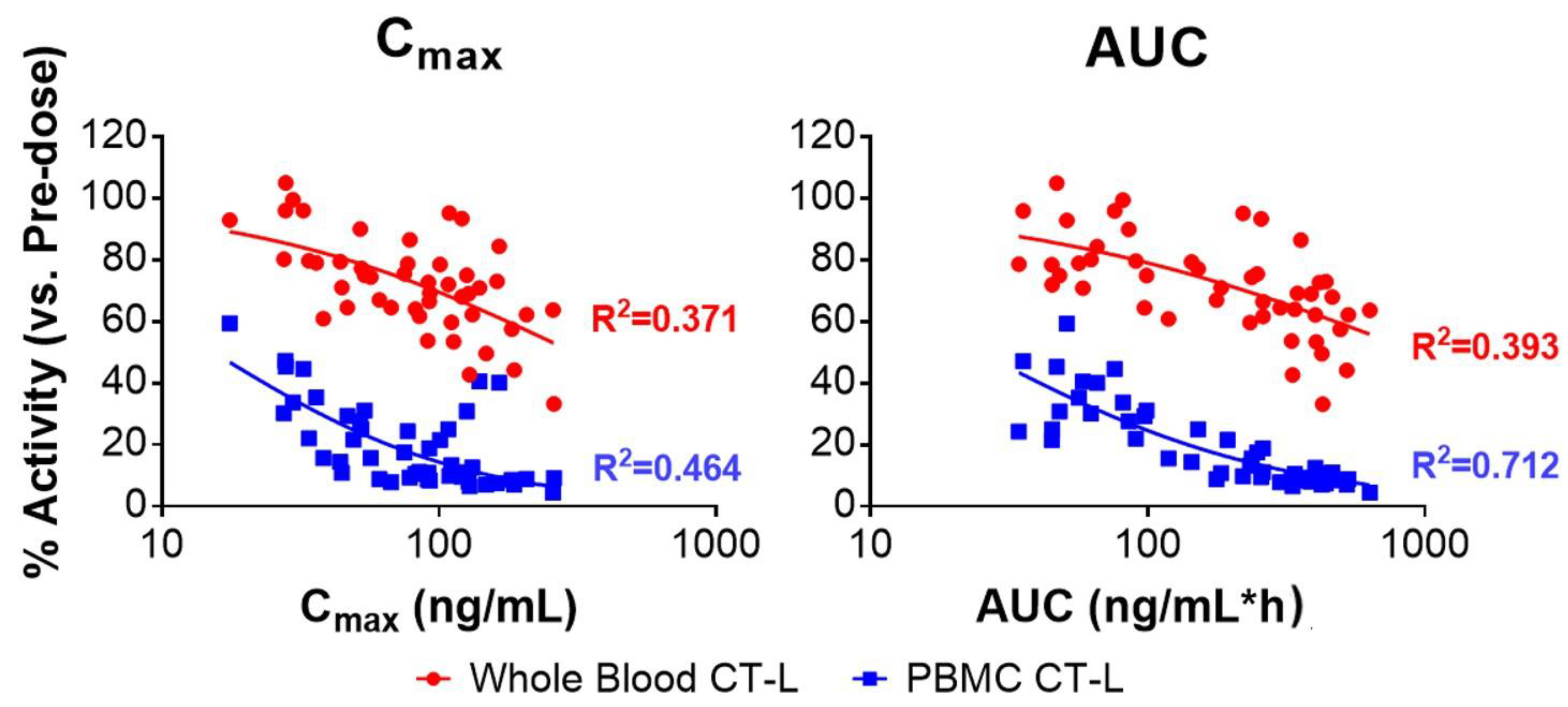
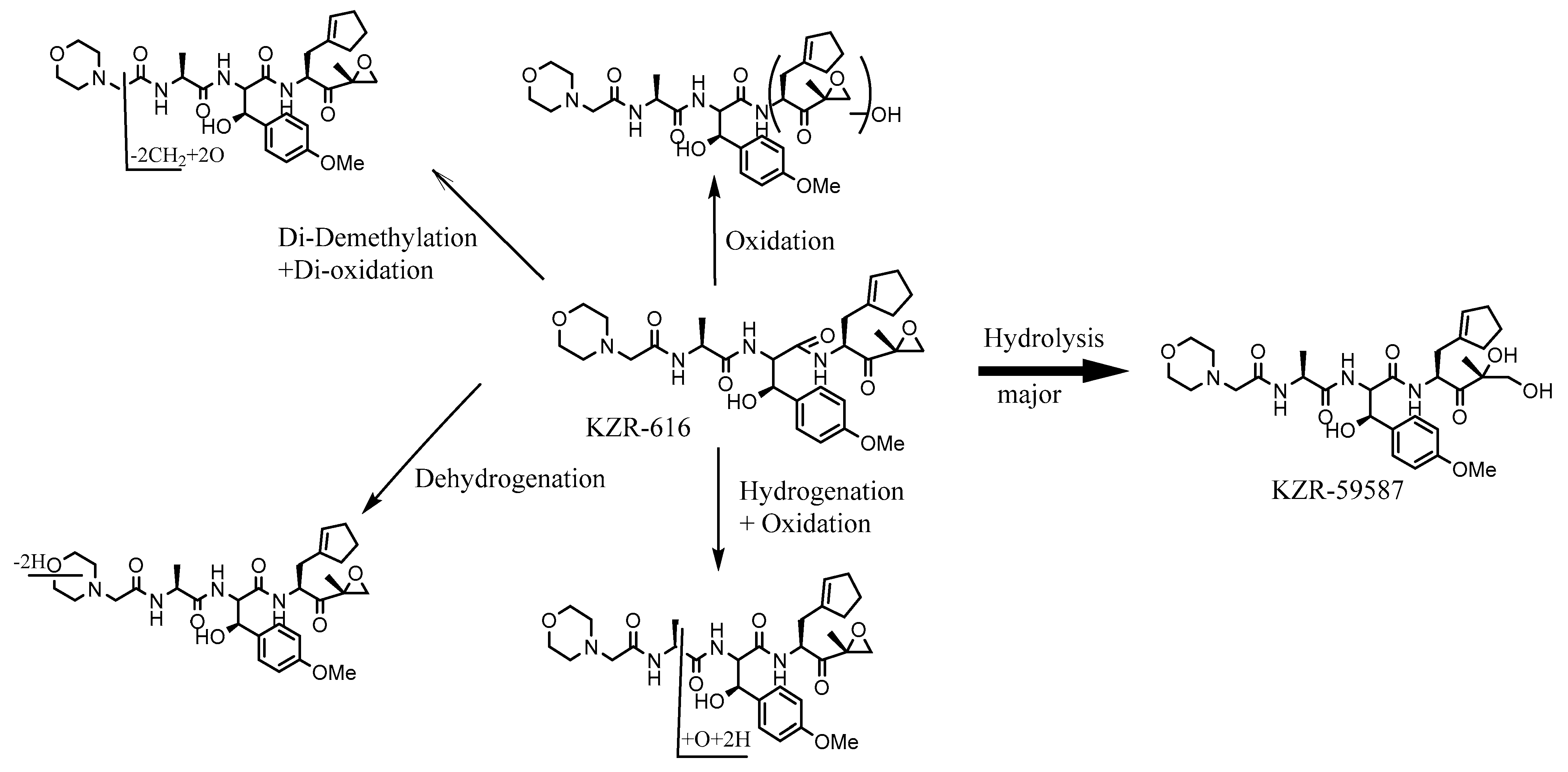
3.3. KZR-616
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Demartino, G.N.; Gillette, T.G. Proteasomes: Machines for all reasons. Cell 2007, 129, 659–662. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Cho, J.; Song, E.J. Ubiquitin-proteasome system (UPS) as a target for anticancer treatment. Arch. Pharm. Res. 2020, 43, 1144–1161. [Google Scholar] [CrossRef]
- Khalil, R. Ubiquitin-proteasome pathway and muscle atrophy. Adv. Exp. Med. Biol. 2018, 1088, 235–248. [Google Scholar] [PubMed]
- Narayanan, S.; Cai, C.Y.; Assaraf, Y.G.; Guo, H.Q.; Cui, Q.; Wei, L.; Huang, J.J.; Ashby, C.R., Jr.; Chen, Z.S. Targeting the ubiquitin-proteasome pathway to overcome anti-cancer drug resistance. Drug Resist Updat. 2020, 48, 100663. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Zhong, M.B.; Toro, C.A.; Zhang, L.; Cai, D. Endo-lysolsomal pathway and ubiquitin-proteasome system dysfunction in Alzheimer’s disease pathogenesis. Neurosci Lett. 2019, 703, 68–78. [Google Scholar] [CrossRef]
- Kudriaeva, A.A.; Belogurov, A.A. Proteasome: A nanomachinery of creative destruction. Biochemistry 2019, 84, 159–192. [Google Scholar] [CrossRef] [PubMed]
- Parlati, F.; Lee, S.J.; Aujay, M.; Suzuki, E.; Levitsky, K.; Lorens, J.B.; Micklem, D.R.; Ruurs, P.; Sylvain, C.; Lu, Y.; et al. Carfilzomib can induce tumor cell death through selective inhibition of the chymotrypsin-like activity of the proteasome. Blood 2009, 114, 3439–3447. [Google Scholar] [CrossRef] [PubMed]
- Cromm, P.M.; Crews, C.M. The proteasome in modern drug discovery: Second life of a highly valuable drug target. ACS Cent Sci. 2017, 3, 830–838. [Google Scholar] [CrossRef] [PubMed]
- Morozov, A.V.; Karpov, V.L. Proteasomes and several aspects of their heterogeneity relevant to cancer. Front. Oncol. 2019, 9, 761. [Google Scholar] [CrossRef]
- Muchamuel, T.; Basler, M.; Aujay, M.A.; Suzuki, E.; Kalim, K.W.; Lauer, C.; Sylvain, C.; Ring, E.R.; Shields, J.; Jiang, J.; et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat. Med. 2009, 15, 781–787. [Google Scholar] [CrossRef]
- Eskandari, S.K.; Seelen, M.A.J.; Lin, G.; Azzi, J.R. The immunoproteasome: An old player with a novel and emerging role in alloimmunity. Am. J. Transplant. 2017, 17, 3033–3039. [Google Scholar] [CrossRef]
- Xi, J.; Zhuang, R.; Kong, L.; He, R.; Zhu, H.; Zhang, J. Immunoproteasome-selective inhibitors: An overview of recent developments as potential drugs for hematologic malignancies and autoimmune diseases. Eur. J. Med. Chem. 2019, 182, 111646. [Google Scholar] [CrossRef] [PubMed]
- Tundo, G.R.; Sbardella, D.; Santoro, A.M.; Coletta, A.; Oddone, F.; Grasso, G.; Milardi, D.; Lacal, P.M.; Marini, S.; Purrello, R.; et al. The proteasome as a druggable target with multiple therapeutic potentialities: Cutting and non-cutting edges. Pharmacol. Ther. 2020, 213, 107579. [Google Scholar] [CrossRef]
- Bross, P.F.; Kane, R.; Farrell, A.T.; Abraham, S.; Benson, K.; Brower, M.E.; Bradley, S.; Gobburu, J.V.; Goheer, A.; Lee, S.L.; et al. Approval summary for bortezomib for injection in the treatment of multiple myeloma. Clin. Cancer Res. 2004, 10, 3954–3964. [Google Scholar] [CrossRef]
- Herndon, T.M.; Deisseroth, A.; Kaminskas, E.; Kane, R.C.; Koti, K.M.; Rothmann, M.D.; Habtemariam, B.; Bullock, J.; Bray, J.D.; Hawes, J. U.S. Food and Drug Administration Approval: Carfilzomib for the Treatment of Multiple. Clin. Cancer Res. 2013, 19, 4559–4563. [Google Scholar] [CrossRef]
- Kumar, S.K.; Jacobus, S.J.; Cohen, A.D.; Weiss, M.; Callander, N.; Singh, A.K.; Parker, T.L.; Menter, A.; Yang, X.; Parsons, B.; et al. Carfilzomib or bortezomib in combination with lenalidomide and dexamethasone for patients with newly diagnosed multiple myeloma without intention for immediate autologous stem-cell transplantation (ENDURANCE): A multicentre, open-label, phase 3, randomized, controlled trial. Lancet Oncol. 2020, 21, 1317–1330. [Google Scholar] [PubMed]
- Dimopoulos, M.A.; Goldschmidt, H.; Niesvizky, R.; Joshua, D.; Chng, W.J.; Oriol, A.; Orlowski, R.Z.; Ludwig, H.; Facon, T.; Hajek, R.; et al. Carfilzomib or bortezomib in relapsed or refractory multiple myeloma (ENDEAVOR): An interim overall survival analysis of an open-label, randomized, phase 3 trial. Lancet Oncol. 2017, 18, 1327–1337. [Google Scholar] [CrossRef]
- Demo, S.D.; Kirk, C.J.; Aujay, M.A.; Buchholz, T.J.; Dajee, M.; Ho, M.N.; Jiang, J.; Laidig, G.J.; Lewis, E.R.; Parlati, F.; et al. Antitumor Activity of PR-171, a Novel Irreversible Inhibitor of the Proteasome. Cancer Res. 2007, 67, 6383–6391. [Google Scholar] [CrossRef]
- Kapur, S.A.; Anderl, J.L.; Kraus, M.; Parlati, F.; Shenk, K.D.; Lee, S.J.; Muchamuel, T.; Bennett, M.K.; Driessen, C., 3rd; Ball, A.J.; et al. Non-proteasomal Targets of the Proteasome Inhibitors Bortezomib and Carfilzomib: A Link to Clinical. Clin. Cancer Res. 2011, 17, 2734–2743. [Google Scholar] [CrossRef] [PubMed]
- Johnson, H.W.B.; Lowe, E.; Anderl, J.L.; Fan, A.; Muchamuel, T.; Bowers, S.; Moebius, D.C.; Kirk, C.; McMinn, D.L. Required Immunoproteasome Subunit Inhibition Profile for Anti-Inflammatory Efficacy and Clinical Candidate KZR-616 ((2S,3R)-N-((S)-3-(Cyclopent-1-en-1-yl)-1-((R)-2-methyloxiran-2-yl)-1oxopropan-2-yl)-3-hydroxy-3-(4-methoxyphenyl)-2-((S)-2-(2morpholinoacetamido)propanamido)propenamide). J. Med. Chem. 2018, 61, 11127–11143. [Google Scholar] [PubMed]
- De Cesco, S.; Kurian, J.; Dufresne, C.; Mittermaier, A.K.; Moitessier, N. Covalent inhibitors design and discovery. Eur. J. Med. Chem. 2017, 138, 96–114. [Google Scholar] [CrossRef]
- De Vita, E. 10 Years into the resurgence of covalent drugs. Future Med. Chem. 2021, 13, 193–210. [Google Scholar] [CrossRef]
- Strelow, J.M. A perspective on the kinetics of covalent and irreversible inhibition. SLAS Discov. 2017, 22, 3–20. [Google Scholar] [CrossRef]
- Groll, M.; Kim, K.B.; Kairies, N.; Huber, R.; Crews, G.M. Crystal structure of epoxomicin: 20S proteasome reveals a molecular basis for selectivity of -epoxyketone proteasome inhibitors. J. Am. Chem. Soc. 2000, 122, 1237–1238. [Google Scholar] [CrossRef]
- Nunes, A.T.; Annunziata, C.M. Proteasome inhibitors: Structure and function. Semin Oncol. 2017, 44, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Schrader, J.; Henneberg, F.; Mata, R.A.; Tittmann, K.; Schneider, T.R.; Stark, H.; Bourenkov, G.; Chari, A. The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design. Science 2016, 353, 594–598. [Google Scholar] [CrossRef] [PubMed]
- Fricker, L.D. Proteasome Inhibitor Drugs. Annu. Rev. Pharmacol. Toxicol. 2020, 60, 457–476. [Google Scholar] [CrossRef] [PubMed]
- Basler, M.; Li, J.; Groettrup, M. On the role of the immunoproteasome in transplant rejection. Iunogenetics 2019, 71, 263–271. [Google Scholar] [CrossRef]
- Muchamuel, T.; Anderl, J.; Fan, R.A.; Johnson, H.W.B.; Kirk, C.J.; Lowe, E. KZR-616, a selective inhibitor of the immunoproteasome, Blocks the disease progression in multiple models of systemic lupus Erythematosus (SLE). In Proceedings of the ACR/ARHP Annual Meeting 2, ACR Poster 2559, New Orleans, LA, USA, 3–8 November 2017. [Google Scholar]
- Huber, E.M.; Basler, M.; Schwab, R.; Heinemeyer, W.; Kirk, C.J. Immuno-and constititive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell 2012, 148, 727–738. [Google Scholar] [CrossRef]
- Groll, M.; Ditzel, L.; Lowe, J.; Stock, D.; Bochtler, M.; Bartunik, H.D.; Huber, R. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature 1997, 386, 463–471. [Google Scholar] [CrossRef]
- Harshbarger, W.; Miller, C.; Diedrich, C.; Sacchettini, J. Crstal structure of the human 20S proteasome in complex with carfilzomib. Structure 2015, 23, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Thibaudeau, T.A.; Smith, D.M. A practical review of proteasome pharmacology. Pharmacol. Rev. 2019, 71, 170–197. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Miller, Z.; Jun, Y.; Lee, W.; Kim, K.B. Next-generation proteasome inhibitors for cancer therapy. Transl. Res. 2018, 198, 1–16. [Google Scholar]
- Moreau, P.; Pylypenko, H.; Grosicki, S.; Karamanesht, L.; Leleu, X.; Grishunina, M.; Pekhtman, G.; Masliak, Z.; Robak, T.; Shubina, A.; et al. Subcutaneous versus intravenous administration of administration of bortezomib in patients with relapsed multiple myeloma: A randomized, phase 3, non-inferiority study. Lancet Oncol. 2011, 12, 431–440. [Google Scholar] [CrossRef]
- Tan, C.R.C.; Abdul-Majeed, S.; Cael, B.; Barta, S.K. Clinical Pharmacokinetics and Pharmacodynamics of Bortezomib. Clin. Pharmacokinet. 2019, 58, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Orlowski, R.Z.; Stinchcombe, T.E.; Mitchell, B.S.; Shea, T.C.; Baldwin, A.S.; Stahl, S.; Adams, J.; Esseltine, D.L.; Elliott, P.J.; Pien, C.S.; et al. Phase I trial of the proteasome inhibitor PS-341 in patients with refractory hematologic malignancies. J. Clin. Oncol. 2002, 20, 4420–4427. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Mager, D.E. Population-based meta-analysis of bortezomib exposure-response relationships in multiple myeloma patients. J. Pharmacokinet. Pharmacodyn. 2020, 47, 77–90. [Google Scholar] [CrossRef]
- Moreau, P.; Karamanesht, L.I.; Domnikova, N.; Kyselyova, M.Y.; Vilchevska, K.V.; Doronin, V.A.; Schmidt, A.; Hulin, C.; Leleu, X.; Esseltine, D.L.; et al. Administration of Bortezomib in patients with relapsed multiple myeloma. Clin. Pharmcokinet. 2012, 51, 823–829. [Google Scholar] [CrossRef]
- Papandreou, C.N.; Daliani, D.D.; Nix, D.; Yang, H.; Madden, T.; Wang, X.; Pien, C.S.; Millikan, R.E.; TU, S.; Pagliaro, L.; et al. Phase I trial of the proteasome inhibitor bortezomib in patients with advanced solid tumors with observations in androgen-independent prostate cancer. J. Clin. Oncol. 2004, 22, 2107–2121. [Google Scholar] [CrossRef]
- Reece, D.E.; Sullivam, D.; Lonial, S.; Mohrbacher, A.F.; Chatta, G.; Shustik, C.; Burris, H.; Venkatakrishnan, K.; Neuwirth, R.W.J.; Karl, M.; et al. Pharmacokinetics and pharmacodynamic study of two doses of bortezomib in patients with relapsed multiple myeloma. Cancer Chemother Pharmocol. 2011, 67, 57–67. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pekol, T.; Daniels, J.S.; Labutti, J.; Parsons, I.; Nix, D.; Baronas, E.; Hsieh, F.; Gan, L.; Miwa, G. Human metabolism of the proteasome inhibitor bortezomib: Identification of circulating metabolites. Drug Metab Dispos. 2005, 33, 771–777. [Google Scholar] [CrossRef]
- Lu, C.; Gallegos, R.; Li, P.; Xia, C.Q.; Pusalkar, S.; Uttamsingh, V.; Nix, D.; Miwa, G.T.; Gan, L. Investigation of drug-drug interaction potential of bortezomib in vivo in female Sprague-dawley rats and in vitro in human microsomes. Drug Metab Dispos. 2006, 34, 702–708. [Google Scholar] [CrossRef]
- Venkatakrishnan, K.; Rader, M.; Ramamathan, R.K.; Ramalingam, S.; Chen, E.; Riordan, W.; Trepicchio, W.; Coope, M.; Karol, M.; Von Moltke, L.; et al. Effect of the CYP3A inhibitor ketoconazole on the pharmacokinetics and pharmacodynamics of bortezomib in patients with advance solid tumors: A prospective, multicenter, open-label, randomized, two-way crossover drug-drug interaction study. Clin. Therapeutics. 2009, 31, 2444–2458. [Google Scholar] [CrossRef]
- Gupta, N.; Hanley, M.J.; Xia, C.; Labotka, R.; Harvey, R.D.; Venkatakrishnan, K. Pharmacology of Ixazomib: The first oral proteasome inhibitor. Clin. Pharmacokinet. 2019, 58, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Assouline, S.E.; Chang, J.; Cheson, B.D.; Rifkin, R.; Hamburg, S.; Reyes, R.; Hui, A.-M.; Yu, J.; Gupta, N.; Di Bacco, A.; et al. Phase I dose-escalation study of IV ixazomib, an investigational proteasome inhibitor, in patients with relapsed/refractory lymphoma. Blood Cancer J. 2014, 4, e251. [Google Scholar] [CrossRef] [PubMed]
- Pusalkar, S.; Plesescu, M.; Gupta, N.; Hanley, M.; Venkakakrishnan, K.; Wu, J.; Xia, C.; Zhang, X.; Chowdhury, S. Biotransformation of [14C]-ixazomib in patients with advanced solid tumors: Characterization of metabolite profiles in plasma, urine, and feces. Cancer Chemother. Pharmacol. 2018, 82, 803–814. [Google Scholar] [CrossRef]
- Kim, K.B.; Crews, C.M. From epoxomicin to carfilzomib: Chemistry, biology, and medical outcomes. Nat. Prod. Rep. 2013, 30, 600–604. [Google Scholar] [CrossRef] [PubMed]
- Dimopoulos, M.A.; Moreau, P.; Palumbo, A.; Joshua, D.; Pour, L.; Hájek, R.; Facon, T.; Ludwig, H.; Oriol, A.; Goldschmidt, H.; et al. Carfilzomib and dexamethasone versus bortezomib and dexamethasone for patients with relapsed or refractory multiple myeloma (ENDEAVOR): A randomized, phase 3, open-label, multicentre study. Lancet Oncol. 2016, 17, 27–38. [Google Scholar] [CrossRef]
- Facon, T.; Lee, J.H.; Moreau, P.; Niesvizky, R.; Dimopoulos, M.; Hajek, R.; Pour, L.; Jurczyszyn, A.; Qiu, L.; Klippel, Z.; et al. Carfilzomib or bortezomib with melphalan-prednisone for transplant-ineligible patients with newly diagnosed multiple myeloma. Blood 2019, 133, 1953–1963. [Google Scholar] [CrossRef]
- Alsina, M.; Trudel, S.; Furman, R.R.; Rosen, P.J.; O’Connor, O.A.; Comezo, R.L.; Wong, A.; Kunkel, L.A.; Molineaux, C.J.; Goy, A. A phase 1 single-agent study of twice-weekly consecutive-day dosing of the proteasome inhibitor carfilzomib in patients with relapsed or refractory multiple myeloma or lymphoma. Clin. Cancer Res. 2012, 18, 4830–4840. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, O.A.; Stewart, A.K.; Vallone, M.; Molineaux, C.J.; Kunkel, L.A.; Gerecitano, J.F.; Orlowski, R.Z. A phase 1 dose escalation study of the safety and pharmacokinetics of the novel proteasome inhibitor carfilzomib (PR-171) in patients with hematologic malignancies. Clin. Cancer Res. 2009, 15, 7085–7091. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Levitsky, K.; Parlati, F.; Bennett, M.K.; Arastu-Kapur, S.; Kellerman, L.; Woo, T.F.; Wong, A.F.; Papadopoulos, K.P.; Nesvizky, R.; et al. Clinical activity of carfilzomib correlates with inhibition of multiple proteasome subunits: Application of a novel pharmacodynamic assay. Br. J. Haematol. 2016, 173, 884–895. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, Z.; Fang, Y.; Jiang, J.; Zhao, F.; Wong, H.; Bennett, M.K.; Molineaux, C.J.; Kirk, C.J. Pharmacokinetics, Pharmacodynamics, Metabolism, Distribution, and Excretion of Carfilzomib in Rat. Drug Metab Dispos. 2011, 39, 1873–1882. [Google Scholar] [CrossRef]
- Brown, J.; Plummer, R.; Bauer, T.M.; Anthony, S.; Sarantopoulos, J.; De Vos, F.; White, M.; Schupp, M.; Ou, Y.; Vaishampayan, U. Pharmacokinetics of carfilzomib in patients with advanced malignancies and varying degrees of hepatic impairment: An open-label, single-arm, phase 1 study. Exp. Hematol. Oncol. 2017, 6, 27. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, J.; Kirk, C.; Fang, Y.; Alsina, M.; Badros, A.; Papadopoulos, K.; Wong, A.; Woo, T.; Bomb, D.; et al. Clinical pharmacokinetics, metabolism, and drug-drug interaction of carfilzomib. Drug Metab Dispos. 2013, 41, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Quach, H.; White, D.; Spencer, A.; Ho, P.J.; Bhutani, D.; White, M.; Inamdar, S.; Morris, C.; Ou, Y.; Gyger, M. Pharmacokinetics and safety of carfilzomib in patients with relapsed multiple myeloma and end-stage renal disease (ESRD): An open-label, single-arm, phase1 study. Cancer Chemother Pharmacol. 2017, 79, 1067–1076. [Google Scholar] [CrossRef]
- Infante, J.R.; Mendelson, D.S.; Burris, H.A., 3rd; Bendell, J.C.; Tolcher, A.W.; Gordon, M.S.; Gillenwater, H.H.; Arastu-Kapur, S.; Wong, H.L.; Papadopoulos, K.P. A first-in-human dose-escalation study of the oral proteasome inhibitor Oprozomib in patients with advanced solid tumors. Investig. New Drugs 2016, 34, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.J.; Aujay, M.A.; Bennett, M.K.; Dajee, M.; Demo, S.D.; Fang, Y.; Ho, M.N.; Jiang, J.; Kirk, C.J.; Laidig, G.J.; et al. Design and synthesis of an orally bioavailable and selective peptide epoxyketone proteasome inhibitor (PR-047). J. Med. Chem. 2009, 52, 3028–3038. [Google Scholar] [CrossRef]
- Ou, Y.; Xu, Y.; Gore, L.; Harvey, R.D.; Mita, A.; Papadopoulos, K.P.; Wang, Z.; Cutler, R.E., Jr.; Pinchasik, D.E.; Tsimberidou, A.M. Physiologically-based pharmacokinetic modelling to predict oprozomib CYP3A drug-drug interaction potential in patients with advanced malignancies. Br. J. Clin. Pharmacol. 2019, 85, 530–539. [Google Scholar] [CrossRef] [PubMed]
- Ghobrial, I.M.; Vij, R.; Siegel, D.; Badros, A.; Kaufman, J.; Raje, N.; Jakubowiak, A.; Savona, M.R.; Obreja, M.; Berdeja, J.G. A phase 1b/II study of oprozomib in patients with advanced multiple myeloma and Waldenström Macroglobulinemia. Clin. Cancer Res. 2019, 25, 4907–4916. [Google Scholar] [CrossRef]
- Wang, Z.; Fang, Y.; Teague, J.; Wong, H.; Morisseau, C.; Hammock, B.D.; Rock, D.A.; Wang, Z. In Vitro Metabolism of Oprozomib, an Oral Proteasome Inhibitor: Role of Epoxide Hydrolases and Cytochrome P450s. Drug Metab Dispos. 2017, 45, 712–720. [Google Scholar] [CrossRef] [PubMed]
- Zerfast, B.L.; Maresh, M.E.; Trader, D.J. The immunoproteasome: Am emerging target in cancer and autoimmune and neurological disorders. J. Med. Chem. 2020, 63, 1841–1858. [Google Scholar] [CrossRef]
- Huber, E.M.; Groll, M. A nut for every bolt: Subunit-selective inhibitors of the immunoproteasome ad their therapeutic potential. Cells 2021, 10, 1929. [Google Scholar] [CrossRef]
- Lickliter, J.; Bomba, D.; Anderl, J.L.; Kirk, C.J.; Wang, J. KZR-616, a selective inhibitor of the immunoproteasome, shows a promising safety and target inhibition profile in a phase I, double-blind, single (SAD) and multiple ascending dose (MAD) study in healthy volunteers. In Proceedings of the ACR/ARHP Auuual Meeting Poster, ACR Poster 2587, New Orleans, LA, USA, 19–24 October 2018. [Google Scholar]
- Fang, Y.; Johnson, H.; Anderl, J.L.; Muchamuel, T.; McMinn, D.; Morisseau, C.; Hammock, B.D.; Kirk, C.; Wang, J. Role of epoxide hydrolases and cytochrome P450s on metabolism of KZR-616, 1 first-in-class selective inhibitor of the immunoproteasome. Drug Metab Dispos. 2021, 49, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Moreau, P.; Coiteux, V.; Hulin, C.; Leleu, X.; van de Velde, H.; Acharya, M.; Harousseau, J.L. Prospective comparison of subcutaneous versus intravenous administration of bortezomib in patients with multiple myeloma. Haematologica 2008, 93, 1908–1911. [Google Scholar] [CrossRef]
- Hellmann, A.; Rule, S.; Walewski, J.; Shpiberg, O.; Feng, H.; van de Velde, H.; Patel, H.; Skee, D.M.; Girgis, S.; Louw, V.J.; et al. Effect of cytochrome P450 3A4 inducers on the pharmacokinetic, pharmacodynamic and safety profiles of bortezomib in patients with multiple myeloma or non-Hodgkin’s lymphoma. Clin. Pharmacokinet. 2011, 50, 781–791. [Google Scholar] [CrossRef]
- Wagner, K.M.; Gomes, A.; McReynolds, C.B.; Hammock, B.D. Soluble epoxide hydrolase regulation of lipid mediators limits pain. Neurotherapeutics 2020, 17, 900–916. [Google Scholar] [CrossRef] [PubMed]
- Kodani, S.D.; Morisseau, C. Role of epoxy-fatty acids and epoxide hydrolases in the pathology of neuro-inflammation. Biochimie 2019, 159, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Gautheron, J.; Jeru, I. The multifaceted role of epoxide hydrolases in human health. Int. J. Mol. Sci. 2021, 22, 13. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xu, X.; Cong, X.; Zeng, Z.; Xu, L.; Luo, X. Interactions between polycyclic aromatic hydrocarbons and epoxide hydrolase 1 play roles in asthma. Environ. Geochem. Health 2019, 41, 191–210. [Google Scholar] [CrossRef]
- Abdull Razis, A.F.; Konsure, N.; Loannides, C. Isothiocynates and xenobiotic detoxifiction. Mol. Nutr. Food Res. 2018, 62, e1700916. [Google Scholar] [CrossRef]
- Coller, J.K.; Fritz, P.; Zanger, U.M.; Siegle, I.; Eichelbaum, M.; Kroemer, H.K.; Mürdter, T.E. Distribution of microsomal epoxide hydrolase in humans: An immunohistochemical study in normal tissues, and benign and malignant tumours. Histol. J. 2001, 33, 329–336. [Google Scholar]
- Larsson, C.; White, I.; Johansson, C.; Stark, A.; Meijer, J. Localization of the human soluble epoxide hydrolase gene (EPHX2) to chromosomal region 8p21-p12. Hum. Genet. 1995, 95, 356–358. [Google Scholar] [CrossRef]
- El-Sherbeni, A.A.; El-Kadi, A.O.S. The role of epoxide hydrolases in health and disease. Arch. Toxicol. 2014, 88, 2013–2032. [Google Scholar] [CrossRef]
- Xia, R.; Sun, L.; Liao, J.; You, X.; Xu, D.; Yang, J.; Hwang, S.H.; Jones, R.D.; Hammock, B.; Yang, G. Inhibition of pancreatic carcinoma growth through enhancing ω-3 epoxy polyunsaturated fatty acid profile by inhibition of soluble epoxide hydrolase. Anticancer Res. 2019, 39, 3651–3660. [Google Scholar] [CrossRef]
- Paigrahy, D.; Gartung, A.; Yang, J.; Yang, H.; Gilligan, M.M.; Sulciner, M.L.; Bhasin, S.S.; Bielenberg, D.R.; Chang, J.; Schmidt, B.A.; et al. Preoperative stimulation of resolution and inflammation blockade eradicates micrometastases. J. Clin. Investig. 2019, 129, 2964–2979. [Google Scholar] [CrossRef]
- Panigraphy, D.; Gilligan, M.M.; Huamg, S.; Gartung, A.; Cortés-Puch, I.; Sime, P.J.; Phipps, R.P.; Serhan, C.N.; Hammock, B.D. Inflammation resolution: A dual-pronged approach to averting cytokine storms in COVID-19? Cancer Metastasis Rev. 2020, 39, 337–340. [Google Scholar] [CrossRef]
- Edin, M.L.; Hamedani, B.G.; Gruzdev, A.; Graves, J.P.; Lih, F.B.; Arbes, S.J.; Singh, R.; Leon, A.C.O.; Bradbury, J.A. Epoxide hydrolase 1 (EPHX1) hydrolyzes epoxyeicosanoids and impairs cardiac recovery after ischemia. J. Biol. Chem. 2018, 293, 3281–3292. [Google Scholar] [CrossRef] [PubMed]
- Saenz-Mendez, P.; Kztz, A.; Perez-Kempner, M.L.; Ventura, O.N.; Vazquez, M. Structural insights into human microsomal epoxide hydrolase by combined homology modeling, molecular dynamics simulations, and molecular docking calculations. Proteins Struct. Funct. Genet. 2017, 85, 720–730. [Google Scholar] [CrossRef]
- Lee, M.J.; Bhattarai, D.; Yoo, J.; Miller, Z.; Park, J.E.; Lee, S.; Lee, W.; Driscoll, J.J.; Kim, K.B. Development of novel epoxyketone-based proteasome inhibitors as a strategy to overcome cancer resistance to carfilzomib and bortezomib. J. Med. Chem. 2019, 62, 4444–4455. [Google Scholar] [CrossRef] [PubMed]



| Proteasome Inhibitors | Warhead | Class | Targets | Administration | Absorption | Clearance | Subunit Inhibition | Metabolic Pathwas |
|---|---|---|---|---|---|---|---|---|
| Bortezomib | Boronate | reversible | Constitutive and immunoproteasome | IV and SC | fast | fast | ß5 and LMP7 | Hydrolases, CYPs |
| Ixazomib | Boronate | reversible | Constitutive and immunoproteasome | IV and Oral | fast | fast | ß5 and LMP7 | Hydrolases, CYPs |
| Carfilzomib | Epoxyketone | Irreversible | Constitutive and immunoproteasome | IV | fast | fast | ß5 and LMP7 | Microsomal epoxide hydrolase peptidases, |
| Oprozomib | Epoxyketone | Irreversible | Constitutive and immunoproteasome | Oral | fast | fast | ß5 and LMP7 | Microsomal epoxide hydrolase peptidases, CYPs |
| KZR-616 | Epoxyketone | Irreversible | Immunoproteasome | SC | fast | fast | LMP2 and LMP7 | Microsomal epoxide hydrolase |
| Component Label | Identification | Retention Time (min) | [M + H]+ (m/z) | Component in Plasma as % of Total aXIC (Extracted Ion Chromatogram) Area | |||
|---|---|---|---|---|---|---|---|
| Subject | |||||||
| 110-004 | 112-021 | 113-002 | 116-012 | ||||
| KZR-616 | Parent | 22.0 | 587.3 | 61.2 | 49.0 | 46.5 | 51.8 |
| KZR-59587 | Epoxide hydrolysis | 16.4 | 605.3 | 33.8 | 45.8 | 47.9 | 43.8 |
| M605_2 | Epoxide hydrolysis (isomer) | 14.5 | 605.3 | 0.270 | 0.270 | 0.270 | 0.280 |
| M603_1 | Oxidation | 11.0 | 603.3 | 0.080 | 0.100 | 0.060 | 0.060 |
| M603_7 | Oxidation | 11.8 | 603.3 | 0.100 | 0.250 | 0.040 | 0.170 |
| M603_8 | Oxidation | 13.3 | 603.3 | 0.150 | 0.190 | 0.150 | 0.190 |
| M603_2 | Oxidation | 15.0 | 603.3 | 0.080 | 0.020 | 0.080 | 0.060 |
| M603_3 | Oxidation | 17.5 | 603.3 | 0.140 | 0.110 | 0.150 | 0.100 |
| M603-4 | Hydrolysis + dehydrogenation | 20.2 | 603.3 | 0.280 | 0.420 | 0.290 | 0.240 |
| M603_5 | Hydrolysis + dehydrogenation | 20.4 | 603.3 | 0.260 | 0.260 | 0.280 | 0.240 |
| M603_6 | Oxidation | 22.8 | 603.3 | 0.390 | 0.340 | 0.410 | 0.330 |
| M601 | Oxidation + dehydrogenation | 14.4 | 601.3 | 0.890 | 0.870 | 0.800 | 0.910 |
| M619_1 | Double Oxidation | 9.71 | 619.3 | 0.280 | 0.280 | 0.160 | 0.190 |
| M619_2 | Double Oxidation | 19.1 | 619.3 | 0.120 | 0.170 | 0.030 | 0.020 |
| ID | KZR-59177 | KZR-616 | KZR-59240 |
|---|---|---|---|
| T1/2 (h) | 1.44 ± 0.420 | 1.05 ± 0.130 | 0.980 ± 0.264 |
| Tmax (h) | 0.792 ± 0.417 | 0.333 ± 0.136 | 0.500 ± 0.334 |
| Cmax (ng/mL) | 182 ± 31.5 | 441 ± 114 | 803 ± 260 |
| AUCinf (h*ng/mL) | 447± 57.3 | 740 ± 188 | 1412 ± 279 |
| AUC Ratio (diol/parent) | 3.35 ± 1.20 | 3.40 ± 0.808 | 0.777 ± 0.464 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Fang, Y.; Fan, R.A.; Kirk, C.J. Proteasome Inhibitors and Their Pharmacokinetics, Pharmacodynamics, and Metabolism. Int. J. Mol. Sci. 2021, 22, 11595. https://doi.org/10.3390/ijms222111595
Wang J, Fang Y, Fan RA, Kirk CJ. Proteasome Inhibitors and Their Pharmacokinetics, Pharmacodynamics, and Metabolism. International Journal of Molecular Sciences. 2021; 22(21):11595. https://doi.org/10.3390/ijms222111595
Chicago/Turabian StyleWang, Jinhai, Ying Fang, R. Andrea Fan, and Christopher J. Kirk. 2021. "Proteasome Inhibitors and Their Pharmacokinetics, Pharmacodynamics, and Metabolism" International Journal of Molecular Sciences 22, no. 21: 11595. https://doi.org/10.3390/ijms222111595
APA StyleWang, J., Fang, Y., Fan, R. A., & Kirk, C. J. (2021). Proteasome Inhibitors and Their Pharmacokinetics, Pharmacodynamics, and Metabolism. International Journal of Molecular Sciences, 22(21), 11595. https://doi.org/10.3390/ijms222111595





