Metabolic Reprogramming in COVID-19
Abstract
:1. Introduction
2. Metabolism Reprogramming in COVID-19 Infection
2.1. Amino Acid Metabolism
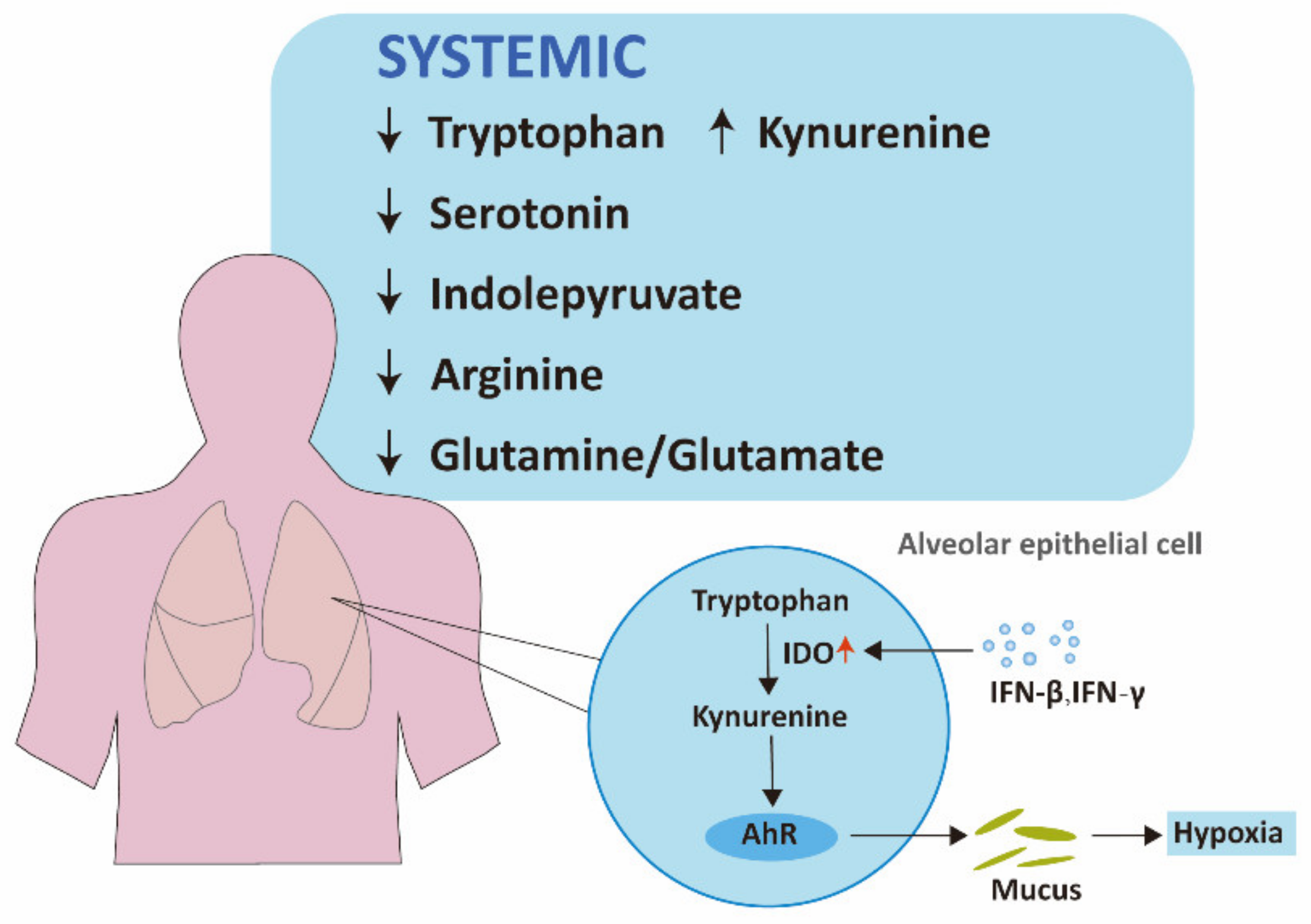
2.1.1. Tryptophan Pathways
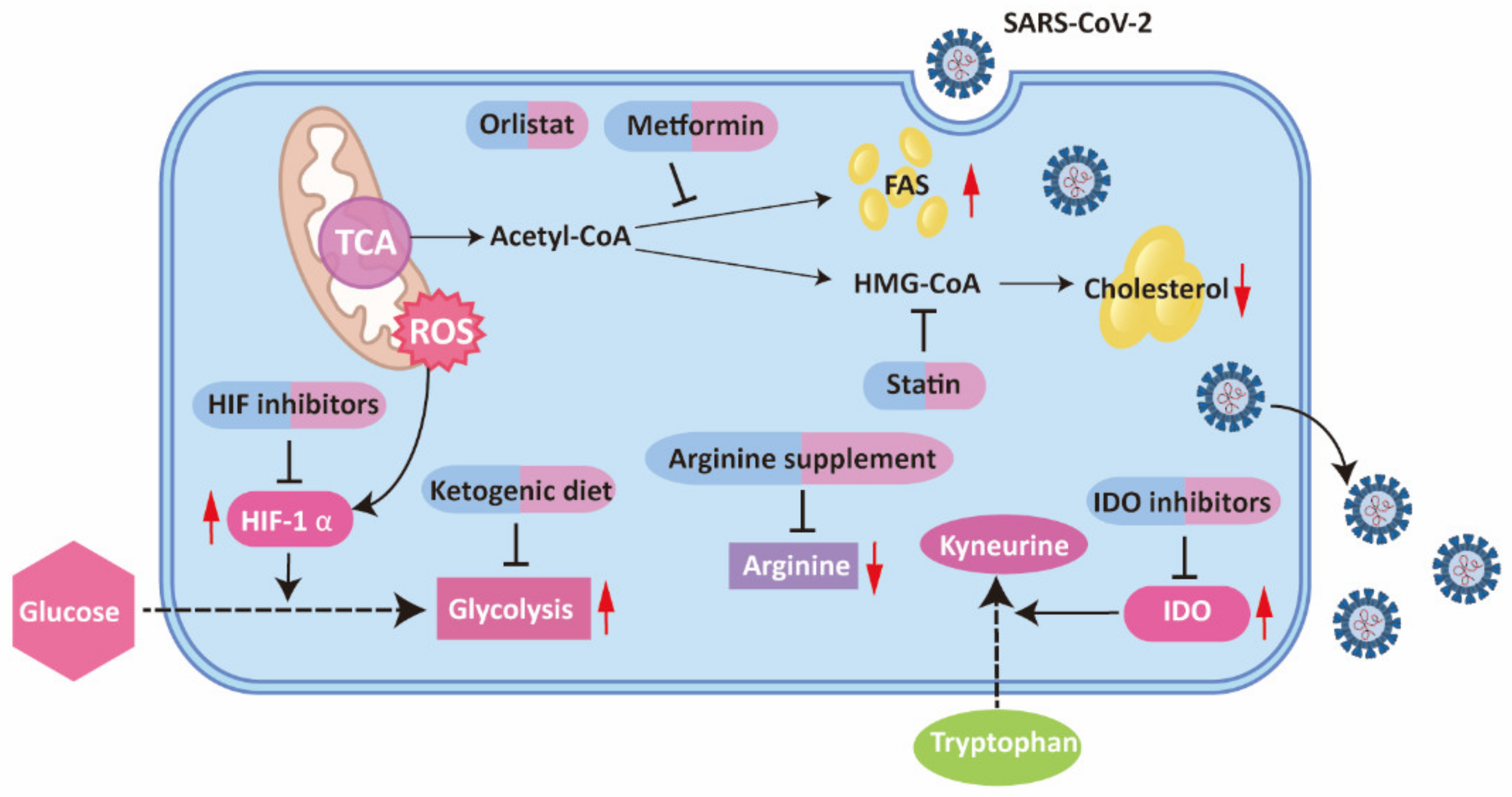
2.1.2. Arginine Reprogramming
2.1.3. Glutamine Metabolism
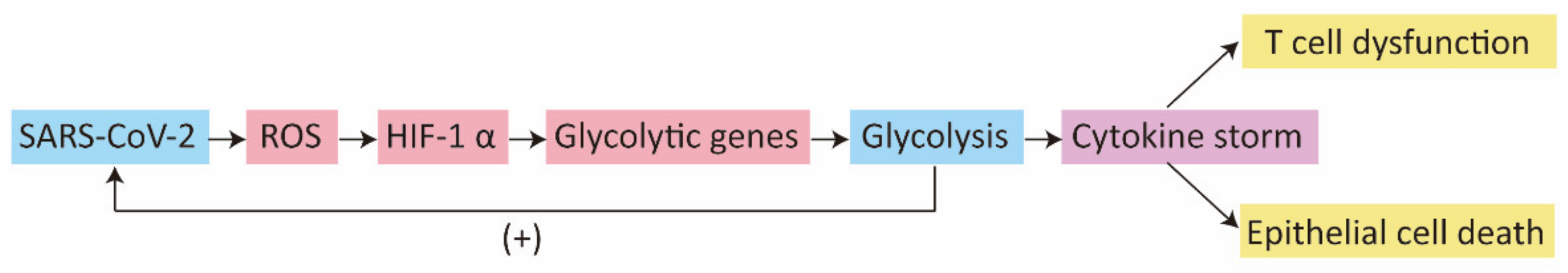
2.2. Glucose Metabolism and HIF
2.3. Cholesterol Metabolism
2.4. Fatty Acid Metabolism
3. Conclusions and Future Direction
Author Contributions
Funding
Conflicts of Interest
References
- Moreno-Altamirano, M.M.B.; Kolstoe, S.E.; Sánchez-García, F.J. Virus Control of Cell Metabolism for Replication and Evasion of Host Immune Responses. Front. Cell. Infect. Microbiol. 2019, 9, 95. [Google Scholar] [CrossRef]
- Thaker, S.K.; Ch’ng, J.; Christofk, H.R. Viral hijacking of cellular metabolism. BMC Biol. 2019, 17, 59. [Google Scholar] [CrossRef]
- Sanchez, E.L.; Lagunoff, M. Viral activation of cellular metabolism. Virology 2015, 479–480, 609–618. [Google Scholar] [CrossRef] [Green Version]
- Goodwin, C.M.; Xu, S.; Munger, J. Stealing the Keys to the Kitchen: Viral Manipulation of the Host Cell Metabolic Network. Trends Microbiol. 2015, 23, 789–798. [Google Scholar] [CrossRef]
- Twomey, J.D.; Luo, S.; Dean, A.Q.; Bozza, W.P.; Nalli, A.; Zhang, B. COVID-19 update: The race to therapeutic development. Drug Resist. Updates 2020, 53, 100733. [Google Scholar] [CrossRef] [PubMed]
- Asselah, T.; Durantel, D.; Pasmant, E.; Lau, G.; Schinazi, R.F. COVID-19: Discovery, diagnostics and drug development. J. Hepatol. 2021, 74, 168–184. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Chi, J.; Lv, W.; Wang, Y. Obesity and diabetes as high-risk factors for severe coronavirus disease 2019 (Covid-19). Diabetes Metab. Res. Rev. 2021, 37, e3377. [Google Scholar] [CrossRef]
- Drucker, D.J. Diabetes, obesity, metabolism, and SARS-CoV-2 infection: The end of the beginning. Cell Metab. 2021, 33, 479–498. [Google Scholar] [CrossRef]
- Loftus, R.M.; Finlay, D.K. Immunometabolism: Cellular Metabolism Turns Immune Regulator. J. Biol. Chem. 2016, 291, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Grohmann, U.; Bronte, V. Control of immune response by amino acid metabolism. Immunol. Rev. 2010, 236, 243–264. [Google Scholar] [CrossRef]
- Tomé, D. Amino acid metabolism and signalling pathways: Potential targets in the control of infection and immunity. Nutr. Diabetes 2021, 11, 20. [Google Scholar] [CrossRef]
- Hirabara, S.M.; Gorjao, R.; Levada-Pires, A.C.; Masi, L.N.; Hatanaka, E.; Cury-Boaventura, M.F.; da Silva, E.B.; Santos-Oliveira, L.C.D.; Sousa Diniz, V.L.; Serdan, T.A.D.; et al. Host cell glutamine metabolism as a potential antiviral target. Clin. Sci. 2021, 135, 305–325. [Google Scholar] [CrossRef]
- Badawy, A.A.B. Tryptophan availability for kynurenine pathway metabolism across the life span: Control mechanisms and focus on aging, exercise, diet and nutritional supplements. Neuropharmacology 2017, 112, 248–263. [Google Scholar] [CrossRef]
- Bender, D.A. Effects of a dietary excess of leucine on the metabolism of tryptophan in the rat: A mechanism for the pellagragenic action of leucine. Br. J. Nutr. 1983, 50, 25–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cervenka, I.; Agudelo, L.Z.; Ruas, J.L. Kynurenines: Tryptophan’s metabolites in exercise, inflammation, and mental health. Science 2017, 357, eaaf9794. [Google Scholar] [CrossRef] [Green Version]
- Platten, M.; Nollen, E.A.A.; Röhrig, U.F.; Fallarino, F.; Opitz, C.A. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 2019, 18, 379–401. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.K.; Park, H.J.; Macleod, M.; Chandler, P.; Munn, D.H.; Mellor, A.L. Tryptophan deprivation sensitizes activated T cells to apoptosis prior to cell division. Immunology 2002, 107, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Munn, D.H.; Mellor, A.L. Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol. 2013, 34, 137–143. [Google Scholar] [CrossRef] [Green Version]
- Lionetto, L.; Ulivieri, M.; Capi, M.; De Bernardini, D.; Fazio, F.; Petrucca, A.; Pomes, L.M.; De Luca, O.; Gentile, G.; Casolla, B.; et al. Increased kynurenine-to-tryptophan ratio in the serum of patients infected with SARS-CoV2: An observational cohort study. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166042. [Google Scholar] [CrossRef]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72. [Google Scholar] [CrossRef]
- Thomas, T.; Stefanoni, D.; Reisz, J.A.; Nemkov, T.; Bertolone, L.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hansen, K.C.; Hod, E.A.; et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Kimhofer, T.; Lodge, S.; Whiley, L.; Gray, N.; Loo, R.L.; Lawler, N.G.; Nitschke, P.; Bong, S.-H.; Morrison, D.L.; Begum, S.; et al. Integrative Modeling of Quantitative Plasma Lipoprotein, Metabolic, and Amino Acid Data Reveals a Multiorgan Pathological Signature of SARS-CoV-2 Infection. J. Proteome Res. 2020, 19, 4442–4454. [Google Scholar] [CrossRef]
- Xiao, N.; Nie, M.; Pang, H.; Wang, B.; Hu, J.; Meng, X.; Li, K.; Ran, X.; Long, Q.; Deng, H.; et al. Integrated cytokine and metabolite analysis reveals immunometabolic reprogramming in COVID-19 patients with therapeutic implications. Nat. Commun. 2021, 12, 1618. [Google Scholar] [CrossRef]
- Soria-Castro, R.; Meneses-Preza, Y.G.; Rodríguez-López, G.M.; Romero-Ramírez, S.; Sosa-Hernández, V.A.; Cervantes-Díaz, R.; Pérez-Fragoso, A.; Torres-Ruíz, J.J.; Gómez-Martín, D.; Campillo-Navarro, M.; et al. Severe COVID-19 is marked by dysregulated serum levels of carboxypeptidase A3 and serotonin. J. Leukoc. Biol. 2021, 110, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Reizine, F.; Lesouhaitier, M.; Gregoire, M.; Pinceaux, K.; Gacouin, A.; Maamar, A.; Painvin, B.; Camus, C.; Le Tulzo, Y.; Tattevin, P.; et al. SARS-CoV-2-Induced ARDS Associates with MDSC Expansion, Lymphocyte Dysfunction, and Arginine Shortage. J. Clin. Immunol. 2021, 41, 515–525. [Google Scholar] [CrossRef]
- Kim, S.; Miller, B.J.; Stefanek, M.E.; Miller, A.H. Inflammation-induced activation of the indoleamine 2,3-dioxygenase pathway: Relevance to cancer-related fatigue. Cancer 2015, 121, 2129–2136. [Google Scholar] [CrossRef] [PubMed]
- Takikawa, O.; Kuroiwa, T.; Yamazaki, F.; Kido, R. Mechanism of interferon-gamma action. Characterization of indoleamine 2,3-dioxygenase in cultured human cells induced by interferon-gamma and evaluation of the enzyme-mediated tryptophan degradation in its anticellular activity. J. Biol. Chem. 1988, 263, 2041–2048. [Google Scholar] [CrossRef]
- Liu, Y.; Lv, J.; Liu, J.; Li, M.; Xie, J.; Lv, Q.; Deng, W.; Zhou, N.; Zhou, Y.; Song, J.; et al. Mucus production stimulated by IFN-AhR signaling triggers hypoxia of COVID-19. Cell Res. 2020, 30, 1078–1087. [Google Scholar] [CrossRef]
- Lemos, H.; Huang, L.; Prendergast, G.C.; Mellor, A.L. Immune control by amino acid catabolism during tumorigenesis and therapy. Nat. Rev. Cancer 2019, 19, 162–175. [Google Scholar] [CrossRef]
- Prendergast, G.C.; Malachowski, W.P.; DuHadaway, J.B.; Muller, A.J. Discovery of IDO1 Inhibitors: From Bench to Bedside. Cancer Res. 2017, 77, 6795–6811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, S.M. Arginine Metabolism Revisited. J. Nutr. 2016, 146, 2579S–2586S. [Google Scholar] [CrossRef]
- Patel, J.J.; Miller, K.R.; Rosenthal, C.; Rosenthal, M.D. When Is It Appropriate to Use Arginine in Critical Illness? Nutr. Clin. Pract. 2016, 31, 438–444. [Google Scholar] [CrossRef]
- Rodriguez, P.C.; Quiceno, D.G.; Ochoa, A.C. L-arginine availability regulates T-lymphocyte cell-cycle progression. Blood 2007, 109, 1568–1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geiger, R.; Rieckmann, J.C.; Wolf, T.; Basso, C.; Feng, Y.; Fuhrer, T.; Kogadeeva, M.; Picotti, P.; Meissner, F.; Mann, M.; et al. L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell 2016, 167, 829–842. [Google Scholar] [CrossRef] [Green Version]
- Rees, C.A.; Rostad, C.A.; Mantus, G.; Anderson, E.J.; Chahroudi, A.; Jaggi, P.; Wrammert, J.; Ochoa, J.B.; Ochoa, A.; Basu, R.K.; et al. Altered amino acid profile in patients with SARS-CoV-2 infection. Proc. Natl. Acad. Sci. USA 2021, 118, e2101708118. [Google Scholar] [CrossRef]
- Rosenthal, M.D.; Carrott, P.W.; Patel, J.; Kiraly, L.; Martindale, R.G. Parenteral or Enteral Arginine Supplementation Safety and Efficacy. J. Nutr. 2016, 146, 2594S–2600S. [Google Scholar] [CrossRef] [Green Version]
- Grimes, J.M.; Khan, S.; Badeaux, M.; Rao, R.M.; Rowlinson, S.W.; Carvajal, R.D. Arginine depletion as a therapeutic approach for patients with COVID-19. Int. J. Infect. Dis. 2021, 102, 566–570. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, S.; Singh, M.; Kirtipal, N.; Kang, S.G. SARS-CoV-2 and Glutamine: SARS-CoV-2 Triggered Pathogenesis Metabolic Reprograming of Glutamine in Host Cells. Front. Mol. Biosci. 2020, 7, 627842. [Google Scholar] [CrossRef] [PubMed]
- Cruzat, V.; Macedo Rogero, M.; Noel Keane, K.; Curi, R.; Newsholme, P. Glutamine: Metabolism and Immune Function, Supplementation and Clinical Translation. Nutrients 2018, 10, 1564. [Google Scholar] [CrossRef] [Green Version]
- Altman, B.J.; Stine, Z.E.; Dang, C.V. From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat. Rev. Cancer 2016, 16, 619–634. [Google Scholar] [CrossRef]
- Lacey, J.M.; Wilmore, D.W. Is glutamine a conditionally essential amino acid? Nutr. Rev. 1990, 48, 297–309. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, R.; Kim, S.H.; Shah, H.; Zhang, S.; Liang, J.H.; Fang, Y.; Gentili, M.; Leary, C.N.O.; Elledge, S.J.; et al. SARS-CoV-2 hijacks folate and one-carbon metabolism for viral replication. Nat. Commun. 2021, 12, 1676. [Google Scholar] [CrossRef]
- Mullen, P.J.; Garcia, G.; Purkayastha, A.; Matulionis, N.; Schmid, E.W.; Momcilovic, M.; Sen, C.; Langerman, J.; Ramaiah, A.; Shackelford, D.B.; et al. SARS-CoV-2 infection rewires host cell metabolism and is potentially susceptible to mTORC1 inhibition. Nat. Commun. 2021, 12, 1876. [Google Scholar] [CrossRef] [PubMed]
- Forman, H.J.; Zhang, H.; Rinna, A. Glutathione: Overview of its protective roles, measurement, and biosynthesis. Mol. Aspects. Med. 2009, 30, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Bartolini, D.; Stabile, A.M.; Bastianelli, S.; Giustarini, D.; Pierucci, S.; Busti, C.; Vacca, C.; Gidari, A.; Francisci, D.; Castronari, R.; et al. SARS-CoV2 infection impairs the metabolism and redox function of cellular glutathione. Redox Biol. 2021, 45, 102041. [Google Scholar] [CrossRef]
- McCaddon, A.; Regland, B. COVID-19: A methyl-group assault? Med. Hypotheses 2021, 149, 110543. [Google Scholar] [CrossRef]
- Khomich, O.A.; Kochetkov, S.N.; Bartosch, B.; Ivanov, A.V. Redox Biology of Respiratory Viral Infections. Viruses 2018, 10, 392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cengiz, M.; Borku Uysal, B.; Ikitimur, H.; Ozcan, E.; Islamoğlu, M.S.; Aktepe, E.; Yavuzer, H.; Yavuzer, S. Effect of oral l-Glutamine supplementation on Covid-19 treatment. Clin. Nutr. Exp. 2020, 33, 24–31. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weidemann, A.; Johnson, R.S. Biology of HIF-1alpha. Cell. Death Differ. 2008, 15, 621–627. [Google Scholar] [CrossRef] [Green Version]
- Warburg, O.; Wind, F.; Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, H.; Liu, C.; Li, D.; Huang, Q.; Liu, D.; Zhang, Y.; Ye, C.; Zhou, D.; Wang, Y.; Tan, Y.; et al. Metabolomic analyses reveals new stage-specific features of the COVID-19. Eur. Respir. J. 2021. [Google Scholar] [CrossRef]
- Bojkova, D.; Klann, K.; Koch, B.; Widera, M.; Krause, D.; Ciesek, S.; Cinatl, J.; Münch, C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature 2020, 583, 469–472. [Google Scholar] [CrossRef] [PubMed]
- Qi, F.; Zhang, W.; Huang, J.; Fu, L.; Zhao, J. Single-Cell RNA Sequencing Analysis of the Immunometabolic Rewiring and Immunopathogenesis of Coronavirus Disease 2019. Front. Immunol. 2021, 12, 651656. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, S.; Xia, H.; Guo, J.; He, K.; Huang, C.; Luo, R.; Chen, Y.; Xu, K.; Gao, H.; et al. Identification of Monocytes Associated with Severe COVID-19 in the PBMCs of Severely Infected patients Through Single-Cell Transcriptome Sequencing. Engineering 2021. [Google Scholar] [CrossRef]
- Ajaz, S.; McPhail, M.J.; Singh, K.K.; Mujib, S.; Trovato, F.M.; Napoli, S.; Agarwal, K. Mitochondrial metabolic manipulation by SARS-CoV-2 in peripheral blood mononuclear cells of patients with COVID-19. Am. J. Physiol. Cell Physiol. 2021, 320, C57–C65. [Google Scholar] [CrossRef]
- Codo, A.C.; Davanzo, G.G.; Monteiro, L.d.B.; de Souza, G.F.; Muraro, S.P.; Virgilio-da-Silva, J.V.; Prodonoff, J.S.; Carregari, V.C.; de Biagi Junior, C.A.O.; Crunfli, F.; et al. Elevated Glucose Levels Favor SARS-CoV-2 Infection and Monocyte Response through a HIF-1α/Glycolysis-Dependent Axis. Cell Metab. 2020, 32, 437–446. [Google Scholar] [CrossRef]
- Semenza, G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 2003, 3, 721–732. [Google Scholar] [CrossRef]
- Appelberg, S.; Gupta, S.; Svensson Akusjärvi, S.; Ambikan, A.T.; Mikaeloff, F.; Saccon, E.; Végvári, Á.; Benfeitas, R.; Sperk, M.; Ståhlberg, M.; et al. Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. Emerg. Microbes Infect. 2020, 9, 1748–1760. [Google Scholar] [CrossRef] [PubMed]
- Palazon, A.; Goldrath, A.W.; Nizet, V.; Johnson, R.S. HIF transcription factors, inflammation, and immunity. Immunity 2014, 41, 518–528. [Google Scholar] [CrossRef] [Green Version]
- Peng, T.; Du, S.-Y.; Son, M.; Diamond, B. HIF-1α is a negative regulator of interferon regulatory factors: Implications for interferon production by hypoxic monocytes. Proc. Natl. Acad. Sci. USA 2021, 118, e2106017118. [Google Scholar] [CrossRef] [PubMed]
- Weber, D.D.; Aminzadeh-Gohari, S.; Tulipan, J.; Catalano, L.; Feichtinger, R.G.; Kofler, B. Ketogenic diet in the treatment of cancer—Where do we stand? Mol. Metab. 2020, 33, 102–121. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.; Shchukina, I.; Youm, Y.-H.; Qing, H.; Hilliard, B.; Dlugos, T.; Zhang, X.; Yasumoto, Y.; Booth, C.J.; Fernández-Hernando, C.; et al. Ketogenic diet restrains aging-induced exacerbation of coronavirus infection in mice. Elife 2021, 10, e66522. [Google Scholar] [CrossRef]
- Goldberg, E.L.; Molony, R.D.; Kudo, E.; Sidorov, S.; Kong, Y.; Dixit, V.D.; Iwasaki, A. Ketogenic diet activates protective γδ T cell responses against influenza virus infection. Sci. Immunol. 2019, 4, eaav2026. [Google Scholar] [CrossRef]
- Sukkar, S.G.; Cogorno, L.; Pisciotta, L.; Pasta, A.; Vena, A.; Gradaschi, R.; Dentone, C.; Guiddo, E.; Martino, E.; Beltramini, S.; et al. Clinical efficacy of eucaloric ketogenic nutrition in the COVID-19 cytokine storm: A retrospective analysis of mortality and intensive care unit admission. Nutrition 2021, 89, 111236. [Google Scholar] [CrossRef]
- Fallah, J.; Rini, B.I. HIF Inhibitors: Status of Current Clinical Development. Curr. Oncol. Rep. 2019, 21, 6. [Google Scholar] [CrossRef]
- Luo, J.; Yang, H.; Song, B.-L. Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 225–245. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhou, S.; Tang, Q.; Xia, H.; Bi, F. Cholesterol metabolism: New functions and therapeutic approaches in cancer. Biochim. Biophys. Acta (BBA) Rev. Cancer 2020, 1874, 188394. [Google Scholar] [CrossRef]
- Kapourchali, F.R.; Surendiran, G.; Goulet, A.; Moghadasian, M.H. The Role of Dietary Cholesterol in Lipoprotein Metabolism and Related Metabolic Abnormalities: A Mini-review. Crit. Rev. Food Sci. Nutr. 2016, 56, 2408–2415. [Google Scholar] [CrossRef]
- Wang, Y.; Yutuc, E.; Griffiths, W.J. Cholesterol metabolism pathways-are the intermediates more important than the products? FEBS J. 2021, 288, 3727–3745. [Google Scholar] [CrossRef]
- Chang, T.-Y.; Li, B.-L.; Chang, C.C.Y.; Urano, Y. Acyl-coenzyme A:cholesterol acyltransferases. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E1–E9. [Google Scholar] [CrossRef] [Green Version]
- Wei, C.; Wan, L.; Yan, Q.; Wang, X.; Zhang, J.; Yang, X.; Zhang, Y.; Fan, C.; Li, D.; Deng, Y.; et al. HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat. Metab. 2020, 2, 1391–1400. [Google Scholar] [CrossRef] [PubMed]
- Sanders, D.W.; Jumper, C.C.; Ackerman, P.J.; Bracha, D.; Donlic, A.; Kim, H.; Kenney, D.; Castello-Serrano, I.; Suzuki, S.; Tamura, T.; et al. SARS-CoV-2 requires cholesterol for viral entry and pathological syncytia formation. Elife 2021, 10, e65962. [Google Scholar] [CrossRef] [PubMed]
- Dias, S.S.G.; Soares, V.C.; Ferreira, A.C.; Sacramento, C.Q.; Fintelman-Rodrigues, N.; Temerozo, J.R.; Teixeira, L.; Nunes da Silva, M.A.; Barreto, E.; Mattos, M.; et al. Lipid droplets fuel SARS-CoV-2 replication and production of inflammatory mediators. PLoS Pathog. 2020, 16, e1009127. [Google Scholar] [CrossRef]
- Nardacci, R.; Colavita, F.; Castilletti, C.; Lapa, D.; Matusali, G.; Meschi, S.; Del Nonno, F.; Colombo, D.; Capobianchi, M.R.; Zumla, A.; et al. Evidences for lipid involvement in SARS-CoV-2 cytopathogenesis. Cell Death Dis. 2021, 12, 263. [Google Scholar] [CrossRef]
- Liu, S.-Y.; Aliyari, R.; Chikere, K.; Li, G.; Marsden, M.D.; Smith, J.K.; Pernet, O.; Guo, H.; Nusbaum, R.; Zack, J.A.; et al. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity 2013, 38. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Li, W.; Hui, H.; Tiwari, S.K.; Zhang, Q.; Croker, B.A.; Rawlings, S.; Smith, D.; Carlin, A.F.; Rana, T.M. Cholesterol 25-Hydroxylase inhibits SARS-CoV-2 and other coronaviruses by depleting membrane cholesterol. EMBO J. 2020, 39, e106057. [Google Scholar] [CrossRef]
- Zang, R.; Case, J.B.; Yutuc, E.; Ma, X.; Shen, S.; Gomez Castro, M.F.; Liu, Z.; Zeng, Q.; Zhao, H.; Son, J.; et al. Cholesterol 25-hydroxylase suppresses SARS-CoV-2 replication by blocking membrane fusion. Proc. Natl. Acad. Sci. USA 2020, 117, 32105–32113. [Google Scholar] [CrossRef]
- Marcello, A.; Civra, A.; Milan Bonotto, R.; Nascimento Alves, L.; Rajasekharan, S.; Giacobone, C.; Caccia, C.; Cavalli, R.; Adami, M.; Brambilla, P.; et al. The cholesterol metabolite 27-hydroxycholesterol inhibits SARS-CoV-2 and is markedly decreased in COVID-19 patients. Redox Biol. 2020, 36, 101682. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Lu, R.; Dai, M.; Shen, M.; Zhang, J.; Cui, Y.; Liu, B.; Lin, F.; Chen, L.; et al. Lipid metabolism changes in patients with severe COVID-19. Clin. Chim. Acta 2021, 517, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Chen, D.; Wu, L.; He, G.; Ye, W. Declined serum high density lipoprotein cholesterol is associated with the severity of COVID-19 infection. Clin. Chim. Acta 2020, 510, 105–110. [Google Scholar] [CrossRef]
- Wei, X.; Zeng, W.; Su, J.; Wan, H.; Yu, X.; Cao, X.; Tan, W.; Wang, H. Hypolipidemia is associated with the severity of COVID-19. J. Clin. Lipidol. 2020, 14, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Zhang, J.; Liu, L.; Yuan, X.; Zang, X.; Lu, F.; He, P.; Wang, Q.; Zhang, X.; Xu, Y.; et al. High-density lipoprotein cholesterol as a factor affecting virus clearance in covid-19 patients. Respir. Med. 2020, 175, 106218. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhang, Q.; Zhao, X.; Dong, H.; Wu, C.; Wu, F.; Yu, B.; Lv, J.; Zhang, S.; Wu, G.; et al. Low high-density lipoprotein level is correlated with the severity of COVID-19 patients: An observational study. Lipids Health Dis. 2020, 19, 204. [Google Scholar] [CrossRef]
- Fan, J.; Wang, H.; Ye, G.; Cao, X.; Xu, X.; Tan, W.; Zhang, Y. Letter to the Editor: Low-density lipoprotein is a potential predictor of poor prognosis in patients with coronavirus disease 2019. Metabolism 2020, 107, 154243. [Google Scholar] [CrossRef]
- Masana, L.; Correig, E.; Ibarretxe, D.; Anoro, E.; Arroyo, J.A.; Jericó, C.; Guerrero, C.; Miret, M.l.; Näf, S.; Pardo, A.; et al. Low HDL and high triglycerides predict COVID-19 severity. Sci. Rep. 2021, 11, 7217. [Google Scholar] [CrossRef]
- Sun, J.T.; Chen, Z.; Nie, P.; Ge, H.; Shen, L.; Yang, F.; Qu, X.L.; Ying, X.Y.; Zhou, Y.; Wang, W.; et al. Lipid Profile Features and Their Associations With Disease Severity and Mortality in Patients With COVID-19. Front. Cardiovasc. Med. 2020, 7, 584987. [Google Scholar] [CrossRef]
- Overmyer, K.A.; Shishkova, E.; Miller, I.J.; Balnis, J.; Bernstein, M.N.; Peters-Clarke, T.M.; Meyer, J.G.; Quan, Q.; Muehlbauer, L.K.; Trujillo, E.A.; et al. Large-Scale Multi-omic Analysis of COVID-19 Severity. Cell Syst. 2021, 12, 23–40. [Google Scholar] [CrossRef] [PubMed]
- Hilser, J.R.; Han, Y.; Biswas, S.; Gukasyan, J.; Cai, Z.; Zhu, R.; Tang, W.H.W.; Deb, A.; Lusis, A.J.; Hartiala, J.A.; et al. Association of serum HDL-cholesterol and apolipoprotein A1 levels with risk of severe SARS-CoV-2 infection. J. Lipid Res. 2021, 62, 100061. [Google Scholar] [CrossRef]
- Zu, S.; Deng, Y.-Q.; Zhou, C.; Li, J.; Li, L.; Chen, Q.; Li, X.-F.; Zhao, H.; Gold, S.; He, J.; et al. 25-Hydroxycholesterol is a potent SARS-CoV-2 inhibitor. Cell Res. 2020, 30, 1043–1045. [Google Scholar] [CrossRef]
- Turgay Yıldırım, Ö.; Kaya, Ş. The atherogenic index of plasma as a predictor of mortality in patients with COVID-19. Heart Lung 2021, 50, 329–333. [Google Scholar] [CrossRef]
- Feingold, K.R.; Grunfeld, C. Effect of inflammation on HDL structure and function. Curr. Opin. Lipidol. 2016, 27, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Pei, R.; Feng, J.; Zhang, Y.; Sun, H.; Li, L.; Yang, X.; He, J.; Xiao, S.; Xiong, J.; Lin, Y.; et al. Host metabolism dysregulation and cell tropism identification in human airway and alveolar organoids upon SARS-CoV-2 infection. Protein Cell 2020, 12, 717–733. [Google Scholar] [CrossRef]
- Brown, M.S.; Goldstein, J.L. The SREBP Pathway: Regulation of Cholesterol Metabolism by Proteolysis of a Membrane-Bound Transcription Factor. Cell 1997, 89, 331–340. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.; Ahn, J.H.; Park, H.H.; Kim, H.N.; Kim, H.; Yoo, Y.; Shin, H.; Hong, K.S.; Jang, J.G.; Park, C.G.; et al. COVID-19-activated SREBP2 disturbs cholesterol biosynthesis and leads to cytokine storm. Signal Trans. Target. Ther. 2020, 5, 186. [Google Scholar] [CrossRef]
- Daniloski, Z.; Jordan, T.X.; Wessels, H.-H.; Hoagland, D.A.; Kasela, S.; Legut, M.; Maniatis, S.; Mimitou, E.P.; Lu, L.; Geller, E.; et al. Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells. Cell 2021, 184, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Hoagland, D.A.; Clarke, D.J.B.; Møller, R.; Han, Y.; Yang, L.; Wojciechowicz, M.L.; Lachmann, A.; Oguntuyo, K.Y.; Stevens, C.; Lee, B.; et al. Modulating the transcriptional landscape of SARS-CoV-2 as an effective method for developing antiviral compounds. bioRxiv 2020. [Google Scholar] [CrossRef]
- Fajgenbaum, D.C.; Rader, D.J. Teaching Old Drugs New Tricks: Statins for COVID-19? Cell Metab. 2020, 32, 145–147. [Google Scholar] [CrossRef] [PubMed]
- Bifulco, M.; Gazzerro, P. Statin therapy in COVID-19 infection: Much more than a single pathway. Eur. Heart J. Cardiovasc. Pharmacother. 2020, 6, 410–411. [Google Scholar] [CrossRef]
- Castiglione, V.; Chiriacò, M.; Emdin, M.; Taddei, S.; Vergaro, G. Statin therapy in COVID-19 infection. Eur. Heart J. Cardiovasc. Pharmacother. 2020, 6, 258–259. [Google Scholar] [CrossRef]
- Ganjali, S.; Bianconi, V.; Penson, P.E.; Pirro, M.; Banach, M.; Watts, G.F.; Sahebkar, A. Commentary: Statins, COVID-19, and coronary artery disease: Killing two birds with one stone. Metabolism 2020, 113, 154375. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-J.; Qin, J.-J.; Cheng, X.; Shen, L.; Zhao, Y.-C.; Yuan, Y.; Lei, F.; Chen, M.-M.; Yang, H.; Bai, L.; et al. In-Hospital Use of Statins Is Associated with a Reduced Risk of Mortality among Individuals with COVID-19. Cell Metab. 2020, 32, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Yetmar, Z.A.; Chesdachai, S.; Kashour, T.; Riaz, M.; Gerberi, D.J.; Badley, A.D.; Berbari, E.F.; Tleyjeh, I.M. Prior Statin Use and Risk of Mortality and Severe Disease From Coronavirus Disease 2019: A Systematic Review and Meta-analysis. Open Forum Infect. Dis. 2021, 8, ofab284. [Google Scholar] [CrossRef]
- Bifulco, M.; Ciccarelli, M.; Bruzzese, D.; Dipasquale, A.; Lania, A.G.; Mazziotti, G.; Gazzerro, P. The benefit of statins in SARS-CoV-2 patients: Further metabolic and prospective clinical studies are needed. Endocrine 2021, 71, 270–272. [Google Scholar] [CrossRef] [PubMed]
- Vahedian-Azimi, A.; Mohammadi, S.M.; Heidari Beni, F.; Banach, M.; Guest, P.C.; Jamialahmadi, T.; Sahebkar, A. Improved COVID-19 ICU admission and mortality outcomes following treatment with statins: A systematic review and meta-analysis. Arch. Med. Sci. 2021, 17, 579–595. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-Y.; Ahn, J.; Park, J.; Kyung Kang, C.; Won, S.-H.; Wook Kim, D.; Park, J.-H.; Chung, K.-H.; Joh, J.-S.; Bang, J.H.; et al. Beneficial Effect of Statins in COVID-19-Related Outcomes-Brief Report: A National Population-Based Cohort Study. Arterioscler. Thromb. Vasc. Biol. 2021, 41, e175–e182. [Google Scholar] [CrossRef]
- Gupta, A.; Madhavan, M.V.; Poterucha, T.J.; DeFilippis, E.M.; Hennessey, J.A.; Redfors, B.; Eckhardt, C.; Bikdeli, B.; Platt, J.; Nalbandian, A.; et al. Association between antecedent statin use and decreased mortality in hospitalized patients with COVID-19. Nat. Commun. 2021, 12, 1325. [Google Scholar] [CrossRef]
- Lohia, P.; Kapur, S.; Benjaram, S.; Mir, T. Association between antecedent statin use and severe disease outcomes in COVID-19: A retrospective study with propensity score matching. J. Clin. Lipidol. 2021, 15, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-C.; Lee, A.-J.; Su, C.-H.; Huang, C.-Y.; Islam, M.M.; Weng, Y.-C. Statin Use Is Associated with a Decreased Risk of Mortality among Patients with COVID-19. J. Clin. Med. 2021, 10, 1450. [Google Scholar] [CrossRef]
- Song, S.L.; Hays, S.B.; Panton, C.E.; Mylona, E.K.; Kalligeros, M.; Shehadeh, F.; Mylonakis, E. Statin Use Is Associated with Decreased Risk of Invasive Mechanical Ventilation in COVID-19 Patients: A Preliminary Study. Pathogens 2020, 9, 759. [Google Scholar] [CrossRef]
- Masana, L.; Correig, E.; Rodríguez-Borjabad, C.; Anoro, E.; Arroyo, J.A.; Jericó, C.; Pedragosa, A.; Miret, M.l.; Näf, S.; Pardo, A.; et al. Effect of statin therapy on SARS-CoV-2 infection-related. Eur. Heart J. Cardiovasc. Pharmacother. 2020. [Google Scholar] [CrossRef]
- Kollias, A.; Kyriakoulis, K.G.; Kyriakoulis, I.G.; Nitsotolis, T.; Poulakou, G.; Stergiou, G.S.; Syrigos, K. Statin use and mortality in COVID-19 patients: Updated systematic review and meta-analysis. Atherosclerosis 2021, 330, 114–121. [Google Scholar] [CrossRef]
- Hariyanto, T.I.; Kurniawan, A. Statin therapy did not improve the in-hospital outcome of coronavirus disease 2019 (COVID-19) infection. Diabetes Metab. Syndr. 2020, 14, 1613–1615. [Google Scholar] [CrossRef]
- Butt, J.H.; Gerds, T.A.; Schou, M.; Kragholm, K.; Phelps, M.; Havers-Borgersen, E.; Yafasova, A.; Gislason, G.H.; Torp-Pedersen, C.; Køber, L.; et al. Association between statin use and outcomes in patients with coronavirus disease 2019 (COVID-19): A nationwide cohort study. BMJ Open 2020, 10, e044421. [Google Scholar] [CrossRef]
- Rubin, R. Could Statins Do More Than Lower Cholesterol in Patients With COVID-19? JAMA 2021, 325, 2424–2425. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, E.L.; Pulliam, T.H.; Dimaio, T.A.; Thalhofer, A.B.; Delgado, T.; Lagunoff, M. Glycolysis, Glutaminolysis, and Fatty Acid Synthesis Are Required for Distinct Stages of Kaposi’s Sarcoma-Associated Herpesvirus Lytic Replication. J. Virol. 2017, 91, e02237-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carracedo, A.; Cantley, L.C.; Pandolfi, P.P. Cancer metabolism: Fatty acid oxidation in the limelight. Nat. Rev. Cancer 2013, 13, 227–232. [Google Scholar] [CrossRef]
- Tong, L. Structure and function of biotin-dependent carboxylases. Cell. Mol. Life Sci. 2013, 70, 863–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, S.F.; Infante, J.R. Molecular Pathways: Fatty Acid Synthase. Clin. Cancer Res. 2015, 21, 5434–5438. [Google Scholar] [CrossRef] [Green Version]
- Eberlé, D.; Hegarty, B.; Bossard, P.; Ferré, P.; Foufelle, F. SREBP transcription factors: Master regulators of lipid homeostasis. Biochimie 2004, 86, 839–848. [Google Scholar] [CrossRef]
- Rinaldo, P.; Matern, D.; Bennett, M.J. Fatty acid oxidation disorders. Annu. Rev. Physiol. 2002, 64, 477–502. [Google Scholar] [CrossRef] [PubMed]
- Tanner, J.E.; Alfieri, C. The Fatty Acid Lipid Metabolism Nexus in COVID-19. Viruses 2021, 13, 90. [Google Scholar] [CrossRef]
- Shaath, H.; Alajez, N.M. Computational and Transcriptome Analyses Revealed Preferential Induction of Chemotaxis and Lipid Synthesis by SARS-CoV-2. Biology 2020, 9, 260. [Google Scholar] [CrossRef]
- Barberis, E.; Timo, S.; Amede, E.; Vanella, V.V.; Puricelli, C.; Cappellano, G.; Raineri, D.; Cittone, M.G.; Rizzi, E.; Pedrinelli, A.R.; et al. Large-Scale Plasma Analysis Revealed New Mechanisms and Molecules Associated with the Host Response to SARS-CoV-2. Int. J. Mol. Sci. 2020, 21, 8623. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-W.; Lam, S.M.; Fan, X.; Cao, W.-J.; Wang, S.-Y.; Tian, H.; Chua, G.H.; Zhang, C.; Meng, F.-P.; Xu, Z.; et al. Omics-Driven Systems Interrogation of Metabolic Dysregulation in COVID-19 Pathogenesis. Cell Metab. 2020, 32, 188–202. [Google Scholar] [CrossRef]
- Williams, C.G.; Jureka, A.S.; Silvas, J.A.; Nicolini, A.M.; Chvatal, S.A.; Carlson-Stevermer, J.; Oki, J.; Holden, K.; Basler, C.F. Inhibitors of VPS34 and fatty-acid metabolism suppress SARS-CoV-2 replication. Cell Rep. 2021, 36, 109479. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Qian, L.; Sun, R.; Huang, B.; Dong, X.; Xiao, Q.; Zhang, Q.; Lu, T.; Yue, L.; Chen, S.; et al. Multi-organ proteomic landscape of COVID-19 autopsies. Cell 2021, 184, 775–791.e14. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Bae, J.H.; Kwon, H.-S.; Nauck, M.A. COVID-19 and diabetes mellitus: From pathophysiology to clinical management. Nat. Rev. Endocrinol. 2021, 17, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Berber, E.; Sumbria, D.; Rouse, B.T. Could targeting immunometabolism be a way to control the burden of COVID-19 infection? Microbes Infect. 2021, 23, 104780. [Google Scholar] [CrossRef]

| Study Population | Lipid Changes in COVID-19 | References |
|---|---|---|
| 519 severe COVID-19 patients from the West Court of Union Hospital in Wuhan, China | Decrease of TC and HDL-C in non-survivors vs survivors | [80] |
| 114 COVID-19 patients from Wenzhou Central Hospital, in Wenzhou, China | Decrease of TC, LDL-C and HDL-C in COVID-19 patients vs healthy controls Decrease of HDL-C in severe vs common groups | [81] |
| 597 COVID-19 patients from the Cancer Center, Union Hospital of Tongji Medical College, Wuhan | Decrease of TC, LDL-C and HDL-C in COVID-19 patients vs healthy controls Decrease of TC and LDL-C in with disease severity | [82] |
| 115 COVID-19 patients from Union Hospital of Tongji Medical College affiliated Huazhong University of Science and Technology | Decrease of HDL-C in COVID-19 patients vs healthy controls | [83] |
| 228 COVID-19 patients from Public Health Treatment Center of Changsha, China | Decrease of TC, LDL-C and HDL-C in COVID-19 patients vs healthy controls Decrease of HDL-C in with disease severity | [84] |
| 17 surviving and 4 non-surviving COVID-19 patients from Zhongnan Hospital of Wuhan University in Wuhan, China | Decrease of TC, LDL-C and HDL-C in non-survivors vs survivors Decrease of LDL-C in with disease severity | [85] |
| 99 COVID-19 patients from Leishenshan Hospital in Wuhan, China. | Decrease of HDL-C in with disease severity | [87] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, T.; Wang, T. Metabolic Reprogramming in COVID-19. Int. J. Mol. Sci. 2021, 22, 11475. https://doi.org/10.3390/ijms222111475
Shen T, Wang T. Metabolic Reprogramming in COVID-19. International Journal of Molecular Sciences. 2021; 22(21):11475. https://doi.org/10.3390/ijms222111475
Chicago/Turabian StyleShen, Tao, and Tingting Wang. 2021. "Metabolic Reprogramming in COVID-19" International Journal of Molecular Sciences 22, no. 21: 11475. https://doi.org/10.3390/ijms222111475
APA StyleShen, T., & Wang, T. (2021). Metabolic Reprogramming in COVID-19. International Journal of Molecular Sciences, 22(21), 11475. https://doi.org/10.3390/ijms222111475





