Comparative Metabolomics Analysis Reveals Sterols and Sphingolipids Play a Role in Cotton Fiber Cell Initiation
Abstract
:1. Introduction
2. Results
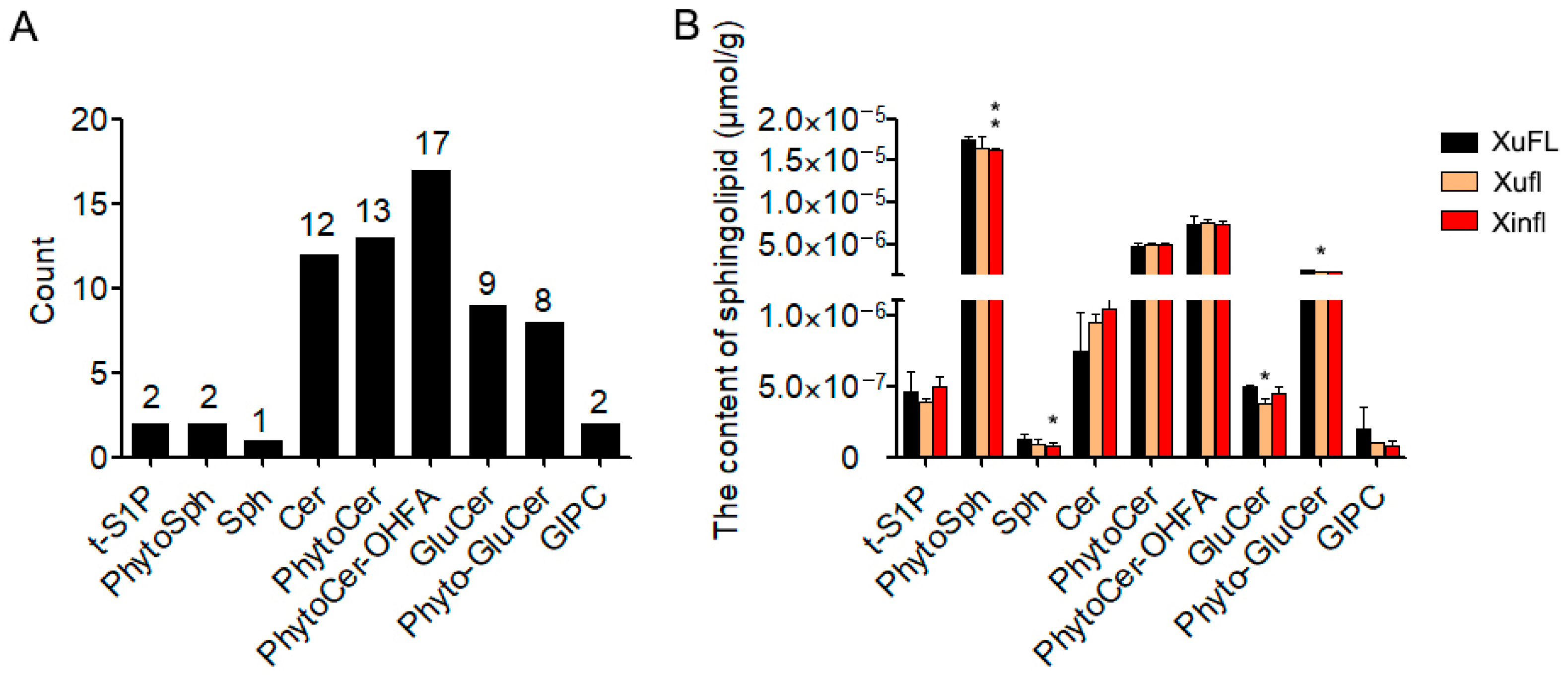
2.1. The Profile and Content of Various Sphingolipids in 0-DPA Ovules of Upland Cotton
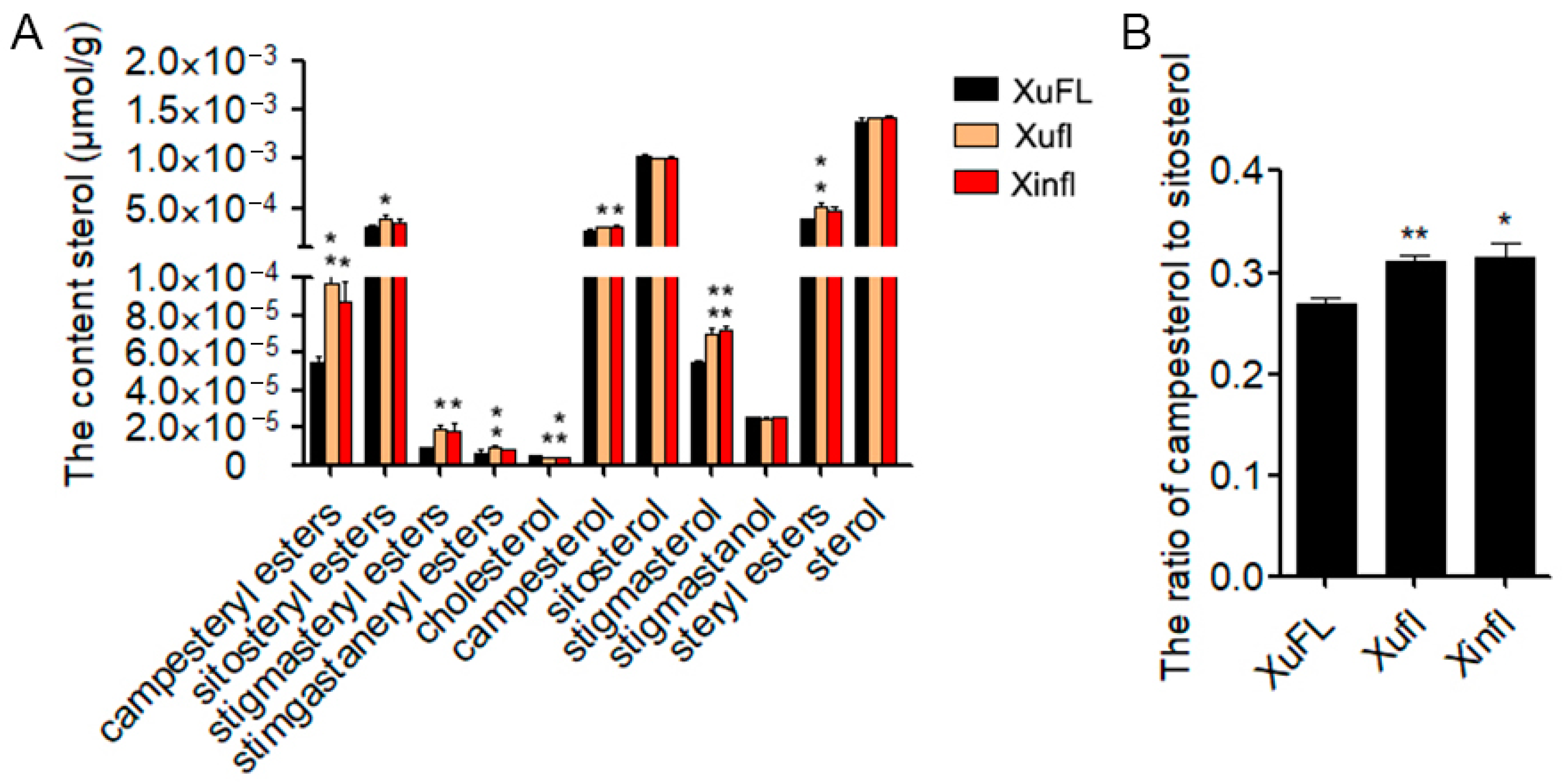
2.2. The Composition and Content of Sterols in 0-DPA Ovules
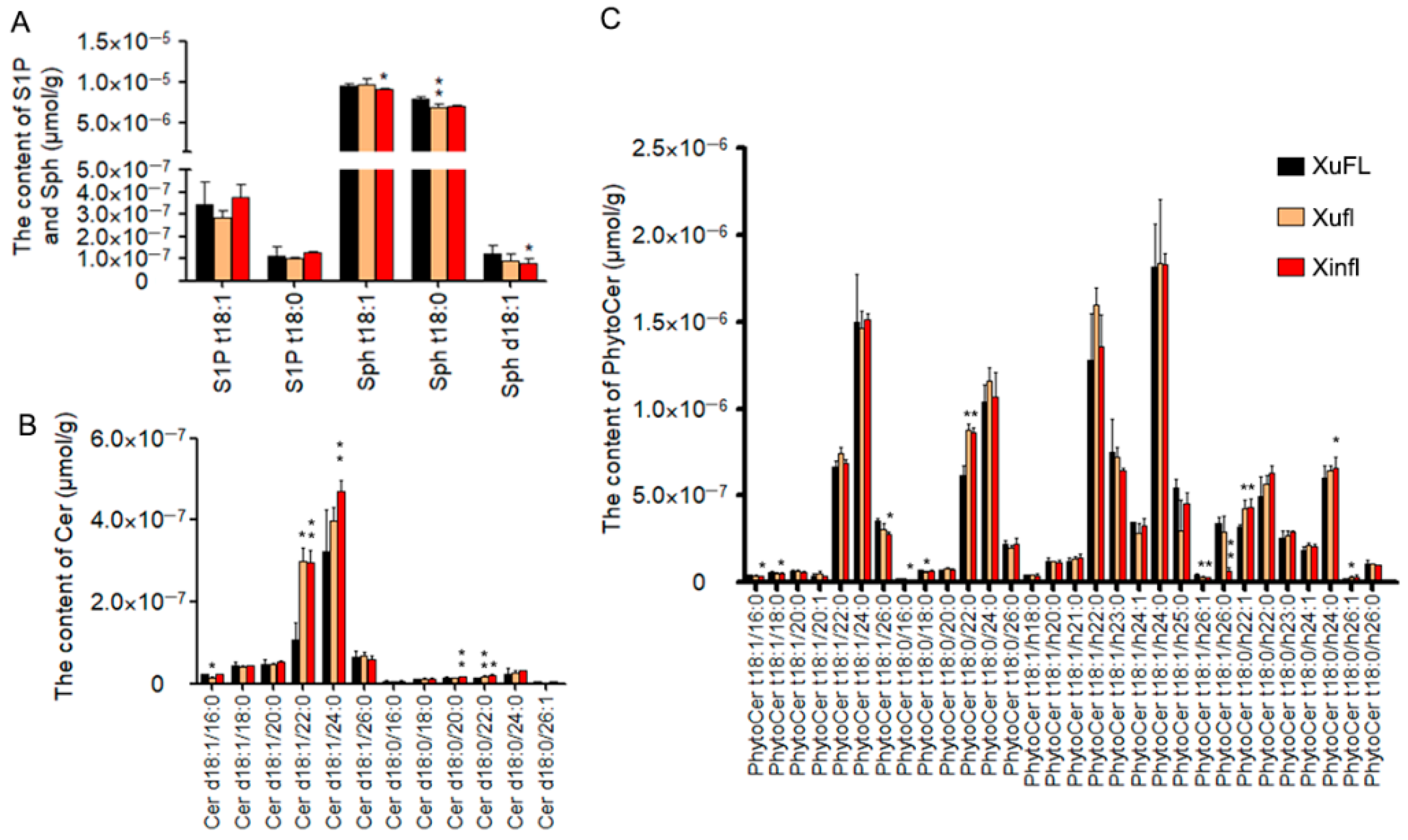
2.3. The Difference of Simple Sphingolipids between Wild-Type and Mutants
2.4. The Difference of Complex Sphingolipids between Wild-Type and Mutants
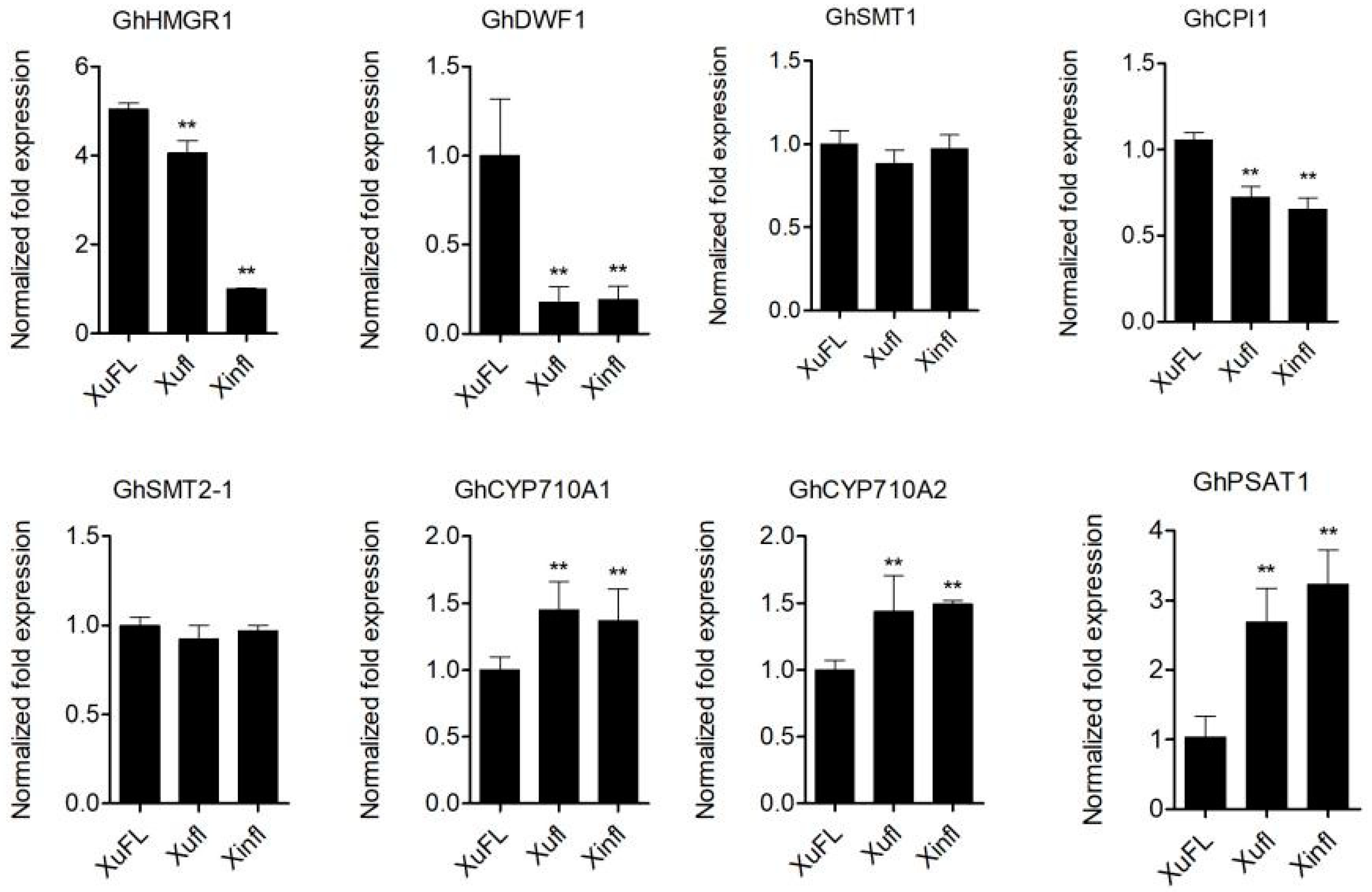
2.5. The Expression Level of GhGCSs and GhIPCSs in Wild-Type and Mutant Ovules
2.6. The Expression of Genes Related to Sterol and Sterol Ester Synthesis in the Two Mutant Ovules
2.7. Exogenous Application of PDMP Inhibited Fiber Cell Initiation and Elongation
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Sample Collection
4.3. Lipid Extraction and Lipidomics
4.4. RNA Extraction and qRT-PCR
4.5. In Vitro Ovule Culture and Scanning Electron Microscopy
4.6. Statistical Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Haigler, C.H.; Betancur, L.; Stiff, M.R.; Tuttle, J.R. Cotton fiber: A powerful single-cell model for cell wall and cellulose research. Front. Plant Sci. 2012, 3, 104. [Google Scholar] [CrossRef] [Green Version]
- Qin, Y.M.; Zhu, Y.X. How cotton fibers elongate: A tale of linear cell-growth mode. Curr. Opin. Plant Biol. 2011, 14, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, T.W. Cotton biotechnology in the next millennium: Yield and fiber duality. Abstr. Pap. Am. Chem. Soc. 2000, 219, U44–U45. [Google Scholar]
- Ruan, Y.L.; Chourey, P.S. A fiberless seed mutation in cotton is associated with lack of fiber cell initiation in ovule epidermis and alterations in sucrose synthase expression and carbon partitioning in developing seeds. Plant Physiol. 1998, 118, 399–406. [Google Scholar] [CrossRef] [Green Version]
- Wilkins, T.A.; Arpat, A.B. The cotton fiber transcriptome. Physiol. Plant. 2005, 124, 295–300. [Google Scholar] [CrossRef]
- Loguerico, L.L.; Zhang, J.Q.; Wilkins, T.A. Differential regulation of six novel MYB-domain genes defines two distinct expression patterns in allotetraploid cotton (Gossypium hirsutum L.). Mol. Gen. Genet. 1999, 261, 660–671. [Google Scholar] [CrossRef]
- Walford, S.A.; Wu, Y.R.; Llewellyn, D.J.; Dennis, E.S. GhMYB25-like: A key factor in early cotton fibre development. Plant J. 2011, 65, 785–797. [Google Scholar] [CrossRef]
- Suo, J.F.; Liang, X.O.; Pu, L.; Zhang, Y.S.; Xue, Y.B. Identification of GhMYB109 encoding a R2R3 MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton (Gossypium Hirsutum L.). Bba-Gene Struct. Expr. 2003, 1630, 25–34. [Google Scholar] [CrossRef]
- Wang, S.; Wang, J.W.; Yu, N.; Li, C.H.; Luo, B.; Gou, J.Y.; Wang, L.J.; Chen, X.Y. Control of plant trichome development by a cotton fiber MYB gene. Plant Cell 2004, 16, 2323–2334. [Google Scholar] [CrossRef] [Green Version]
- Li, C.H.; Zhu, Y.Q.; Meng, Y.L.; Wang, J.W.; Xu, K.X.; Zhang, T.Z.; Chen, X.Y. Isolation of genes preferentially expressed in cotton fibers by cDNA filter arrays and RT-PCR. Plant Sci. 2002, 163, 1113–1120. [Google Scholar] [CrossRef] [Green Version]
- Humphries, J.A.; Walker, A.R.; Timmis, J.N.; Orford, S.J. Two WD-repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant Mol. Biol. 2005, 57, 67–81. [Google Scholar] [CrossRef]
- Wu, Y.; Llewellyn, D.J.; White, R.; Ruggiero, K.; Al-Ghazi, Y.; Dennis, E.S. Laser capture microdissection and cDNA microarrays used to generate gene expression profiles of the rapidly expanding fibre initial cells on the surface of cotton ovules. Planta 2007, 226, 1475–1490. [Google Scholar] [CrossRef]
- Guan, X.-Y.; Li, Q.-J.; Shan, C.-M.; Wang, S.; Mao, Y.-B.; Wang, L.-J.; Chen, X.-Y. The HD-Zip IV gene GaHOX1 from cotton is a functional homologue of the Arabidopsis GLABRA2. Physiol. Plant. 2008, 134, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Padmalatha, K.V.; Patil, D.P.; Kumar, K.; Dhandapani, G.; Kanakachari, M.; Phanindra, M.L.V.; Kumar, S.; Mohan, T.C.; Jain, N.; Prakash, A.H.; et al. Functional genomics of fuzzless-lintless mutant of Gossypium hirsutum L. cv. MCU5 reveal key genes and pathways involved in cotton fibre initiation and elongation. BMC Genom. 2012, 13, 624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shangguan, X.X.; Yang, C.Q.; Zhang, X.F.; Wang, L.J. Functional characterization of a basic helix-loop-helix (bHLH) transcription factor GhDEL65 from cotton (Gossypium hirsutum). Physiol. Plant. 2016, 158, 200–212. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Wang, M.; Ding, Y.; Zhu, S.; Zhao, G.; Tu, L.; Zhang, X. Transcriptomic repertoires depict the initiation of lint and fuzz fibres in cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 2018, 16, 1002–1012. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.C.; Li, Q.; Jin, X.; Xiao, G.H.; Liu, G.J.; Liu, N.J.; Qin, Y.M. Quantitative proteomics and transcriptomics reveal key metabolic processes associated with cotton fiber initiation. J. Proteom. 2015, 114, 16–27. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, T.; Guo, W. Effect of H2O2 on fiber initiation using fiber retardation initiation mutants in cotton (Gossypium hirsutum). J. Plant Physiol. 2010, 167, 393–399. [Google Scholar] [CrossRef]
- Liu, K.; Han, M.; Zhang, C.; Yao, L.; Sun, J.; Zhang, T. Comparative proteomic analysis reveals the mechanisms governing cotton fiber differentiation and initiation. J. Proteom. 2012, 75, 845–856. [Google Scholar] [CrossRef]
- Taliercio, E.W.; Boykin, D. Analysis of gene expression in cotton fiber initials. BMC Plant Biol. 2007, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Hoekstra, D.; Maier, O.; van der Wouden, J.M.; Slimane, T.A.; van Ijzendoorn, S.C.D. Membrane dynamics and cell polarity: The role of sphingolipids. J. Lipid Res. 2003, 44, 869–877. [Google Scholar] [CrossRef] [Green Version]
- Zäuner, S.; Ternes, P.; Warnecke, D. Biosynthesis of sphingolipids in plants (and some of their functions). Adv. Exp. Med. Biol. 2010, 688, 249–263. [Google Scholar] [CrossRef] [PubMed]
- Benveniste, P. Biosynthesis and Accumulation of Sterols. Annu. Rev. Plant Biol. 2004, 55, 429–457. [Google Scholar] [CrossRef] [PubMed]
- Worrall, D.; Ng, C.K.Y.; Hetherington, A.M. Sphingolipids, new players in plant signaling. Trends Plant Sci. 2003, 8, 317–320. [Google Scholar] [CrossRef]
- Chen, M.; Cahoon, E.B.; Saucedo-García, M.; Plasencia, J.; Gavilanes-Ruíz, M. Plant Sphingolipids: Structure, Synthesis and Function. In Lipids in Photosynthesis: Essential and Regulatory Functions; Wada, H., Murata, N., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 77–115. [Google Scholar]
- Lynch, D.V.; Dunn, T.M. An introduction to plant sphingolipids and a review of recent advances in understanding their metabolism and function. New Phytol. 2004, 161, 677–702. [Google Scholar] [CrossRef]
- Markham, J.E.; Jaworski, J.G. Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectometry 2007, 21, 1304–1314. [Google Scholar] [CrossRef] [PubMed]
- Tarkowska, D.; Strnad, M. Isoprenoid-derived plant signaling molecules: Biosynthesis and biological importance. Planta 2018, 247, 1051–1066. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Li, R.-L.; Zhang, L.; Wang, Q.-L.; Niehaus, K.; Baluška, F.; Šamaj, J.; Lin, J.-X. Lipid microdomain polarization is required for NADPH oxidase-dependent ROS signaling in Picea meyeri pollen tube tip growth. Plant J. 2009, 60, 303–313. [Google Scholar] [CrossRef]
- Markham, J.E.; Molino, D.; Gissot, L.; Bellec, Y.; Hématy, K.; Marion, J.; Belcram, K.; Palauqui, J.-C.; Satiat-JeuneMaître, B.; Faure, J.-D. Sphingolipids Containing Very-Long-Chain Fatty Acids Define a Secretory Pathway for Specific Polar Plasma Membrane Protein Targeting in Arabidopsis. Plant Cell 2011, 23, 2362–2378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borner, G.H.H.; Lilley, K.S.; Stevens, T.J.; Dupree, P. Identification of Glycosylphosphatidylinositol-Anchored Proteins in Arabidopsis. A Proteomic and Genomic Analysis. Plant Physiol. 2003, 132, 568–577. [Google Scholar] [CrossRef] [Green Version]
- Schindelman, G.; Morikami, A.; Jung, J.; Baskin, T.I.; Carpita, N.C.; Derbyshire, P.; McCann, M.C.; Benfey, P.N. COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 2001, 15, 1115–1127. [Google Scholar] [CrossRef] [Green Version]
- Willemsen, V.; Friml, J.; Grebe, M.; van den Toorn, A.; Palme, K.; Scheres, B. Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 2003, 15, 612–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Men, S.; Boutte, Y.; Ikeda, Y.; Li, X.; Palme, K.; Stierhof, Y.D.; Hartmann, M.A.; Moritz, T.; Grebe, M. Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat. Cell Biol. 2008, 10, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.Z.; Pan, J.J. Genetic analysis of fuzzless-lintless mutant in upland cotton. Jiangsu J. Agr. Sci. 1991, 7, 13–16. [Google Scholar]
- Yu, X.; Zhu, Y.; Lu, S.; Zhang, T.; Chen, X.; Xu, Z. A comparative analysis ofafuzzless-lintless mutant of Gossypium hirsutum L. cv. Xu-142. Sci. China Ser. C Life Sci. 2000, 43, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.Q.; Liu, F.; Chen, X.S.; Ma, X.J.; Zeng, H.Q.; Yang, Z.M. Transcriptome profiling of early developing cotton fiber by deep-sequencing reveals significantly differential expression of genes in a fuzzless/lintless mutant. Genomics 2010, 96, 369–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.H.; Du, X.M. SSR Fingerprinting Analysis on Distinct Mutants of Fiber Development in Gossypium hisutum. Sci. Agric. Sin. 2005, 38, 2139–2146. [Google Scholar]
- Tang, W.X.; Tu, L.L.; Yang, X.Y.; Tan, J.F.; Deng, F.L.; Hao, J.; Guo, K.; Lindsey, K.; Zhang, X.L. The calcium sensor GhCaM7 promotes cotton fiber elongation by modulating reactive oxygen species (ROS) production. New Phytol. 2014, 202, 509–520. [Google Scholar] [CrossRef]
- Schaeffer, A.; Bronner, R.; Benveniste, P.; Schaller, H. The ratio of campesterol to sitosterol that modulates growth in Arabidopsis is controlled by STEROL METHYLTRANSFERASE 2;1. Plant J. Cell Mol. Biol. 2001, 25, 605–615. [Google Scholar] [CrossRef]
- Deng, S.; Wei, T.; Tan, K.; Hu, M.; Li, F.; Zhai, Y.; Ye, S.; Xiao, Y.; Hou, L.; Pei, Y.; et al. Phytosterol content and the campesterol:sitosterol ratio influence cotton fiber development: Role of phytosterols in cell elongation. Sci. China. Life Sci. 2016, 59, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Goñi, F.M.; Alonso, A. Effects of ceramide and other simple sphingolipids on membrane lateral structure. Biochim. Et Biophys. Acta (BBA)–Biomembr. 2009, 1788, 169–177. [Google Scholar] [CrossRef] [Green Version]
- Markham, J.E.; Lynch, D.V.; Napier, J.A.; Dunn, T.M.; Cahoon, E.B. Plant sphingolipids: Function follows form. Curr. Opin. Plant Biol. 2013, 16, 350–357. [Google Scholar] [CrossRef]
- Mina, J.G.; Okada, Y.; Wansadhipathi-Kannangara, N.K.; Pratt, S.; Shams-Eldin, H.; Schwarz, R.T.; Steel, P.G.; Fawcett, T.; Denny, P.W. Functional analyses of differentially expressed isoforms of the Arabidopsis inositol phosphorylceramide synthase. Plant Mol. Biol. 2010, 73, 399–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valitova, J.N.; Sulkarnayeva, A.G.; Minibayeva, F.V. Plant sterols: Diversity, biosynthesis, and physiological functions. Biochemistry 2016, 81, 819–834. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, A.; Altabella, T.; Arró, M.; Boronat, A. Emerging roles for conjugated sterols in plants. Prog. Lipid Res. 2017, 67, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Banaś, A.; Carlsson, A.S.; Huang, B.; Lenman, M.; Banaś, W.; Lee, M.; Noiriel, A.; Benveniste, P.; Schaller, H.; Bouvier-Navé, P.; et al. Cellular Sterol Ester Synthesis in Plants Is Performed by an Enzyme (Phospholipid:Sterol Acyltransferase) Different from the Yeast and Mammalian Acyl-CoA:Sterol Acyltransferases*. J. Biol. Chem. 2005, 280, 34626–34634. [Google Scholar] [CrossRef] [Green Version]
- Sonawane, P.D.; Pollier, J.; Panda, S.; Szymanski, J.; Massalha, H.; Yona, M.; Unger, T.; Malitsky, S.; Arendt, P.; Pauwels, L.; et al. Plant cholesterol biosynthetic pathway overlaps with phytosterol metabolism. Nat. Plants 2016, 3, 16205. [Google Scholar] [CrossRef]
- Fujioka, S.; Yokota, T. Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 2003, 54, 137–164. [Google Scholar] [CrossRef] [PubMed]
- Clouse, S.D. Arabidopsis mutants reveal multiple roles for sterols in plant development. Plant Cell 2002, 14, 1995–2000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nolan, T.; Chen, J.N.; Yin, Y.H. Cross-talk of Brassinosteroid signaling in controlling growth and stress responses. Biochem. J. 2017, 474, 2641–2661. [Google Scholar] [CrossRef]
- Xia, X.-J.; Zhou, Y.-H.; Shi, K.; Zhou, J.; Foyer, C.; Yu, J.-Q. Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J. Exp. Bot. 2015, 66, 2839–2856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, Q.; Tan, K.L.; Zang, Z.L.; Xiao, Z.Y.; Chen, K.J.; Hu, M.Y.; Luo, M. Modification of phytosterol composition influences cotton fiber cell elongation and secondary cell wall deposition. BMC Plant Biol. 2019, 19, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Suo, X.; Li, F.; Bao, C.; He, S.; Huang, L.; Luo, M. Membrane lipid raft organization during cotton fiber development. J. Cotton Res. 2020, 3, 13. [Google Scholar] [CrossRef]
- Suo, X.; Xu, F.; Tan, K.; Huang, L.; Bao, C.; Luo, M. Functions of phytosterols in seed development of upland cotton (Gossypium hirsutum L.). Ind. Crop Prod. 2021, 170, 113802. [Google Scholar] [CrossRef]
- Xu, F.; Chen, Q.; Huang, L.; Luo, M. Advances about the Roles of Membranes in Cotton Fiber Development. Membranes 2021, 11, 471. [Google Scholar] [CrossRef]
- Boutte, Y.; Grebe, M. Cellular processes relying on sterol function in plants. Curr. Opin. Plant Biol. 2009, 12, 705–713. [Google Scholar] [CrossRef]
- Beasley, C.A.; Ting, I.P. Effects of Plant Growth Substances on in Vitro Fiber Development from Unfertilized Cotton Ovules. Am. J. Bot. 1974, 61, 188–194. [Google Scholar] [CrossRef]
- Beasley, C.A. Hormonal Regulation of Growth in Unfertilized Cotton Ovules. Science 1973, 179, 1003–1005. [Google Scholar] [CrossRef]
- Zhang, M.; Zheng, X.L.; Song, S.Q.; Zeng, Q.W.; Hou, L.; Li, D.M.; Zhao, J.; Wei, Y.; Li, X.B.; Luo, M.; et al. Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat. Biotechnol. 2011, 29, 453–458. [Google Scholar] [CrossRef]
- Senthil-Kumar, M.; Wang, K.; Mysore, K.S. AtCYP710A1 gene-mediated stigmasterol production plays a role in imparting temperature stress tolerance in Arabidopsis thaliana. Plant Signal. Behav. 2013, 8, e23142. [Google Scholar] [CrossRef] [Green Version]
- Griebel, T.; Zeier, J. A role for β-sitosterol to stigmasterol conversion in plant–pathogen interactions. Plant J. 2010, 63, 254–268. [Google Scholar] [CrossRef] [PubMed]
- Aboobucker, S.I.; Suza, W.P. Why Do Plants Convert Sitosterol to Stigmasterol? Front. Plant Sci. 2019, 10, 354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, S.J.; Dong, C.J.; Zhang, B.; Lai, T.F.; Du, X.M.; Liu, J.Y. Comparative proteomic analysis reveals differentially expressed proteins correlated with fuzz fiber initiation in diploid cotton (Gossypium arbareum L.). J. Proteom. 2013, 82, 113–129. [Google Scholar] [CrossRef] [PubMed]
- Gondet, L.; Weber, T.; Maillot-Vernier, P.; Benveniste, P.; Bach, T.J. Regulatory role of microsomal 3-hydroxy-3-methylglutaryl-coenzyme a reductase in a tobacco mutant that overproduces sterols. Biochem. Biophys. Res. Commun. 1992, 186, 888–893. [Google Scholar] [CrossRef]
- Chappell, J.; Wolf, F.; Proulx, J.; Cuellar, R.; Saunders, C. Is the Reaction Catalyzed by 3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase a Rate-Limiting Step for Isoprenoid Biosynthesis in Plants? Plant Physiol. 1995, 109, 1337–1343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimada, T.L.; Shimada, T.; Okazaki, Y.; Higashi, Y.; Saito, K.; Kuwata, K.; Oyama, K.; Kato, M.; Ueda, H.; Nakano, A.; et al. HIGH STEROL ESTER 1 is a key factor in plant sterol homeostasis. Nat. Plants 2019, 5, 1154–1166. [Google Scholar] [CrossRef]
- Nomura, T.; Kitasaka, Y.; Takatsuto, S.; Reid, J.B.; Fukami, M.; Yokota, T. Brassinosteroid/sterol synthesis and plant growth as affected by Ika and Ikb mutations of pea. Plant Physiol. 1999, 119, 1517–1526. [Google Scholar] [CrossRef] [Green Version]
- Diener, A.C.; Li, H.; Zhou, W.-x.; Whoriskey, W.J.; Nes, W.D.; Fink, G.R. STEROL METHYLTRANSFERASE 1 Controls the Level of Cholesterol in Plants. Plant Cell 2000, 12, 853–870. [Google Scholar] [CrossRef] [Green Version]
- Luo, M.; Xiao, Z.-y.; Xiao, Y.-H.; Li, X.; Hou, L.; Zhou, J.-p.; Hu, M.-y.; Pei, Y. Cloning and Expression Analysis of a Brassinosteroid Biosynthetic Enzyme Gene, GhDWF1, from Cotton (Gossypium hirsuturm L.). Agric. Sci. China 2007, 6, 1297–1305. [Google Scholar] [CrossRef]
- Ormancey, M.; Thuleau, P.; van der Hoorn, R.A.L.; Grat, S.; Testard, A.; Kamal, K.Y.; Boudsocq, M.; Cotelle, V.; Mazars, C. Sphingolipid-induced cell death in Arabidopsis is negatively regulated by the papain-like cysteine protease RD21. Plant Sci. 2019, 280, 12–17. [Google Scholar] [CrossRef]
- Liang, H.; Yao, N.; Song, L.T.; Luo, S.; Lu, H.; Greenberg, L.T. Ceramides modulate programmed cell death in plants. Genes Dev. 2003, 17, 2636–2641. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.H.; Bielawski, J.; Mu, J.Y.; Dong, H.L.; Teng, C.; Zhang, J.; Yang, X.H.; Tomishige, N.; Hanada, K.; Hannun, Y.A.; et al. Involvement of sphingoid bases in mediating reactive oxygen intermediate production and programmed cell death in Arabidopsis. Cell Res. 2007, 17, 1030–1040. [Google Scholar] [CrossRef]
- Spassieva, S.D.; Markham, J.E.; Hille, J. The plant disease resistance gene Asc-1 prevents disruption of sphingolipid metabolism during AAL-toxin-induced programmed cell death. Plant J. 2002, 32, 561–572. [Google Scholar] [CrossRef] [Green Version]
- Cantrel, C.; Vazquez, T.; Puyaubert, J.; Reze, N.; Lesch, M.; Kaiser, W.M.; Dutilleul, C.; Guillas, I.; Zachowski, A.; Baudouin, E. Nitric oxide participates in cold-responsive phosphosphingolipid formation and gene expression in Arabidopsis thaliana. New Phytol. 2011, 189, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Dutilleul, C.; Chavarria, H.; Reze, N.; Sotta, B.; Baudouin, E.; Guillas, I. Evidence for ACD5 ceramide kinase activity involvement in Arabidopsis response to cold stress. Plant Cell Environ. 2015, 38, 2688–2697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutilleul, C.; Benhassaine-Kesri, G.; Demandre, C.; Reze, N.; Launay, A.; Pelletier, S.; Renou, J.P.; Zachowski, A.; Baudouin, E.; Guillas, I. Phytosphingosine-phosphate is a signal for AtMPK6 activation and Arabidopsis response to chilling. New Phytol. 2012, 194, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Peer, M.; Stegmann, M.; Mueller, M.J.; Waller, F. Pseudomonas syringae infection triggers de novo synthesis of phytosphingosine from sphinganine in Arabidopsis thaliana. FEBS Lett. 2010, 584, 4053–4056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coursol, S.; Fromentin, J.; Noirot, E.; Briere, C.; Robert, F.; Morel, J.; Liang, Y.K.; Lherminier, J.; Simon-Plas, F. Long-chain bases and their phosphorylated derivatives differentially regulate cryptogein-induced production of reactive oxygen species in tobacco (Nicotiana tabacum) BY-2 cells. New Phytol. 2015, 205, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Coursol, S.; Fan, L.M.; Le Stunff, H.; Spiegel, S.; Gilroy, S.; Assmann, S.M. Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 2003, 423, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.K.Y.; Carr, K.; McAinsh, M.R.; Powell, B.; Hetherington, A.M. Drought-induced guard cell signal transduction involves sphingosine-1-phosphate. Nature 2001, 410, 596–599. [Google Scholar] [CrossRef]
- Worrall, D.; Liang, Y.-K.; Alvarez, S.; Holroyd, G.H.; Spiegel, S.; Panagopulos, M.; Gray, J.E.; Hetherington, A.M. Involvement of sphingosine kinase in plant cell signalling. Plant J. 2008, 56, 64–72. [Google Scholar] [CrossRef] [Green Version]
- Sperling, P.; Heinz, E. Plant sphingolipids: Structural diversity, biosynthesis, first genes and functions. Biochim. Et Biophys. Acta-Mol. Cell Biol. Lipids 2003, 1632, 1–15. [Google Scholar] [CrossRef]
- Wang, L.; Suo, X.; Liu, Y.; Liu, C.; Luo, M. Sphingosine Promotes Embryo Biomass in Upland Cotton: A Biochemical and Transcriptomic Analysis. Biomolecules 2021, 11, 525. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, C.; Liu, Y.; Luo, M. Fumonisin B1-Induced Changes in Cotton Fiber Elongation Revealed by Sphingolipidomics and Proteomics. Biomolecules 2020, 10, 1258. [Google Scholar] [CrossRef]
- Welti, R.; Li, W.; Li, M.; Sang, Y.; Biesiada, H.; Zhou, H.-E.; Rajashekar, C.B.; Williams, T.D.; Wang, X. Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 2002, 277, 31994–32002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.; Sun, Y.; Ma, Z.; Ke, M.; Cui, Y.; Chen, Z.; Chen, C.; Ji, C.; Tran, T.M.; Yang, L.; et al. Salicylic acid-mediated plasmodesmal closure via Remorin-dependent lipid organization. Proc. Natl. Acad. Sci. USA 2019, 116, 21274–21284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shui, G.; Cheong, W.F.; Jappar, I.A.; Hoi, A.; Xue, Y.; Fernandis, A.Z.; Tan, B.K.; Wenk, M.R. Derivatization-independent cholesterol analysis in crude lipid extracts by liquid chromatography/mass spectrometry: Applications to a rabbit model for atherosclerosis. J. Chromatogr. A 2011, 1218, 4357–4365. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Meng, Q.; Xu, F.; Chen, Q.; Ma, C.; Huang, L.; Li, G.; Luo, M. Comparative Metabolomics Analysis Reveals Sterols and Sphingolipids Play a Role in Cotton Fiber Cell Initiation. Int. J. Mol. Sci. 2021, 22, 11438. https://doi.org/10.3390/ijms222111438
Wang Q, Meng Q, Xu F, Chen Q, Ma C, Huang L, Li G, Luo M. Comparative Metabolomics Analysis Reveals Sterols and Sphingolipids Play a Role in Cotton Fiber Cell Initiation. International Journal of Molecular Sciences. 2021; 22(21):11438. https://doi.org/10.3390/ijms222111438
Chicago/Turabian StyleWang, Qiaoling, Qian Meng, Fan Xu, Qian Chen, Caixia Ma, Li Huang, Guiming Li, and Ming Luo. 2021. "Comparative Metabolomics Analysis Reveals Sterols and Sphingolipids Play a Role in Cotton Fiber Cell Initiation" International Journal of Molecular Sciences 22, no. 21: 11438. https://doi.org/10.3390/ijms222111438
APA StyleWang, Q., Meng, Q., Xu, F., Chen, Q., Ma, C., Huang, L., Li, G., & Luo, M. (2021). Comparative Metabolomics Analysis Reveals Sterols and Sphingolipids Play a Role in Cotton Fiber Cell Initiation. International Journal of Molecular Sciences, 22(21), 11438. https://doi.org/10.3390/ijms222111438





