Nuclear mRNA Quality Control and Cytoplasmic NMD Are Linked by the Guard Proteins Gbp2 and Hrb1
Abstract
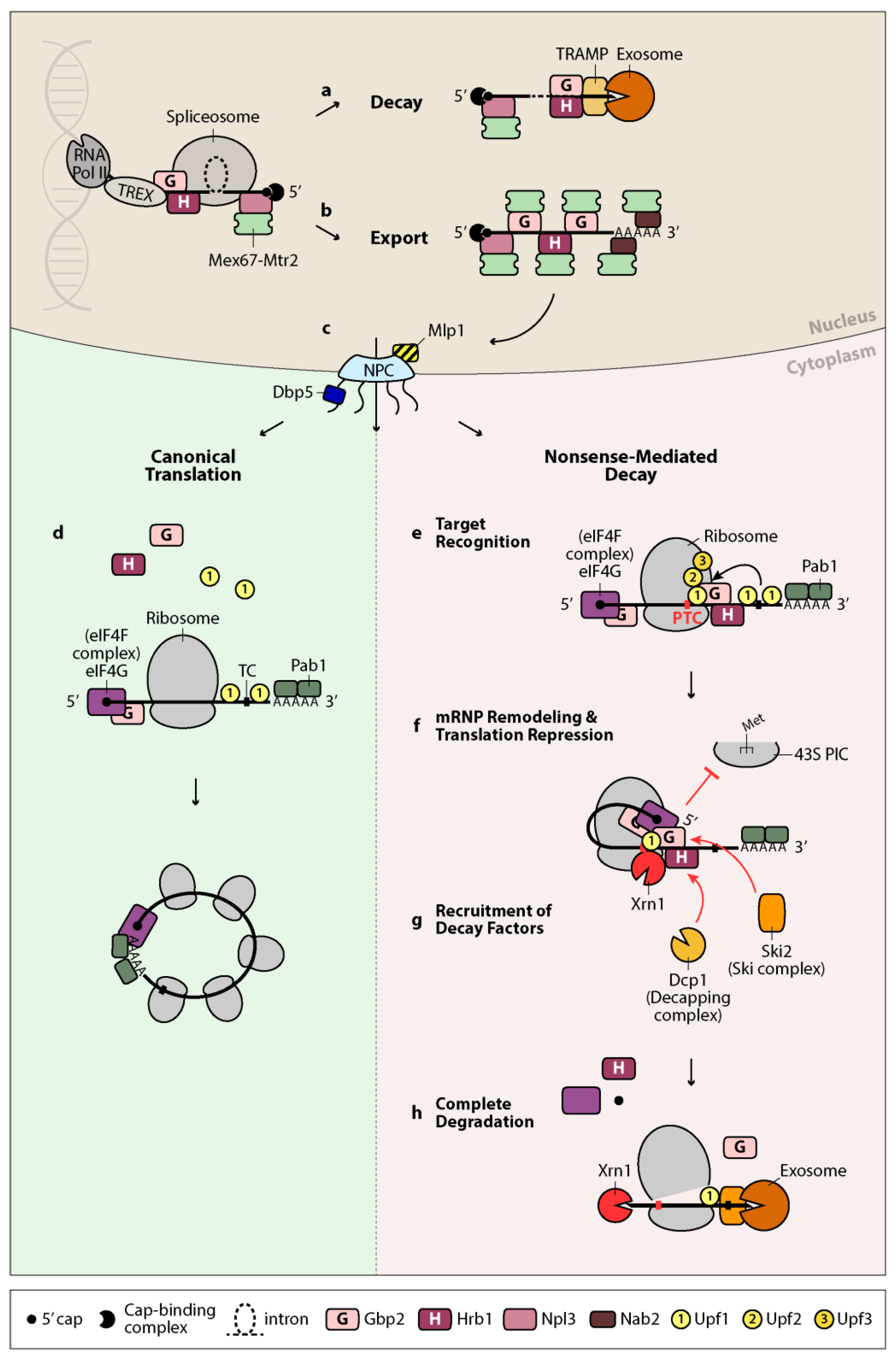
1. Introduction
2. Quality Control of Splicing in the Nucleus—Retention or Export of mRNA
3. Further Surveillance in the Cytoplasm—Detection of Errors via Nonsense-Mediated Decay
4. Gbp2 and Hrb1 in Nuclear Quality Control of Splicing—Decay or Export of mRNA
5. Gbp2 and Hrb1 in Nonsense-Mediated mRNA Decay—New Cytoplasmic Roles
6. Gbp2 and Hrb1 as Prototypes of Human Proteins
7. Closing Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jacob, A.G.; Smith, C.W.J. Intron retention as a component of regulated gene expression programs. Hum. Genet. 2017, 136, 1043–1057. [Google Scholar] [CrossRef] [PubMed]
- Kelemen, O.; Convertini, P.; Zhang, Z.; Wen, Y.; Shen, M.; Falaleeva, M.; Stamm, S. Function of alternative splicing. Gene 2013, 514, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Rio, D.C. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef] [PubMed]
- Long, J.C.; Caceres, J.F. The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 2009, 417, 15–27. [Google Scholar] [CrossRef]
- Zhou, Z.; Fu, X.D. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 2013, 122, 191–207. [Google Scholar] [CrossRef]
- Wende, W.; Friedhoff, P.; Strasser, K. Mechanism and Regulation of Co-transcriptional mRNP Assembly and Nuclear mRNA Export. Adv. Exp. Med. Biol. 2019, 1203, 1–31. [Google Scholar] [CrossRef]
- De Almeida, S.F.; Garcia-Sacristan, A.; Custodio, N.; Carmo-Fonseca, M. A link between nuclear RNA surveillance, the human exosome and RNA polymerase II transcriptional termination. Nucleic Acids Res. 2010, 38, 8015–8026. [Google Scholar] [CrossRef]
- Martins, S.B.; Rino, J.; Carvalho, T.; Carvalho, C.; Yoshida, M.; Klose, J.M.; de Almeida, S.F.; Carmo-Fonseca, M. Spliceosome assembly is coupled to RNA polymerase II dynamics at the 3′ end of human genes. Nat. Struct. Mol. Biol. 2011, 18, 1115–1123. [Google Scholar] [CrossRef]
- Martinson, H.G. An active role for splicing in 3′-end formation. Wiley Interdiscip. Rev. RNA 2011, 2, 459–470. [Google Scholar] [CrossRef]
- Custodio, N.; Carmo-Fonseca, M.; Geraghty, F.; Pereira, H.S.; Grosveld, F.; Antoniou, M. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 1999, 18, 2855–2866. [Google Scholar] [CrossRef]
- Dower, K.; Kuperwasser, N.; Merrikh, H.; Rosbash, M. A synthetic A tail rescues yeast nuclear accumulation of a ribozyme-terminated transcript. RNA 2004, 10, 1888–1899. [Google Scholar] [CrossRef]
- Rigo, F.; Martinson, H.G. Polyadenylation releases mRNA from RNA polymerase II in a process that is licensed by splicing. RNA 2009, 15, 823–836. [Google Scholar] [CrossRef][Green Version]
- Eberle, A.B.; Visa, N. Quality control of mRNP biogenesis: Networking at the transcription site. Semin. Cell Dev. Biol. 2014, 32, 37–46. [Google Scholar] [CrossRef]
- Schmid, M.; Jensen, T.H. Controlling nuclear RNA levels. Nat. Rev. Genet. 2018, 19, 518–529. [Google Scholar] [CrossRef]
- Davidson, L.; Kerr, A.; West, S. Co-transcriptional degradation of aberrant pre-mRNA by Xrn2. EMBO J. 2012, 31, 2566–2578. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, T.; Martins, S.; Rino, J.; Marinho, S.; Carmo-Fonseca, M. Pharmacological inhibition of the spliceosome subunit SF3b triggers exon junction complex-independent nonsense-mediated decay. J. Cell Sci. 2017, 130, 1519–1531. [Google Scholar] [CrossRef] [PubMed]
- Girard, C.; Will, C.L.; Peng, J.; Makarov, E.M.; Kastner, B.; Lemm, I.; Urlaub, H.; Hartmuth, K.; Luhrmann, R. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat. Commun. 2012, 3, 994. [Google Scholar] [CrossRef] [PubMed]
- Hett, A.; West, S. Inhibition of U4 snRNA in human cells causes the stable retention of polyadenylated pre-mRNA in the nucleus. PLoS ONE 2014, 9, e96174. [Google Scholar] [CrossRef]
- Galganski, L.; Urbanek, M.O.; Krzyzosiak, W.J. Nuclear speckles: Molecular organization, biological function and role in disease. Nucleic Acids Res. 2017, 45, 10350–10368. [Google Scholar] [CrossRef]
- Wegener, M.; Muller-McNicoll, M. Nuclear retention of mRNAs—quality control, gene regulation and human disease. Semin. Cell Dev. Biol. 2018, 79, 131–142. [Google Scholar] [CrossRef]
- Dias, A.P.; Dufu, K.; Lei, H.; Reed, R. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat. Commun. 2010, 1, 97. [Google Scholar] [CrossRef]
- Ishihama, Y.; Tadakuma, H.; Tani, T.; Funatsu, T. The dynamics of pre-mRNAs and poly(A)+ RNA at speckles in living cells revealed by iFRAP studies. Exp. Cell Res. 2008, 314, 748–762. [Google Scholar] [CrossRef]
- Boothby, T.C.; Zipper, R.S.; van der Weele, C.M.; Wolniak, S.M. Removal of retained introns regulates translation in the rapidly developing gametophyte of Marsilea vestita. Dev. Cell 2013, 24, 517–529. [Google Scholar] [CrossRef]
- Majewska, K.; Wroblewska-Ankiewicz, P.; Rudzka, M.; Hyjek-Skladanowska, M.; Golebiewski, M.; Smolinski, D.J.; Kolowerzo-Lubnau, A. Different Patterns of mRNA Nuclear Retention during Meiotic Prophase in Larch Microsporocytes. Int. J. Mol. Sci. 2021, 22, 8501. [Google Scholar] [CrossRef] [PubMed]
- Naro, C.; Jolly, A.; Di Persio, S.; Bielli, P.; Setterblad, N.; Alberdi, A.J.; Vicini, E.; Geremia, R.; De la Grange, P.; Sette, C. An Orchestrated Intron Retention Program in Meiosis Controls Timely Usage of Transcripts during Germ Cell Differentiation. Dev. Cell 2017, 41, 82–93 e84. [Google Scholar] [CrossRef]
- Paci, G.; Caria, J.; Lemke, E.A. Cargo transport through the nuclear pore complex at a glance. J. Cell Sci. 2021, 134, jcs247874. [Google Scholar] [CrossRef] [PubMed]
- Soheilypour, M.; Mofrad, M.R.K. Quality control of mRNAs at the entry of the nuclear pore: Cooperation in a complex molecular system. Nucleus 2018, 9, 202–211. [Google Scholar] [CrossRef]
- Le Hir, H.; Izaurralde, E.; Maquat, L.E.; Moore, M.J. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000, 19, 6860–6869. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Kucukural, A.; Cenik, C.; Leszyk, J.D.; Shaffer, S.A.; Weng, Z.; Moore, M.J. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 2012, 151, 750–764. [Google Scholar] [CrossRef] [PubMed]
- Botti, V.; McNicoll, F.; Steiner, M.C.; Richter, F.M.; Solovyeva, A.; Wegener, M.; Schwich, O.D.; Poser, I.; Zarnack, K.; Wittig, I.; et al. Cellular differentiation state modulates the mRNA export activity of SR proteins. J. Cell Biol. 2017, 216, 1993–2009. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.C.; Tarn, W.Y. Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J. Biol. Chem. 2004, 279, 31745–31749. [Google Scholar] [CrossRef]
- Müller-McNicoll, M.; Botti, V.; de Jesus Domingues, A.M.; Brandl, H.; Schwich, O.D.; Steiner, M.C.; Curk, T.; Poser, I.; Zarnack, K.; Neugebauer, K.M. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 2016, 30, 553–566. [Google Scholar] [CrossRef]
- Hackmann, A.; Wu, H.; Schneider, U.M.; Meyer, K.; Jung, K.; Krebber, H. Quality control of spliced mRNAs requires the shuttling SR proteins Gbp2 and Hrb1. Nat. Commun. 2014, 5, 3123. [Google Scholar] [CrossRef] [PubMed]
- Fasken, M.B.; Corbett, A.H. Links between mRNA Splicing, mRNA Quality Control, and Intellectual Disability. RNA Dis. 2016, 3. [Google Scholar] [CrossRef]
- Soucek, S.; Zeng, Y.; Bellur, D.L.; Bergkessel, M.; Morris, K.J.; Deng, Q.; Duong, D.; Seyfried, N.T.; Guthrie, C.; Staley, J.P.; et al. The Evolutionarily-conserved Polyadenosine RNA Binding Protein, Nab2, Cooperates with Splicing Machinery to Regulate the Fate of pre-mRNA. Mol. Cell. Biol. 2016, 36, 2697–2714. [Google Scholar] [CrossRef] [PubMed]
- Galy, V.; Gadal, O.; Fromont-Racine, M.; Romano, A.; Jacquier, A.; Nehrbass, U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 2004, 116, 63–73. [Google Scholar] [CrossRef]
- Coyle, J.H.; Bor, Y.C.; Rekosh, D.; Hammarskjold, M.L. The Tpr protein regulates export of mRNAs with retained introns that traffic through the Nxf1 pathway. RNA 2011, 17, 1344–1356. [Google Scholar] [CrossRef]
- Rajanala, K.; Nandicoori, V.K. Localization of nucleoporin Tpr to the nuclear pore complex is essential for Tpr mediated regulation of the export of unspliced RNA. PLoS ONE 2012, 7, e29921. [Google Scholar] [CrossRef]
- Green, D.M.; Johnson, C.P.; Hagan, H.; Corbett, A.H. The C-terminal domain of myosin-like protein 1 (Mlp1p) is a docking site for heterogeneous nuclear ribonucleoproteins that are required for mRNA export. Proc. Natl. Acad. Sci. USA 2003, 100, 1010–1015. [Google Scholar] [CrossRef]
- Vinciguerra, P.; Iglesias, N.; Camblong, J.; Zenklusen, D.; Stutz, F. Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. EMBO J. 2005, 24, 813–823. [Google Scholar] [CrossRef]
- Fasken, M.B.; Corbett, A.H. Mechanisms of nuclear mRNA quality control. RNA Biol. 2009, 6, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Skruzny, M.; Schneider, C.; Racz, A.; Weng, J.; Tollervey, D.; Hurt, E. An endoribonuclease functionally linked to perinuclear mRNP quality control associates with the nuclear pore complexes. PLoS Biol. 2009, 7, e8. [Google Scholar] [CrossRef] [PubMed]
- Sayani, S.; Janis, M.; Lee, C.Y.; Toesca, I.; Chanfreau, G.F. Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol. Cell 2008, 31, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Zander, G.; Hackmann, A.; Bender, L.; Becker, D.; Lingner, T.; Salinas, G.; Krebber, H. mRNA quality control is bypassed for immediate export of stress-responsive transcripts. Nature 2016, 540, 593–596. [Google Scholar] [CrossRef] [PubMed]
- Karousis, E.D.; Muhlemann, O. Nonsense-Mediated mRNA Decay Begins Where Translation Ends. Cold Spring Harb. Perspect. Biol. 2019, 11, a032862. [Google Scholar] [CrossRef]
- Kurosaki, T.; Popp, M.W.; Maquat, L.E. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. 2019, 20, 406–420. [Google Scholar] [CrossRef]
- Celik, A.; Baker, R.; He, F.; Jacobson, A. High-resolution profiling of NMD targets in yeast reveals translational fidelity as a basis for substrate selection. RNA 2017, 23, 735–748. [Google Scholar] [CrossRef]
- He, F.; Peltz, S.W.; Donahue, J.L.; Rosbash, M.; Jacobson, A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc. Natl. Acad. Sci. USA 1993, 90, 7034–7038. [Google Scholar] [CrossRef]
- Jaillon, O.; Bouhouche, K.; Gout, J.F.; Aury, J.M.; Noel, B.; Saudemont, B.; Nowacki, M.; Serrano, V.; Porcel, B.M.; Segurens, B.; et al. Translational control of intron splicing in eukaryotes. Nature 2008, 451, 359–362. [Google Scholar] [CrossRef]
- Green, R.E.; Lewis, B.P.; Hillman, R.T.; Blanchette, M.; Lareau, L.F.; Garnett, A.T.; Rio, D.C.; Brenner, S.E. Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics 2003, 19 (Suppl. 1), i118–i121. [Google Scholar] [CrossRef]
- Lewis, B.P.; Green, R.E.; Brenner, S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. USA 2003, 100, 189–192. [Google Scholar] [CrossRef] [PubMed]
- Barberan-Soler, S.; Lambert, N.J.; Zahler, A.M. Global analysis of alternative splicing uncovers developmental regulation of nonsense-mediated decay in C. elegans. RNA 2009, 15, 1652–1660. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.D.; Lareau, L.F.; Blanchette, M.; Green, R.E.; Meng, Q.; Rehwinkel, J.; Gallusser, F.L.; Izaurralde, E.; Rio, D.C.; Dudoit, S.; et al. Genome-wide identification of alternative splice forms down-regulated by nonsense-mediated mRNA decay in Drosophila. PLoS Genet. 2009, 5, e1000525. [Google Scholar] [CrossRef] [PubMed]
- McIlwain, D.R.; Pan, Q.; Reilly, P.T.; Elia, A.J.; McCracken, S.; Wakeham, A.C.; Itie-Youten, A.; Blencowe, B.J.; Mak, T.W. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc. Natl. Acad. Sci. USA 2010, 107, 12186–12191. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Moreno, J.F.; Romao, L. Perspective in Alternative Splicing Coupled to Nonsense-Mediated mRNA Decay. Int. J. Mol. Sci. 2020, 21, 9424. [Google Scholar] [CrossRef]
- Andjus, S.; Morillon, A.; Wery, M. From Yeast to Mammals, the Nonsense-Mediated mRNA Decay as a Master Regulator of Long Non-Coding RNAs Functional Trajectory. Non-Coding RNA 2021, 7, 44. [Google Scholar] [CrossRef] [PubMed]
- Nasif, S.; Contu, L.; Muhlemann, O. Beyond quality control: The role of nonsense-mediated mRNA decay (NMD) in regulating gene expression. Semin. Cell Dev. Biol. 2018, 75, 78–87. [Google Scholar] [CrossRef]
- Nickless, A.; Bailis, J.M.; You, Z. Control of gene expression through the nonsense-mediated RNA decay pathway. Cell Biosci. 2017, 7, 26. [Google Scholar] [CrossRef]
- Gupta, P.; Li, Y.R. Upf proteins: Highly conserved factors involved in nonsense mRNA mediated decay. Mol. Biol. Rep. 2018, 45, 39–55. [Google Scholar] [CrossRef]
- Czaplinski, K.; Ruiz-Echevarria, M.J.; Paushkin, S.V.; Han, X.; Weng, Y.; Perlick, H.A.; Dietz, H.C.; Ter-Avanesyan, M.D.; Peltz, S.W. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998, 12, 1665–1677. [Google Scholar] [CrossRef]
- Ivanov, P.V.; Gehring, N.H.; Kunz, J.B.; Hentze, M.W.; Kulozik, A.E. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008, 27, 736–747. [Google Scholar] [CrossRef] [PubMed]
- Chamieh, H.; Ballut, L.; Bonneau, F.; Le Hir, H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 2008, 15, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Czaplinski, K.; Weng, Y.; Hagan, K.W.; Peltz, S.W. Purification and characterization of the Upf1 protein: A factor involved in translation and mRNA degradation. RNA 1995, 1, 610–623. [Google Scholar] [PubMed]
- Fiorini, F.; Bagchi, D.; Le Hir, H.; Croquette, V. Human Upf1 is a highly processive RNA helicase and translocase with RNP remodelling activities. Nat. Commun. 2015, 6, 7581. [Google Scholar] [CrossRef]
- Franks, T.M.; Singh, G.; Lykke-Andersen, J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense- mediated mRNA decay. Cell 2010, 143, 938–950. [Google Scholar] [CrossRef]
- Serdar, L.D.; Whiteside, D.L.; Baker, K.E. ATP hydrolysis by UPF1 is required for efficient translation termination at premature stop codons. Nat. Commun. 2016, 7, 14021. [Google Scholar] [CrossRef]
- Serdar, L.D.; Whiteside, D.L.; Nock, S.L.; McGrath, D.; Baker, K.E. Inhibition of post-termination ribosome recycling at premature termination codons in UPF1 ATPase mutants. eLife 2020, 9, e57834. [Google Scholar] [CrossRef]
- Colombo, M.; Karousis, E.D.; Bourquin, J.; Bruggmann, R.; Mühlemann, O. Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways. RNA 2017, 23, 189–201. [Google Scholar] [CrossRef]
- Nagy, E.; Maquat, L.E. A rule for termination-codon position within intron-containing genes: When nonsense affects RNA abundance. Trends Biochem. Sci. 1998, 23, 198–199. [Google Scholar] [CrossRef]
- Schlautmann, L.P.; Gehring, N.H. A Day in the Life of the Exon Junction Complex. Biomolecules 2020, 10, 866. [Google Scholar] [CrossRef] [PubMed]
- Woodward, L.A.; Mabin, J.W.; Gangras, P.; Singh, G. The exon junction complex: A lifelong guardian of mRNA fate. Wiley Interdiscip. Rev. RNA 2017, 8, e1411. [Google Scholar] [CrossRef]
- Lejeune, F.; Ishigaki, Y.; Li, X.; Maquat, L.E. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: Dynamics of mRNP remodeling. EMBO J. 2002, 21, 3536–3545. [Google Scholar] [CrossRef] [PubMed]
- Buchwald, G.; Ebert, J.; Basquin, C.; Sauliere, J.; Jayachandran, U.; Bono, F.; Le Hir, H.; Conti, E. Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC-UPF3b complex. Proc. Natl. Acad. Sci. USA 2010, 107, 10050–10055. [Google Scholar] [CrossRef]
- Gehring, N.H.; Neu-Yilik, G.; Schell, T.; Hentze, M.W.; Kulozik, A.E. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell 2003, 11, 939–949. [Google Scholar] [CrossRef]
- Wen, J.; He, M.; Petrj, M.; Marzi, L.; Wang, J.; Piechocki, K.; McLeod, T.; Singh, A.K.; Dwivedi, V.; Brogna, S. An intron proximal to a PTC enhances NMD in Saccharomyces cerevisiae. bioRxiv 2020, 149245. [Google Scholar] [CrossRef]
- Cao, D.; Parker, R. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell 2003, 113, 533–545. [Google Scholar] [CrossRef][Green Version]
- Hagan, K.W.; Ruiz-Echevarria, M.J.; Quan, Y.; Peltz, S.W. Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol. Cell. Biol. 1995, 15, 809–823. [Google Scholar] [CrossRef]
- He, F.; Li, X.; Spatrick, P.; Casillo, R.; Dong, S.; Jacobson, A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 2003, 12, 1439–1452. [Google Scholar] [CrossRef]
- Muhlrad, D.; Parker, R. Premature translational termination triggers mRNA decapping. Nature 1994, 370, 578–581. [Google Scholar] [CrossRef]
- Nissan, T.; Rajyaguru, P.; She, M.; Song, H.; Parker, R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol. Cell 2010, 39, 773–783. [Google Scholar] [CrossRef]
- Mitchell, P.; Tollervey, D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′-->5′ degradation. Mol. Cell 2003, 11, 1405–1413. [Google Scholar] [CrossRef]
- Yamashita, A.; Izumi, N.; Kashima, I.; Ohnishi, T.; Saari, B.; Katsuhata, Y.; Muramatsu, R.; Morita, T.; Iwamatsu, A.; Hachiya, T.; et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009, 23, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Eberle, A.B.; Lykke-Andersen, S.; Mühlemann, O.; Jensen, T.H. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat. Struct. Mol. Biol. 2009, 16, 49–55. [Google Scholar] [CrossRef]
- Gatfield, D.; Izaurralde, E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 2004, 429, 575–578. [Google Scholar] [CrossRef]
- Huntzinger, E.; Kashima, I.; Fauser, M.; Saulière, J.; Izaurralde, E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 2008, 14, 2609–2617. [Google Scholar] [CrossRef]
- Ohnishi, T.; Yamashita, A.; Kashima, I.; Schell, T.; Anders, K.R.; Grimson, A.; Hachiya, T.; Hentze, M.W.; Anderson, P.; Ohno, S. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell 2003, 12, 1187–1200. [Google Scholar] [CrossRef]
- Okada-Katsuhata, Y.; Yamashita, A.; Kutsuzawa, K.; Izumi, N.; Hirahara, F.; Ohno, S. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 2012, 40, 1251–1266. [Google Scholar] [CrossRef]
- Loh, B.; Jonas, S.; Izaurralde, E. The SMG5-SMG7 heterodimer directly recruits the CCR4-NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev. 2013, 27, 2125–2138. [Google Scholar] [CrossRef]
- Muhlrad, D.; Parker, R. Recognition of yeast mRNAs as “nonsense containing” leads to both inhibition of mRNA translation and mRNA degradation: Implications for the control of mRNA decapping. Mol. Biol. Cell 1999, 10, 3971–3978. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Isken, O.; Kim, Y.K.; Hosoda, N.; Mayeur, G.L.; Hershey, J.W.; Maquat, L.E. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 2008, 133, 314–327. [Google Scholar] [CrossRef] [PubMed]
- Egecioglu, D.E.; Chanfreau, G. Proofreading and spellchecking: A two-tier strategy for pre-mRNA splicing quality control. RNA 2011, 17, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Sayani, S.; Chanfreau, G.F. Sequential RNA degradation pathways provide a fail-safe mechanism to limit the accumulation of unspliced transcripts in Saccharomyces cerevisiae. RNA 2012, 18, 1563–1572. [Google Scholar] [CrossRef] [PubMed]
- Hacker, S.; Krebber, H. Differential Export Requirements for Shuttling Serine/Arginine-type mRNA-binding Proteins. J. Biol. Chem. 2004, 279, 5049–5052. [Google Scholar] [CrossRef]
- Windgassen, M.; Krebber, H. Identification of Gbp2 as a novel poly(A)+ RNA-binding protein involved in the cytoplasmic delivery of messenger RNAs in yeast. EMBO Rep. 2003, 4, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Bossie, M.A.; Silver, P.A. Movement of macromolecules between the cytoplasm and the nucleus in yeast. Curr. Opin. Genet. Dev. 1992, 2, 768–774. [Google Scholar] [CrossRef]
- Birney, E.; Kumar, S.; Krainer, A.R. Analysis of the RNA-recognition motif and RS and RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993, 21, 5803–5816. [Google Scholar] [CrossRef]
- Wegener, M.; Muller-McNicoll, M. View from an mRNP: The Roles of SR Proteins in Assembly, Maturation and Turnover. Adv. Exp. Med. Biol 2019, 1203, 83–112. [Google Scholar] [CrossRef] [PubMed]
- Baierlein, C.; Hackmann, A.; Gross, T.; Henker, L.; Hinz, F.; Krebber, H. Monosome formation during translation initiation requires the serine/arginine-rich protein Npl3. Mol. Cell. Biol. 2013, 33, 4811–4823. [Google Scholar] [CrossRef]
- Bucheli, M.E.; Buratowski, S. Npl3 is an antagonist of mRNA 3′ end formation by RNA polymerase II. EMBO J. 2005, 24, 2150–2160. [Google Scholar] [CrossRef]
- Dermody, J.L.; Dreyfuss, J.M.; Villén, J.; Ogundipe, B.; Gygi, S.P.; Park, P.J.; Ponticelli, A.S.; Moore, C.L.; Buratowski, S.; Bucheli, M.E. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS ONE 2008, 3, e3273. [Google Scholar] [CrossRef]
- Estrella, L.A.; Wilkinson, M.F.; Gonzalez, C.I. The Shuttling Protein Npl3 Promotes Translation Termination Accuracy in Saccharomyces cerevisiae. J. Mol. Biol. 2009. [Google Scholar] [CrossRef]
- Kress, T.L.; Krogan, N.J.; Guthrie, C. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol. Cell 2008, 32, 727–734. [Google Scholar] [CrossRef]
- Lee, M.S.; Henry, M.; Pamela, A. A protein that shuttles between the nucleus and the cvtoplasm is an important mediator of RNA export. Genes Dev. 1996, 10, 1233–1246. [Google Scholar] [CrossRef] [PubMed]
- Rajyaguru, P.; She, M.; Parker, R. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol. Cell 2012, 45, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Windgassen, M.; Sturm, D.; Cajigas, I.J.; Gonzalez, C.I.; Seedorf, M.; Bastians, H.; Krebber, H. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol. Cell. Biol. 2004, 24, 10479–10491. [Google Scholar] [CrossRef] [PubMed]
- Caceres, J.F.; Screaton, G.R.; Krainer, A.R. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998, 12, 55–66. [Google Scholar] [CrossRef]
- Huang, Y.; Steitz, J.A. SRprises along a messenger’s journey. Mol. Cell 2005, 17, 613–615. [Google Scholar] [CrossRef] [PubMed]
- Hurt, E.; Luo, M.-J.; Röther, S.; Reed, R.; Strässer, K. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc. Natl. Acad. Sci. USA 2004, 101, 1858–1862. [Google Scholar] [CrossRef] [PubMed]
- Meinel, D.M.; Burkert-Kautzsch, C.; Kieser, A.; O’Duibhir, E.; Siebert, M.; Mayer, A.; Cramer, P.; Söding, J.; Holstege, F.C.P.; Sträßer, K. Recruitment of TREX to the Transcription Machinery by Its Direct Binding to the Phospho-CTD of RNA Polymerase II. PLoS Genet. 2013, 9, e1003914. [Google Scholar] [CrossRef] [PubMed]
- Meinel, D.M.; Sträßer, K. Co-transcriptional mRNP formation is coordinated within a molecular mRNP packaging station in S. cerevisiae. BioEssays 2015, 37, 666–677. [Google Scholar] [CrossRef]
- Abruzzi, K.C.; Lacadie, S.; Rosbash, M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004, 23, 2620–2631. [Google Scholar] [CrossRef]
- Chanarat, S.; Seizl, M.; Strasser, K. The Prp19 complex is a novel transcription elongation factor required for TREX occupancy at transcribed genes. Genes Dev. 2011, 25, 1147–1158. [Google Scholar] [CrossRef]
- Gromadzka, A.M.; Steckelberg, A.L.; Singh, K.K.; Hofmann, K.; Gehring, N.H. A short conserved motif in ALYREF directs cap- and EJC-dependent assembly of export complexes on spliced mRNAs. Nucleic Acids Res. 2016, 44, 2348–2361. [Google Scholar] [CrossRef] [PubMed]
- Lardelli, R.M.; Thompson, J.X.; Yates, J.R., 3rd; Stevens, S.W. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 2010, 16, 516–528. [Google Scholar] [CrossRef] [PubMed]
- Masuda, S.; Das, R.; Cheng, H.; Hurt, E.; Dorman, N.; Reed, R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005, 19, 1512–1517. [Google Scholar] [CrossRef]
- Warkocki, Z.; Odenwalder, P.; Schmitzova, J.; Platzmann, F.; Stark, H.; Urlaub, H.; Ficner, R.; Fabrizio, P.; Luhrmann, R. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat. Struct. Mol. Biol. 2009, 16, 1237–1243. [Google Scholar] [CrossRef]
- Tuck, A.C.; Tollervey, D. A Transcriptome-wide Atlas of RNP Composition Reveals Diverse Classes of mRNAs and lncRNAs. Cell 2013, 154, 996–1009. [Google Scholar] [CrossRef]
- Mourier, T.; Jeffares, D.C. Eukaryotic intron loss. Science 2003, 300, 1393. [Google Scholar] [CrossRef] [PubMed]
- Neuveglise, C.; Marck, C.; Gaillardin, C. The intronome of budding yeasts. Comptes Rendus Biol. 2011, 334, 662–670. [Google Scholar] [CrossRef]
- Baejen, C.; Torkler, P.; Gressel, S.; Essig, K.; Söding, J.; Cramer, P. Transcriptome Maps of mRNP Biogenesis Factors Define Pre-mRNA Recognition. Mol. Cell 2014, 55, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Bretes, H.; Rouviere, J.O.; Leger, T.; Oeffinger, M.; Devaux, F.; Doye, V.; Palancade, B. Sumoylation of the THO complex regulates the biogenesis of a subset of mRNPs. Nucleic Acids Res. 2014, 42, 5043–5058. [Google Scholar] [CrossRef] [PubMed]
- Hackmann, A.; Gross, T.; Baierlein, C.; Krebber, H. The mRNA export factor Npl3 mediates the nuclear export of large ribosomal subunits. EMBO Rep. 2011, 12, 1024–1031. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.Y.E.; Tang, H.M.V.; Pan, K.; Huang, Z.; Lee, T.H.J.; Hinnebusch, A.G.; Jin, D.Y.; Wong, C.M. Cotranscriptional recruitment of yeast TRAMP complex to intronic sequences promotes optimal pre-mRNA splicing. Nucleic Acids Res. 2014, 42, 643–660. [Google Scholar] [CrossRef] [PubMed]
- Bresson, S.; Tollervey, D. Surveillance-ready transcription: Nuclear RNA decay as a default fate. Open Biol. 2018, 8, 170270. [Google Scholar] [CrossRef] [PubMed]
- Jensen, T.H.; Dower, K.; Libri, D.; Rosbash, M. Early formation of mRNP: License for export or quality control? Mol. Cell 2003, 11, 1129–1138. [Google Scholar] [CrossRef]
- Saguez, C.; Olesen, J.R.; Jensen, T.H. Formation of export-competent mRNP: Escaping nuclear destruction. Curr. Opin. Cell Biol. 2005, 17, 287–293. [Google Scholar] [CrossRef]
- Stewart, M. Nuclear export of mRNA. Trends Biochem. Sci. 2010, 35, 609–617. [Google Scholar] [CrossRef]
- Merrick, W.C.; Pavitt, G.D. Protein Synthesis Initiation in Eukaryotic Cells. Cold Spring Harb. Perspect. Biol. 2018, 10, a033092. [Google Scholar] [CrossRef]
- Lund, M.K.; Guthrie, C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol. Cell 2005, 20, 645–651. [Google Scholar] [CrossRef]
- Tieg, B.; Krebber, H. Dbp5—From nuclear export to translation. Biochim. Biophys. Acta 2013, 1829, 791–798. [Google Scholar] [CrossRef]
- Tran, E.J.; Zhou, Y.; Corbett, A.H.; Wente, S.R. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell 2007, 28, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Grosse, S.; Lu, Y.Y.; Coban, I.; Neumann, B.; Krebber, H. Nuclear SR-protein mediated mRNA quality control is continued in cytoplasmic nonsense-mediated decay. RNA Biol. 2021, 18, 1390–1407. [Google Scholar] [CrossRef]
- Johnson, S.J.; Jackson, R.N. Ski2-like RNA helicase structures: Common themes and complex assemblies. RNA Biol. 2013, 10, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Poornima, G.; Srivastava, G.; Roy, B.; Kuttanda, I.A.; Kurbah, I.; Rajyaguru, P.I. RGG-motif containing mRNA export factor Gbp2 acts as a translation repressor. RNA Biol. 2021, 1–12. [Google Scholar] [CrossRef]
- He, F.; Jacobson, A. Nonsense-Mediated mRNA Decay: Degradation of Defective Transcripts Is Only Part of the Story. Annu Rev. Genet. 2015, 49, 339–366. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Krainer, A.R. Involvement of SR proteins in mRNA surveillance. Mol. Cell 2004, 16, 597–607. [Google Scholar] [CrossRef]
- Aznarez, I.; Nomakuchi, T.T.; Tetenbaum-Novatt, J.; Rahman, M.A.; Fregoso, O.; Rees, H.; Krainer, A.R. Mechanism of Nonsense-Mediated mRNA Decay Stimulation by Splicing Factor SRSF1. Cell Rep. 2018, 23, 2186–2198. [Google Scholar] [CrossRef]
- Rahman, M.A.; Lin, K.T.; Bradley, R.K.; Abdel-Wahab, O.; Krainer, A.R. Recurrent SRSF2 mutations in MDS affect both splicing and NMD. Genes Dev. 2020, 34, 413–427. [Google Scholar] [CrossRef]
- Kim, J.; Park, R.Y.; Chen, J.K.; Kim, J.; Jeong, S.; Ohn, T. Splicing factor SRSF3 represses the translation of programmed cell death 4 mRNA by associating with the 5′-UTR region. Cell Death Differ. 2014, 21, 481–490. [Google Scholar] [CrossRef]
- Swartz, J.E.; Bor, Y.C.; Misawa, Y.; Rekosh, D.; Hammarskjold, M.L. The shuttling SR protein 9G8 plays a role in translation of unspliced mRNA containing a constitutive transport element. J. Biol. Chem. 2007, 282, 19844–19853. [Google Scholar] [CrossRef]
- Siebel, C.W.; Feng, L.; Guthrie, C.; Fu, X.D. Conservation in budding yeast of a kinase specific for SR splicing factors. Proc. Natl. Acad. Sci. USA 1999, 96, 5440–5445. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, N.; Bachorik, J.L.; Dreyfuss, G. Transportin-SR, a nuclear import receptor for SR proteins. J. Cell Biol. 1999, 145, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Saudemont, B.; Popa, A.; Parmley, J.L.; Rocher, V.; Blugeon, C.; Necsulea, A.; Meyer, E.; Duret, L. The fitness cost of mis-splicing is the main determinant of alternative splicing patterns. Genome Biol. 2017, 18, 208. [Google Scholar] [CrossRef] [PubMed]
- Anna, A.; Monika, G. Splicing mutations in human genetic disorders: Examples, detection, and confirmation. J. Appl. Genet. 2018, 59, 253–268. [Google Scholar] [CrossRef]
- Scotti, M.M.; Swanson, M.S. RNA mis-splicing in disease. Nat. Rev. Genet. 2016, 17, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Urbanski, L.M.; Leclair, N.; Anczukow, O. Alternative-splicing defects in cancer: Splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip. Rev. RNA 2018, 9, e1476. [Google Scholar] [CrossRef]
- Bhadra, M.; Howell, P.; Dutta, S.; Heintz, C.; Mair, W.B. Alternative splicing in aging and longevity. Hum. Genet. 2020, 139, 357–369. [Google Scholar] [CrossRef]
- Buchan, J.R.; Muhlrad, D.; Parker, R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 2008, 183, 441–455. [Google Scholar] [CrossRef]
- Twyffels, L.; Gueydan, C.; Kruys, V. Shuttling SR proteins: More than splicing factors. FEBS J. 2011, 278, 3246–3255. [Google Scholar] [CrossRef]
- Cai, B.; Li, Z.; Ma, M.; Zhang, J.; Kong, S.; Abdalla, B.A.; Xu, H.; Jebessa, E.; Zhang, X.; Lawal, R.A.; et al. Long noncoding RNA SMUL suppresses SMURF2 production-mediated muscle atrophy via nonsense-mediated mRNA decay. Mol. Nucleic Acids 2021, 23, 512–526. [Google Scholar] [CrossRef]
- Wery, M.; Descrimes, M.; Vogt, N.; Dallongeville, A.S.; Gautheret, D.; Morillon, A. Nonsense-Mediated Decay Restricts LncRNA Levels in Yeast Unless Blocked by Double-Stranded RNA Structure. Mol. Cell 2016, 61, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Ajiro, M.; Jia, R.; Yang, Y.; Zhu, J.; Zheng, Z.M. A genome landscape of SRSF3-regulated splicing events and gene expression in human osteosarcoma U2OS cells. Nucleic Acids Res. 2016, 44, 1854–1870. [Google Scholar] [CrossRef]
- Li, H.; Guo, S.; Zhang, M.; Li, L.; Wang, F.; Song, B. Long non-coding RNA AGAP2-AS1 accelerates cell proliferation, migration, invasion and the EMT process in colorectal cancer via regulating the miR-4,668-3p/SRSF1 axis. J. Gene Med. 2020, 22, e3250. [Google Scholar] [CrossRef] [PubMed]
- Paz, S.; Ritchie, A.; Mauer, C.; Caputi, M. The RNA binding protein SRSF1 is a master switch of gene expression and regulation in the immune system. Cytokine Growth Factor Rev. 2021, 57, 19–26. [Google Scholar] [CrossRef]
- Sokol, E.; Kedzierska, H.; Czubaty, A.; Rybicka, B.; Rodzik, K.; Tanski, Z.; Boguslawska, J.; Piekielko-Witkowska, A. microRNA-mediated regulation of splicing factors SRSF1, SRSF2 and hnRNP A1 in context of their alternatively spliced 3′UTRs. Exp. Cell Res. 2018, 363, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.F.; Xu, X.; Gin, A.; Nshimiyimana, J.D.; Mooers, B.H.M.; Caputi, M.; Hannafon, B.N.; Ding, W.Q. SRSF1 regulates exosome microRNA enrichment in human cancer cells. Cell Commun. Signal. 2020, 18, 130. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.-Y.; Krebber, H. Nuclear mRNA Quality Control and Cytoplasmic NMD Are Linked by the Guard Proteins Gbp2 and Hrb1. Int. J. Mol. Sci. 2021, 22, 11275. https://doi.org/10.3390/ijms222011275
Lu Y-Y, Krebber H. Nuclear mRNA Quality Control and Cytoplasmic NMD Are Linked by the Guard Proteins Gbp2 and Hrb1. International Journal of Molecular Sciences. 2021; 22(20):11275. https://doi.org/10.3390/ijms222011275
Chicago/Turabian StyleLu, Yen-Yun, and Heike Krebber. 2021. "Nuclear mRNA Quality Control and Cytoplasmic NMD Are Linked by the Guard Proteins Gbp2 and Hrb1" International Journal of Molecular Sciences 22, no. 20: 11275. https://doi.org/10.3390/ijms222011275
APA StyleLu, Y.-Y., & Krebber, H. (2021). Nuclear mRNA Quality Control and Cytoplasmic NMD Are Linked by the Guard Proteins Gbp2 and Hrb1. International Journal of Molecular Sciences, 22(20), 11275. https://doi.org/10.3390/ijms222011275





