Progranulin A Promotes Compensatory Hepatocyte Proliferation via HGF/c-Met Signaling after Partial Hepatectomy in Zebrafish
Abstract
:1. Introduction
2. Results
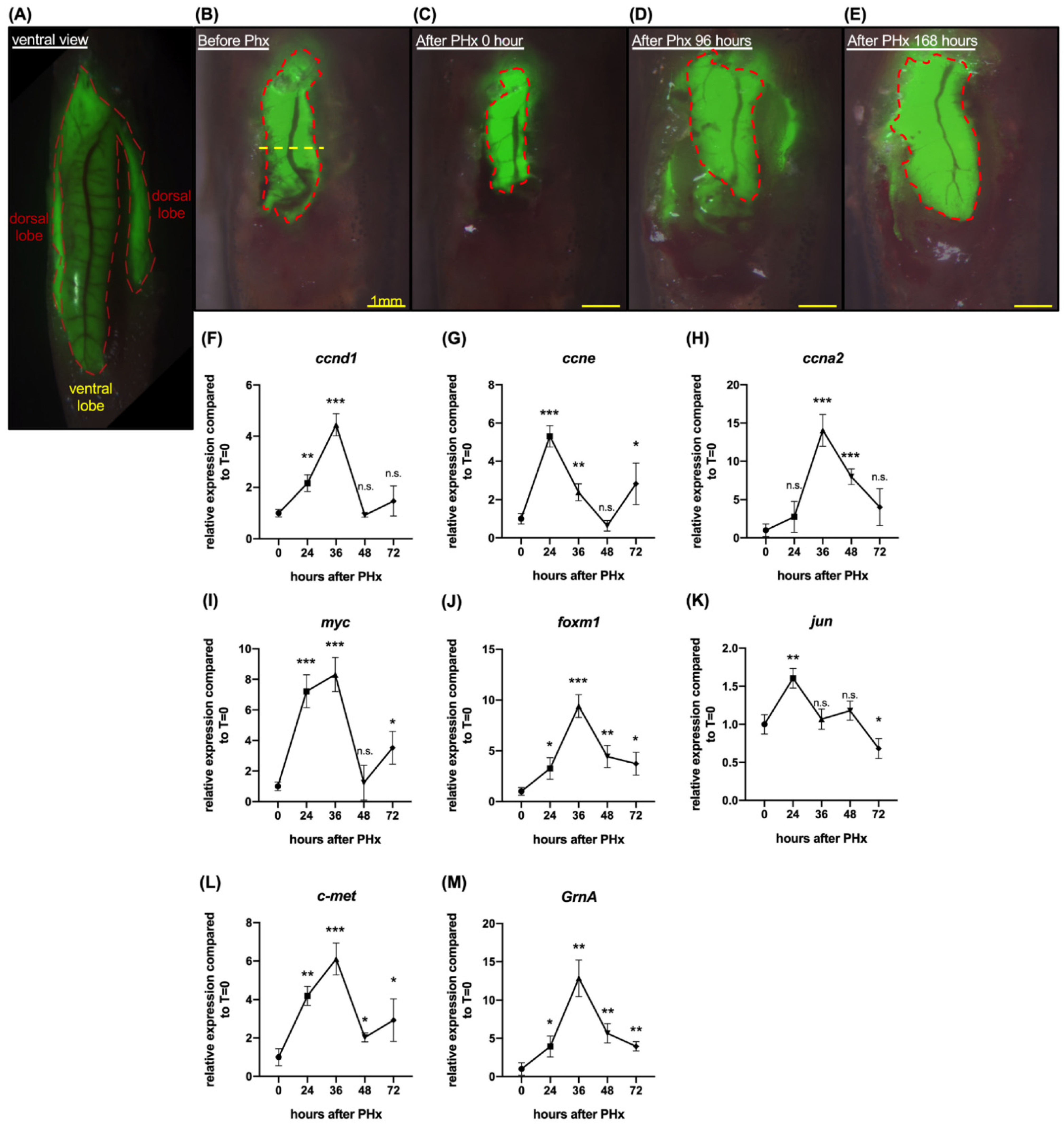
2.1. Zebrafish Progranulin A Is Involved in Liver Regeneration of Hepatectomized Zebrafish
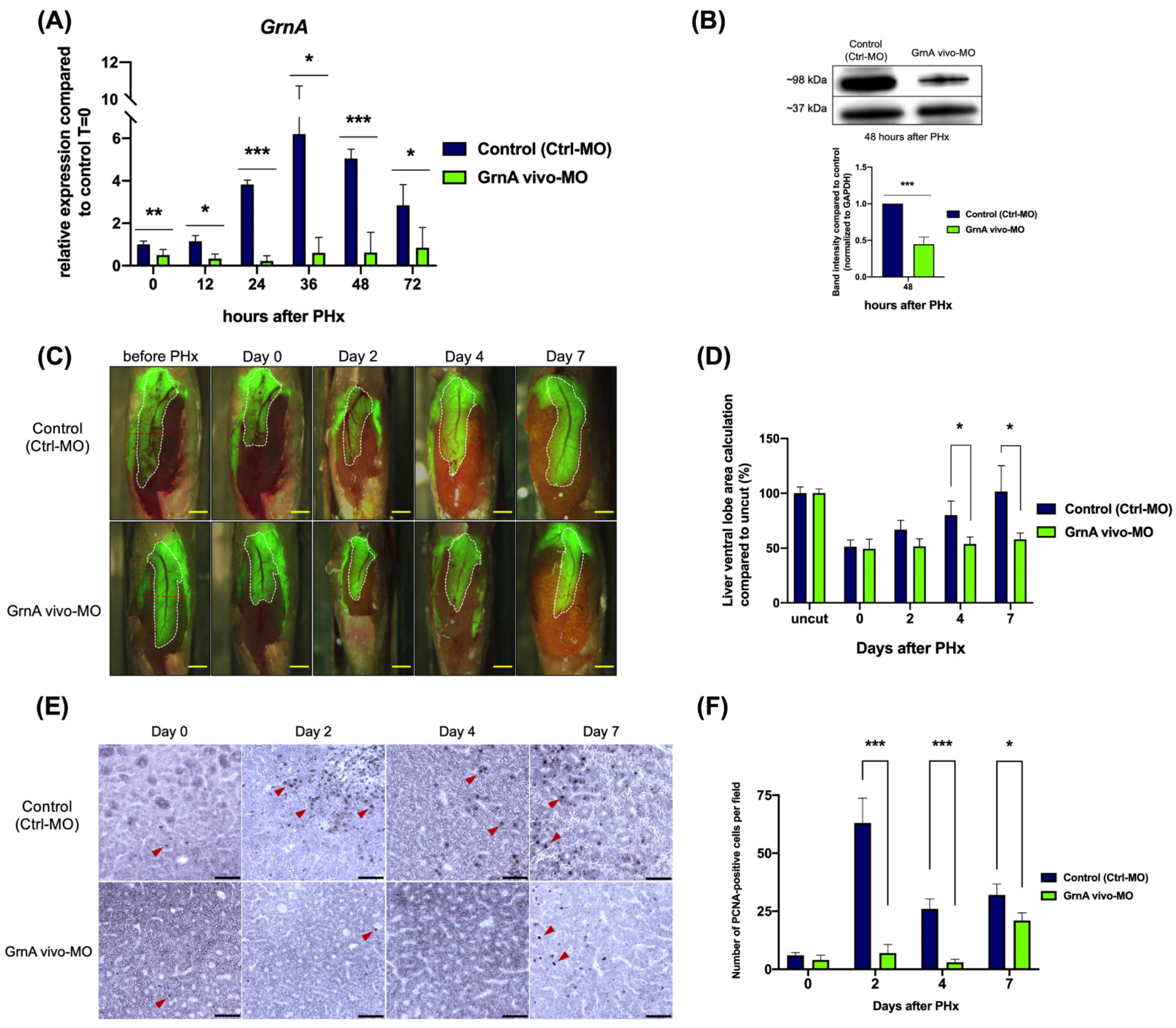
2.2. Knockdown of GrnA Delayed Liver Regeneration after Partial Hepatectomy
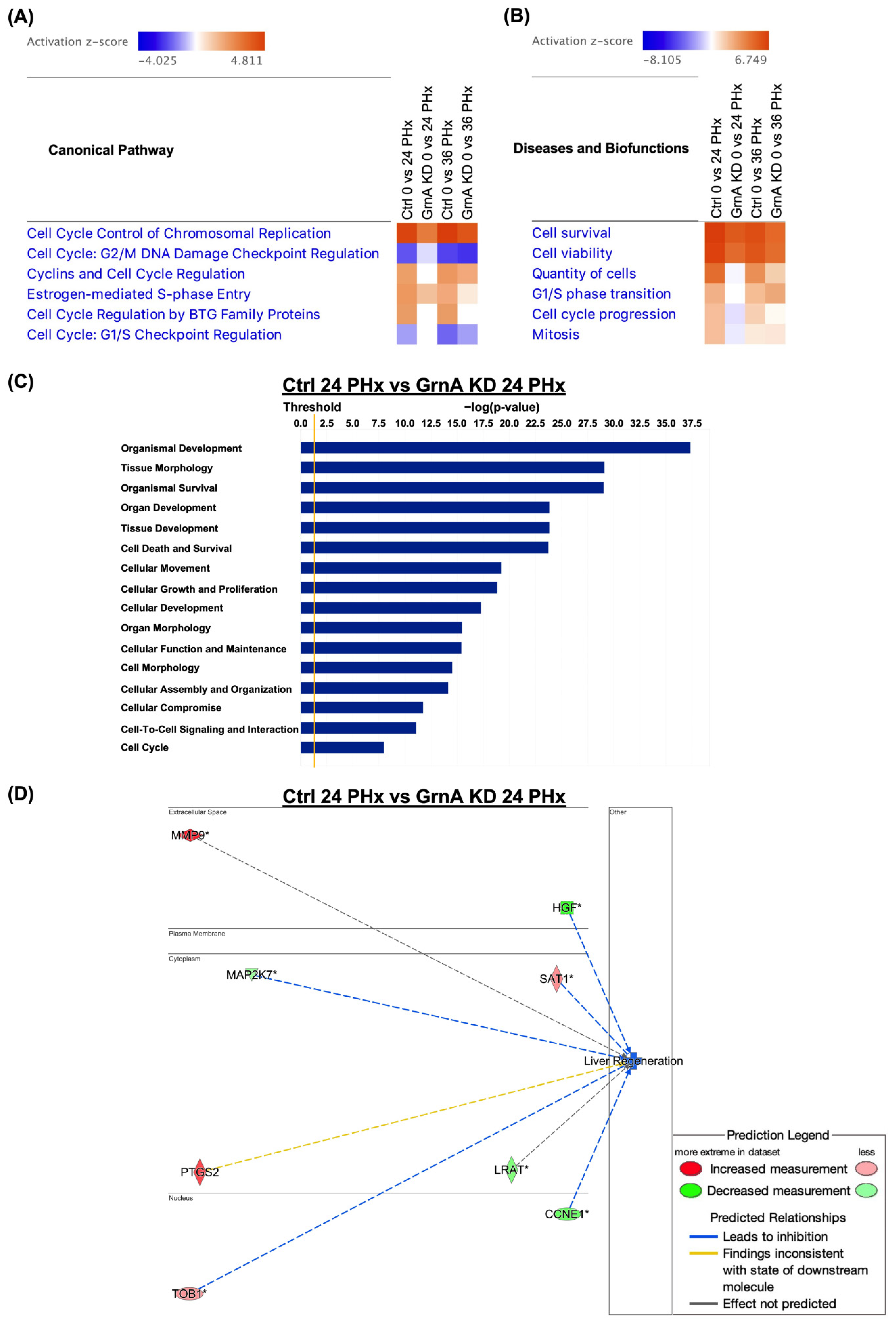
2.3. Transcriptome Analysis
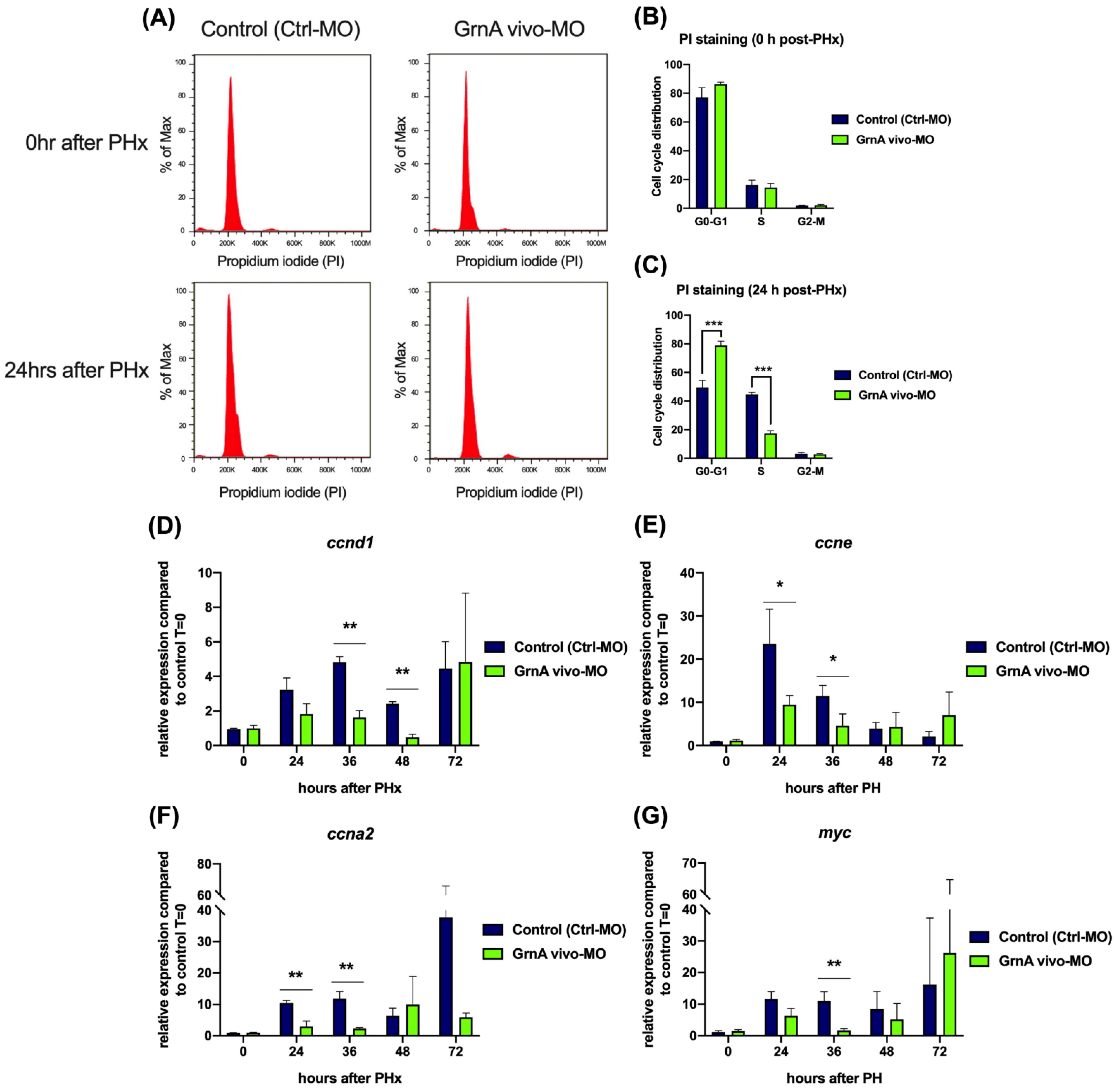
2.4. GrnA Morphants Exhibited Impaired Liver Regeneration with Suppressed Cell Cycle and Cell Proliferation
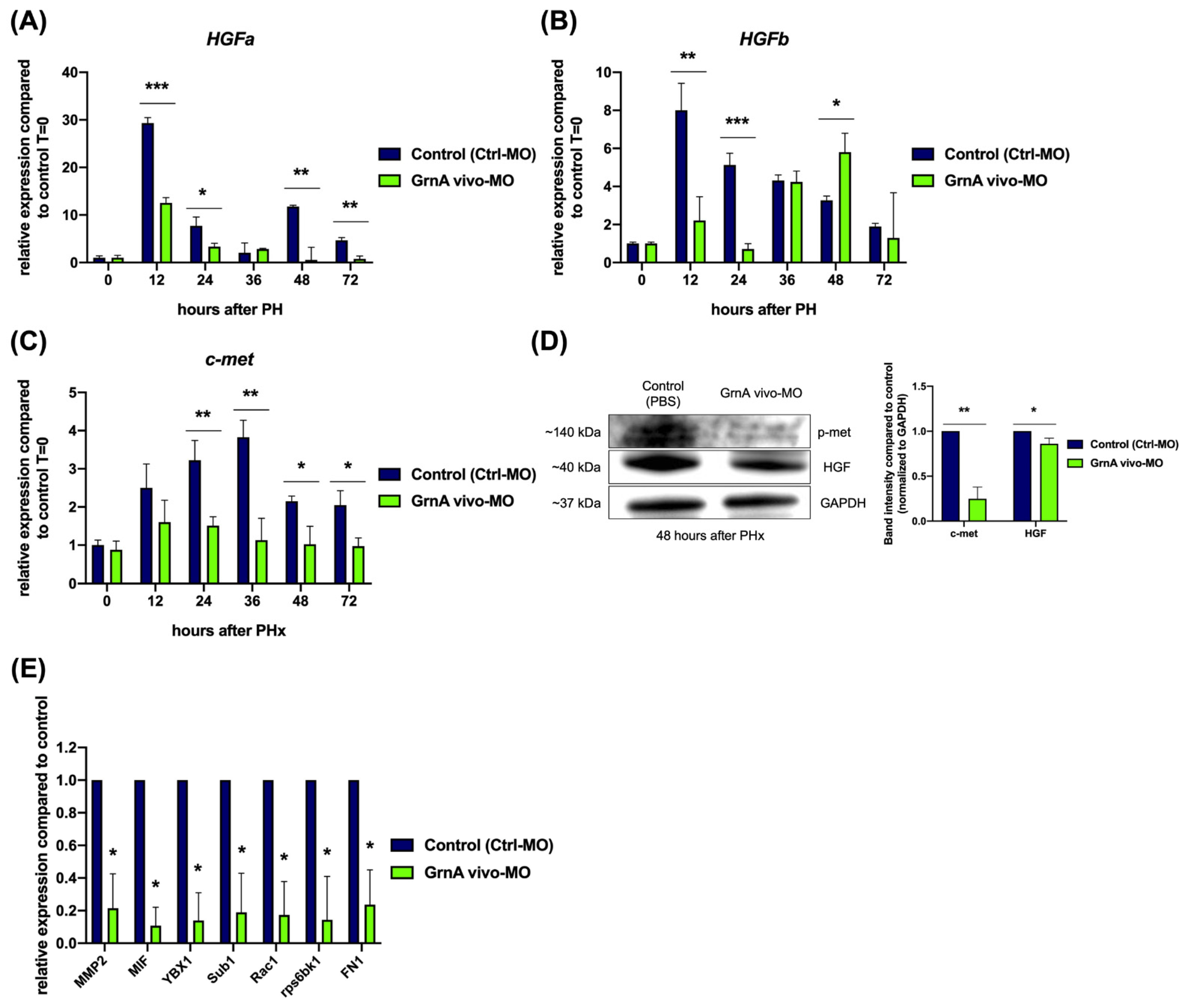
2.5. Knockdown of GrnA Prevented Signaling through the HGF/C-Met Axis after Partial Hepatectomy
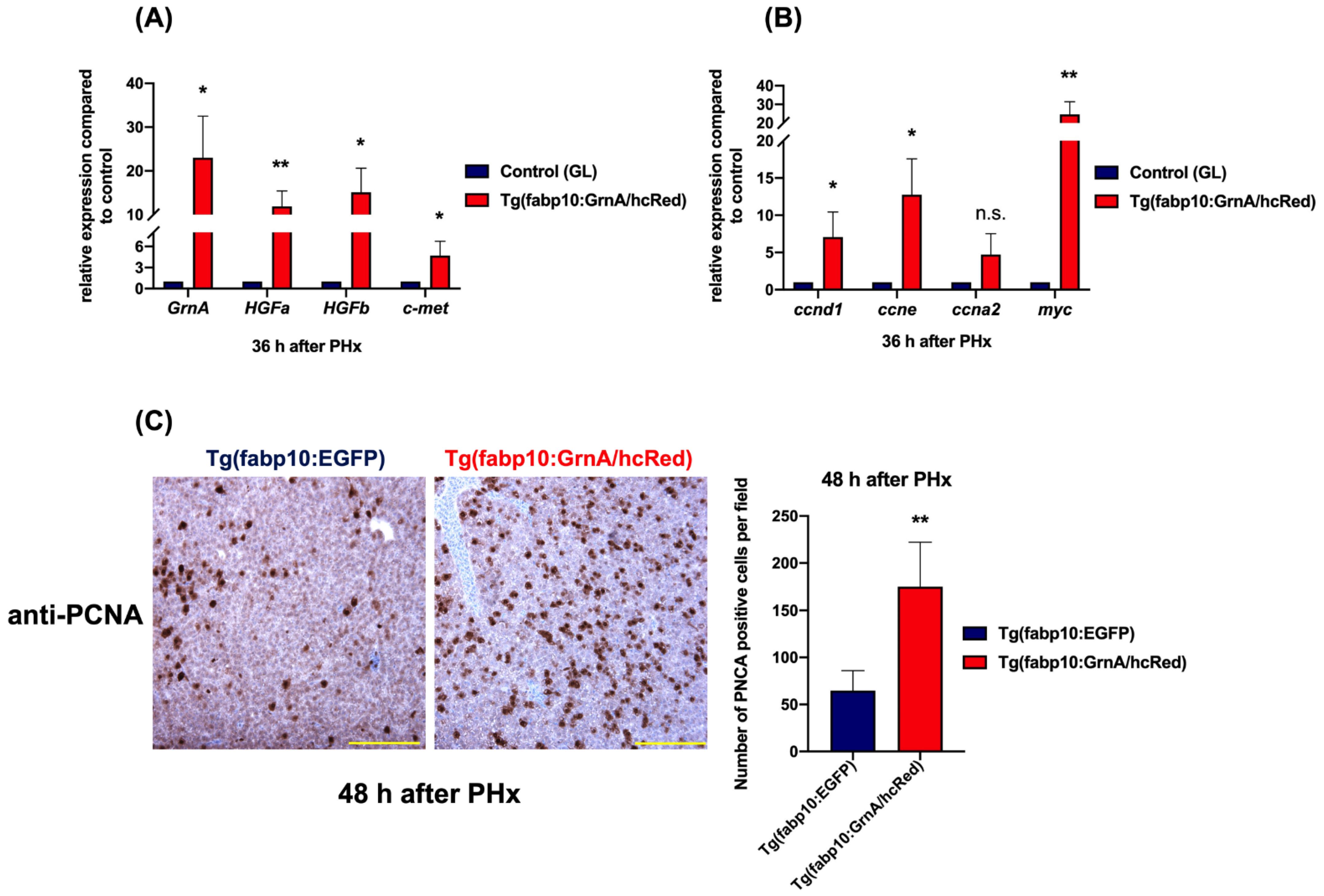
2.6. Liver-Specific Expression of GrnA in Transgenic Zebrafish Promoted Hepatocyte Proliferation after Partial Hepatectomy
3. Discussion
4. Materials and Methods
4.1. Zebrafish
4.2. Morpholino Design and Intraperitoneal Injection
4.3. Partial Hepatectomy
4.4. RNA Sequencing and Analysis
4.5. Cell Cycle Analysis by Flow Cytometry
4.6. Quantitative Real-Time PCR
4.7. Immunohistochemistry and Western Blots
4.8. Imaging
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wasserman, D.H. Four grams of glucose. Am. J. Physiol. Metab. 2009, 296, E11–E21. [Google Scholar] [CrossRef] [PubMed]
- Grant, D.M. Detoxification pathways in the liver. J. Inherit. Metab. Dis. 1991, 14, 421–430. [Google Scholar] [CrossRef]
- Boyer, J.L. Bile Formation and Secretion. Compr. Physiol. 2013, 3, 1035–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaret, K.S. Regulatory phases of early liver development: Paradigms of organogenesis. Nat. Rev. Genet. 2002, 3, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Taub, R. Liver regeneration: From myth to mechanism. Nat. Rev. Mol. Cell Biol. 2004, 5, 836–847. [Google Scholar] [CrossRef] [PubMed]
- Fausto, N.; Campbell, J.S.; Riehle, K.J. Liver regeneration. Hepatol. 2006, 43, S45–S53. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.-Y.; Chen, K.-F.; Chen, P.-J. Treatment of Liver Cancer. Cold Spring Harb. Perspect. Med. 2015, 5, a021535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuppan, D.; Afdhal, N.H. Liver cirrhosis. Lancet 2008, 371, 838–851. [Google Scholar] [CrossRef]
- Böhm, F.; Köhler, U.A.; Speicher, T.; Werner, S. Regulation of liver regeneration by growth factors and cytokines. EMBO Mol. Med. 2010, 2, 294–305. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, T.; Yamaguchi, S.; Mitsui, S.; Emi, A.; Shimoda, F.; Okamura, H. Control mechanism of the circadian clock for timing of cell division in vivo. Science 2003, 302, 255–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schibler, U. Liver Regeneration Clocks On. Sci. 2003, 302, 234–235. [Google Scholar] [CrossRef] [PubMed]
- Weglarz, T.C.; Sandgren, E.P. Timing of hepatocyte entry into DNA synthesis after partial hepatectomy is cell autonomous. Proc. Natl. Acad. Sci. USA 2000, 97, 12595–12600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borowiak, M.; Garratt, A.; Wüstefeld, T.; Strehle, M.; Trautwein, C.; Birchmeier, C. Met provides essential signals for liver regeneration. Proc. Natl. Acad. Sci. USA 2004, 101, 10608–10613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Factor, V.M.; Seo, D.; Ishikawa, T.; Kaposi-Novak, P.; Marquardt, J.; Andersen, J.B.; Conner, E.A.; Thorgeirsson, S.S. Loss of c-Met Disrupts Gene Expression Program Required for G2/M Progression during Liver Regeneration in Mice. PLoS ONE 2010, 5, e12739. [Google Scholar] [CrossRef] [PubMed]
- Benvenuti, S.; Comoglio, P.M. The MET receptor tyrosine kinase in invasion and metastasis. J. Cell. Physiol. 2007, 213, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Huh, C.-G.; Factor, V.M.; Sánchez, A.; Uchida, K.; Conner, E.A.; Thorgeirsson, S.S. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proc. Natl. Acad. Sci. USA 2004, 101, 4477–4482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stolz, D.B.; Mars, W.M.; Petersen, B.E.; Kim, T.H.; Michalopoulos, G.K. Growth factor signal transduction immediately after two-thirds partial hepatectomy in the rat. Cancer Res. 1999, 59, 3954–3960. [Google Scholar] [PubMed]
- Bell, A.; Chen, Q.; DeFrances, M.C.; Michalopoulos, G.K.; Zarnegar, R. The five amino acid-deleted isoform of hepatocyte growth factor promotes carcinogenesis in transgenic mice. Oncogene 1999, 18, 887–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michalopoulos, G.K. Liver regeneration. J. Cell. Physiol. 2007, 213, 286–300. [Google Scholar] [CrossRef]
- He, Z.; Bateman, A. Progranulin (granulin-epithelin precursor, PC-cell-derived growth factor, acrogranin) mediates tissue repair and tumorigenesis. J. Mol. Med. 2003, 81, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-H.; Chen, M.H.-C.; Gong, H.-Y.; Hu, S.-Y.; Li, Y.-W.; Lin, G.-H.; Lin, C.-C.; Liu, W.; Wu, J.-L. Progranulin A-mediated MET Signaling Is Essential for Liver Morphogenesis in Zebrafish. J. Biol. Chem. 2010, 285, 41001–41009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kung, J.W.C.; Currie, I.S.; Forbes, S.J.; Ross, J.A. Liver Development, Regeneration, and Carcinogenesis. J. Biomed. Biotechnol. 2010, 2010, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otu, H.; Naxerova, K.; Ho, K.; Can, H.; Nesbitt, N.; Libermann, T.; Karp, S.J. Restoration of Liver Mass after Injury Requires Proliferative and Not Embryonic Transcriptional Patterns. J. Biol. Chem. 2007, 282, 11197–11204. [Google Scholar] [CrossRef] [Green Version]
- Kuse, Y.; Tsuruma, K.; Sugitani, S.; Izawa, H.; Ohno, Y.; Shimazawa, M.; Hara, H. Progranulin promotes the retinal precursor cell proliferation and the photoreceptor differentiation in the mouse retina. Sci. Rep. 2016, 6, 23811. [Google Scholar] [CrossRef] [Green Version]
- Frampton, G.; Invernizzi, P.; Bernuzzi, F.; Pae, H.Y.; Quinn, M.; Horvat, D.; Galindo, C.; Huang, L.; McMillin, M.; Cooper, B.; et al. Interleukin-6-driven progranulin expression increases cholangiocarcinoma growth by an Akt-dependent mechanism. Gut 2011, 61, 268–277. [Google Scholar] [CrossRef] [Green Version]
- Alvaro, D. Progranulin and cholangiocarcinoma: Another bad boy on the block! Gut 2011, 61, 170–171. [Google Scholar] [CrossRef]
- Raya, A.; Koth, C.M.; Buscher, D.; Kawakami, Y.; Itoh, T.; Raya, R.M.; Sternik, G.; Tsai, H.-J.; Rodríguez-Esteban, C.; Izpisúa-Belmonte, J.C. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc. Natl. Acad. Sci. USA 2003, 100, 11889–11895. [Google Scholar] [CrossRef] [Green Version]
- Thatcher, E.J.; Paydar, I.; Anderson, K.K.; Patton, J.G. Regulation of zebrafish fin regeneration by microRNAs. Proc. Natl. Acad. Sci. USA 2008, 105, 18384–18389. [Google Scholar] [CrossRef] [Green Version]
- Rojas-Muñoz, A.; Rajadhyksha, S.; Gilmour, D.; van Bebber, F.; Antos, C.; Esteban, C.R.; Nüsslein-Volhard, C.; Belmonte, J.C.I. ErbB2 and ErbB3 regulate amputation-induced proliferation and migration during vertebrate regeneration. Dev. Biol. 2009, 327, 177–190. [Google Scholar] [CrossRef] [Green Version]
- Kishimoto, N.; Shimizu, K.; Sawamoto, K. Neuronal regeneration in a zebrafish model of adult brain injury. Dis. Model. Mech. 2012, 5, 200–209. [Google Scholar] [CrossRef] [Green Version]
- Sadler, K.C.; Krahn, K.N.; Gaur, N.; Ukomadu, C. Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proc. Natl. Acad. Sci. USA 2007, 104, 1570–1575. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, C.; Bladt, F.; Goedecke, S.; Brinkmann, V.; Zschiesche, W.; Sharpe, M.; Gherardi, E.; Birchmeler, C. Scatter factor/hepatocyte growth factor is essential for liver development. Nat. Cell Biol. 1995, 373, 699–702. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, W.; Yang, F.; Feng, T.; Zhou, M.; Yu, Y.; Yu, X.; Zhao, W.; Yi, F.; Tang, W.; et al. Interleukin-6-stimulated progranulin expression contributes to the malignancy of hepatocellular carcinoma cells by activating mTOR signaling. Sci. Rep. 2016, 6, 21260. [Google Scholar] [CrossRef] [PubMed]
- Daya, M.; Loilome, W.; Techasen, A.; Thanee, M.; Sa-Ngiamwibool, P.; Titapun, A.; Yongvanit, P.; Namwat, N. Progranulin modulates cholangiocarcinoma cell proliferation, apoptosis, and motility via the PI3K/pAkt pathway. Oncol. Targets Ther. 2018, 11, 395–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, S.A.; Davidson, B.R.; Goldin, R.; Heaton, N.; Karani, J.; Pereira, S.; Rosenberg, W.; Tait, P.; Taylor-Robinson, S.D.; Thillainayagam, A.V.; et al. Guidelines for the diagnosis and treatment of cholangiocarcinoma: An update. Gut 2012, 61, 1657–1669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, C.D.; Pinson, C.W.; Berlin, J.; Chari, R.S. Diagnosis and Treatment of Cholangiocarcinoma. Oncogene 2004, 9, 43–57. [Google Scholar] [CrossRef]
- Her, G.M.; Yeh, Y.-H.; Wu, J.-L. 435-bp liver regulatory sequence in the liver fatty acid binding protein (L-FABP) gene is sufficient to modulate liver regional expression in transgenic zebrafish. Dev. Dyn. 2003, 227, 347–356. [Google Scholar] [CrossRef]
- Her, G.M.; Chiang, C.-C.; Chen, W.-Y.; Wu, J.-L. In vivo studies of liver-type fatty acid binding protein (L-FABP) gene expression in liver of transgenic zebrafish (Danio rerio). FEBS Lett. 2003, 538, 125–133. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Radhakrishnan, U.P.; Rajpurohit, S.K.; Kulkarni, V.; Jagadeeswaran, P. Vivo-Morpholino knockdown of αIIb: A novel approach to inhibit thrombocyte function in adult zebrafish. Blood Cells Mol. Dis. 2010, 44, 169–174. [Google Scholar] [CrossRef] [Green Version]
- Robu, M.E.; Larson, J.D.; Nasevicius, A.; Beiraghi, S.; Brenner, C.; Farber, S.; Ekker, S.C. p53 Activation by Knockdown Technologies. PLoS Genet. 2007, 3, e78. [Google Scholar] [CrossRef]
- Hu, S.-Y.; Tai, C.-C.; Li, Y.-H.; Wu, J.-L. Progranulin compensates for blocked IGF-1 signaling to promote myotube hypertrophy in C2C12 myoblasts via the PI3K/Akt/mTOR pathway. FEBS Lett. 2012, 586, 3485–3492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beyer, T.A.; Xu, W.; Teupser, D.; Keller, U.A.D.; Bugnon, P.; Hildt, E.; Thiery, J.; Kan, Y.W.; Werner, S. Impaired liver regeneration in Nrf2 knockout mice: Role of ROS-mediated insulin/IGF-1 resistance. EMBO J. 2007, 27, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.M.; Delous, M.; Bosch, J.A.; Ye, L.; Robertson, M.A.; Hesselson, D.; Stainier, D.Y.R. Hepatocyte Growth Factor Signaling in Intrapancreatic Ductal Cells Drives Pancreatic Morphogenesis. PLoS Genet. 2013, 9, e1003650. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.E.; Yue, B.; Serrero, G. Signaling Pathway of GP88 (Progranulin) in Breast Cancer Cells: Upregulation and Phosphorylation of c-myc by GP88/Progranulin in Her2-Overexpressing Breast Cancer Cells. Breast Cancer Basic Clin. Res. 2015, 92, 71–77. [Google Scholar] [CrossRef]
- Seol, D.-W.; Chen, Q.; Zarnegar, R. Transcriptional activation of the Hepatocyte Growth Factor receptor (c-met) gene by its ligand (Hepatocyte Growth Factor) is mediated through AP-1. Oncogene 2000, 19, 1132–1137. [Google Scholar] [CrossRef] [Green Version]
- Garnett, J.; Chumbalkar, V.; Vaillant, B.; Gururaj, A.E.; Hill, K.; Latha, K.; Yao, J.; Priebe, W.; Colman, H.; Elferink, L.A.; et al. Regulation of HGF Expression by ΔEGFR-Mediated c-Met Activation in Glioblastoma Cells. Neoplasia 2013, 15, 73. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.-H.; Chen, H.-Y.; Li, Y.-W.; Wu, S.-Y.; Liu, W.-; Lin, G.-H.; Hu, S.-Y.; Chang, Z.-K.; Gong, H.-Y.; Liao, C.-H.; et al. Progranulin regulates zebrafish muscle growth and regeneration through maintaining the pool of myogenic progenitor cells. Sci. Rep. 2013, 3, srep01176. [Google Scholar] [CrossRef] [Green Version]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Kinkel, M.D.; Eames, S.C.; Philipson, L.H.; Prince, V. Intraperitoneal Injection into Adult Zebrafish. J. Vis. Exp. 2010, 91, e2126. [Google Scholar] [CrossRef]
- Ormerod, M.G. Flow Cytometric Analysis of Cells Using an Antibody to a Surface Antigen. Immunochem. Protoc. 2003, 10, 373–380. [Google Scholar] [CrossRef]
| Vivo-Morpholino Name | Sequence 5′ to 3′ |
|---|---|
| GrnA vivo-MO | GGCTTAACTCTGCTCAATACCTTTT |
| Ctrl-MO | GCCTTAAATCTCCTAAATACATTTT |
| GrnA vivo-MO sense | AAAAGGTATTGAGCAGAGTTAAGCC |
| Primer Name | Gene | Species | Sequence 5′-3′ |
|---|---|---|---|
| zf-ccnd1-F′ | ccnd1 | Danio rerio | CTGTGCGACAGACGTCAACT |
| zf-ccnd1-R′ | Danio rerio | GGTGAGGTTCTGGGATGAGA | |
| zf-ccne-F′ | ccne1 | Danio rerio | TCTTCAACCCAAAATGAGAGC |
| zf-ccne-R′ | Danio rerio | CCCAAATAAAATGTTTCTCTGTGTAA | |
| zf-ccna2-F′ | ccna2 | Danio rerio | CCAATAACTGAAGCCATAGCCTC |
| zf-ccna2-R′ | Danio rerio | TACAAATATCTGGCTGAATCAAGC | |
| zf-myc-F′ | myc | Danio rerio | TGACTGTGGAAAAGCGACAG |
| zf-myc-R′ | Danio rerio | GCTGCTGTTGATGCTGTGAT | |
| zf-foxm1-F′ | foxm1 | Danio rerio | TCAGCCTGTGACCTCATCTG |
| zf-foxm1-R′ | Danio rerio | AAGAGAGTGCTGTCGGGGTA | |
| zf-jun-F′ | jun | Danio rerio | AAGACCCTGAAGTCGCAAAA |
| zf-jun-R′ | Danio rerio | CAAAATGTCCTTCGGCTCTC | |
| zf-c-met-F′ | met | Danio rerio | GCGCCATCAAGTCCTTAAAC |
| zf-c-met-R′ | Danio rerio | GATGGCTGAAATCCTTCACG | |
| zf-GrnA Mo-F′ | grna | Danio rerio | GCATGTTCGGATGGGAAA |
| zf-GrnA Mo-R′ | Danio rerio | TGTCCCGTTTCCACAAAGA | |
| zf-HGFa-F′ | hgfa | Danio rerio | GATTCATACACTCCGCACAAC |
| zf-HGFa-R′ | Danio rerio | GTGGTGAAACACCAGGGAAT | |
| zf-HGFb-F′ | hgfb | Danio rerio | AAGTCATTGTTGGTGTGAGCA |
| zf-HGFb-R′ | Danio rerio | AAAATGGCTGGTCGGTTATG | |
| zf-MMP2-F′ | mmp2 | Danio rerio | CCCTCTGGAAGAGAGGACTGT |
| zf-MMP2-R′ | Danio rerio | TTCCAGGGTACTGGCAGAAT | |
| zf-MIF-F′ | mif | Danio rerio | TGTTCGTAAAGACTCGGTTCC |
| zf-MIF-R′ | Danio rerio | TCTGATCCGCGACAACCT | |
| zf-YBX1-F′ | ybx1 | Danio rerio | TGTGGAGTTCGACGTGGTAG |
| zf-YBX1-R′ | Danio rerio | GGGCCGGTAACATTTGCT | |
| zf-Sub1-F′ | sub1 | Danio rerio | TTGTAGACCACTGCTCAACCTTAC |
| zf-Sub1-R′ | Danio rerio | GCTTCTTTCTTTTAACCTTGGTTTC | |
| zf-Rac1-F′ | rac1 | Danio rerio | CTCCCATCACCTACCCTCAA |
| zf-Rac1-R′ | Danio rerio | GGCAGAGCACTCCAGGTACT | |
| zf-rps6bk1-F′ | rps6bk1 | Danio rerio | GAGAATGTGTCCGATGACGA |
| zf-rps6bk1-R′ | Danio rerio | GCCACTGCACTGGTCCAT | |
| zf-FN1-F′ | fn1 | Danio rerio | AGAGGGATCACCTGCCATC |
| zf-FN1-R′ | Danio rerio | GTGGTGCCCACATCAGAGA | |
| zf-EF1a-F′ | ef1a | Danio rerio | CCTCTTTCTGTTACCTGGCAAA |
| zf-EF1a-R′ | Danio rerio | CTTTTCCTTTCCCATGATTGA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiang, K.-Y.; Li, Y.-W.; Li, Y.-H.; Huang, S.-J.; Wu, C.-L.; Gong, H.-Y.; Wu, J.-L. Progranulin A Promotes Compensatory Hepatocyte Proliferation via HGF/c-Met Signaling after Partial Hepatectomy in Zebrafish. Int. J. Mol. Sci. 2021, 22, 11217. https://doi.org/10.3390/ijms222011217
Chiang K-Y, Li Y-W, Li Y-H, Huang S-J, Wu C-L, Gong H-Y, Wu J-L. Progranulin A Promotes Compensatory Hepatocyte Proliferation via HGF/c-Met Signaling after Partial Hepatectomy in Zebrafish. International Journal of Molecular Sciences. 2021; 22(20):11217. https://doi.org/10.3390/ijms222011217
Chicago/Turabian StyleChiang, Keng-Yu, Ya-Wen Li, Yen-Hsing Li, Shin-Jie Huang, Chih-Lu Wu, Hong-Yi Gong, and Jen-Leih Wu. 2021. "Progranulin A Promotes Compensatory Hepatocyte Proliferation via HGF/c-Met Signaling after Partial Hepatectomy in Zebrafish" International Journal of Molecular Sciences 22, no. 20: 11217. https://doi.org/10.3390/ijms222011217
APA StyleChiang, K.-Y., Li, Y.-W., Li, Y.-H., Huang, S.-J., Wu, C.-L., Gong, H.-Y., & Wu, J.-L. (2021). Progranulin A Promotes Compensatory Hepatocyte Proliferation via HGF/c-Met Signaling after Partial Hepatectomy in Zebrafish. International Journal of Molecular Sciences, 22(20), 11217. https://doi.org/10.3390/ijms222011217







