Transcriptomic and Coexpression Network Analyses Revealed Pine Chalcone Synthase Genes Associated with Pine Wood Nematode Infection
Abstract
1. Introduction
2. Results
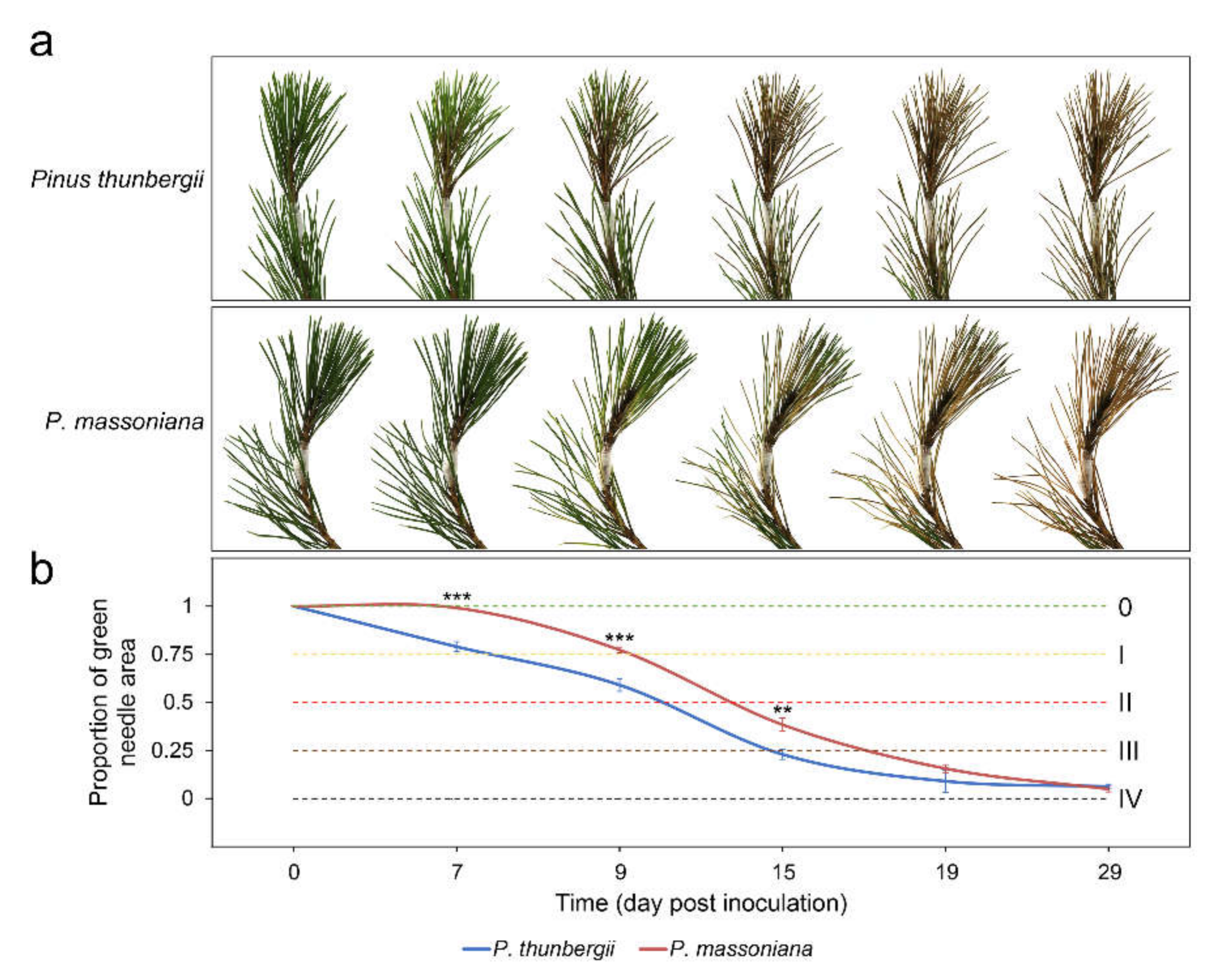
2.1. Changes in Pines and Nematode Populations after PWN Inoculation
2.2. Transcriptome Sequencing
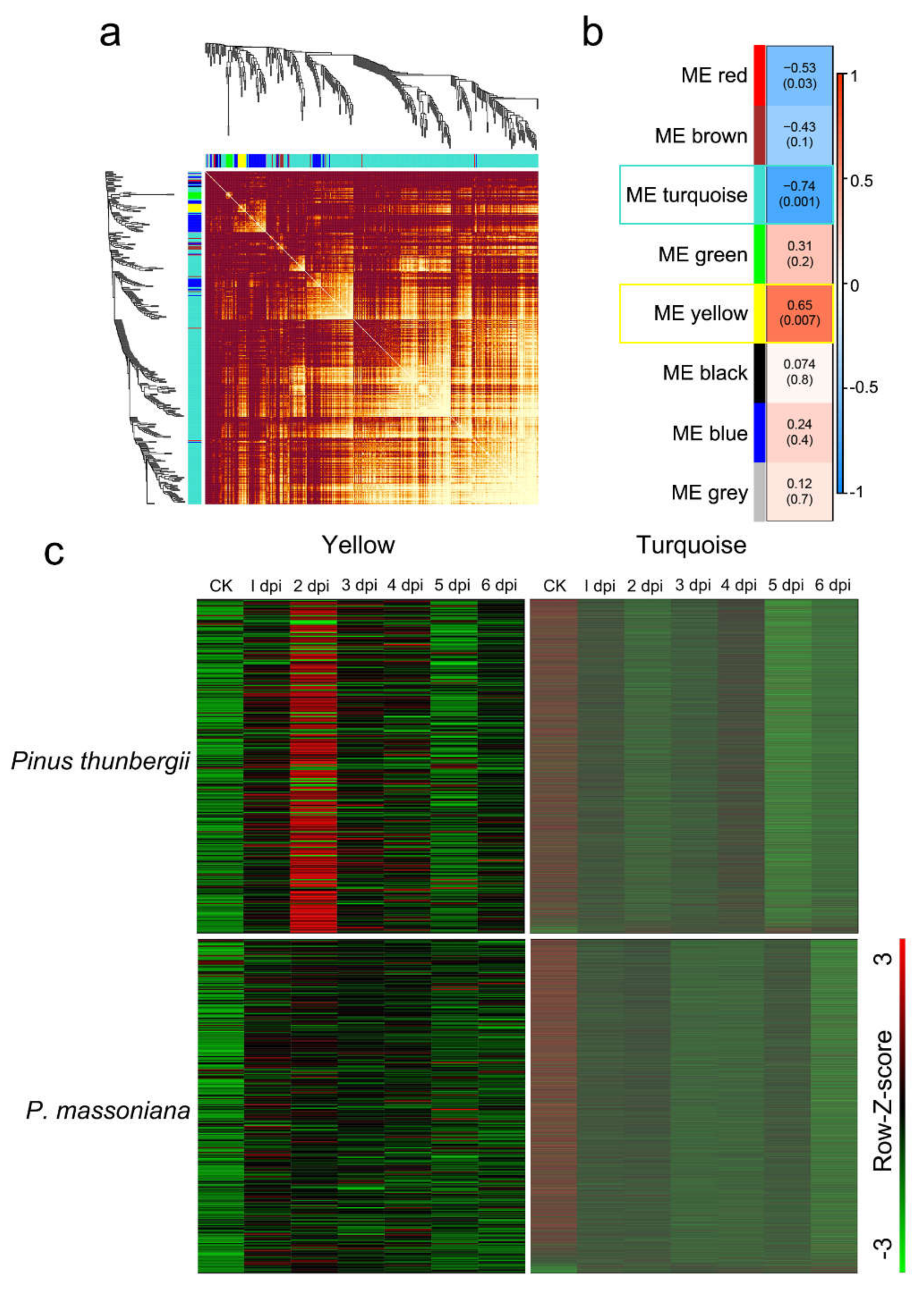
2.3. WGCNA Revealed Modules Related to the Nematode Population
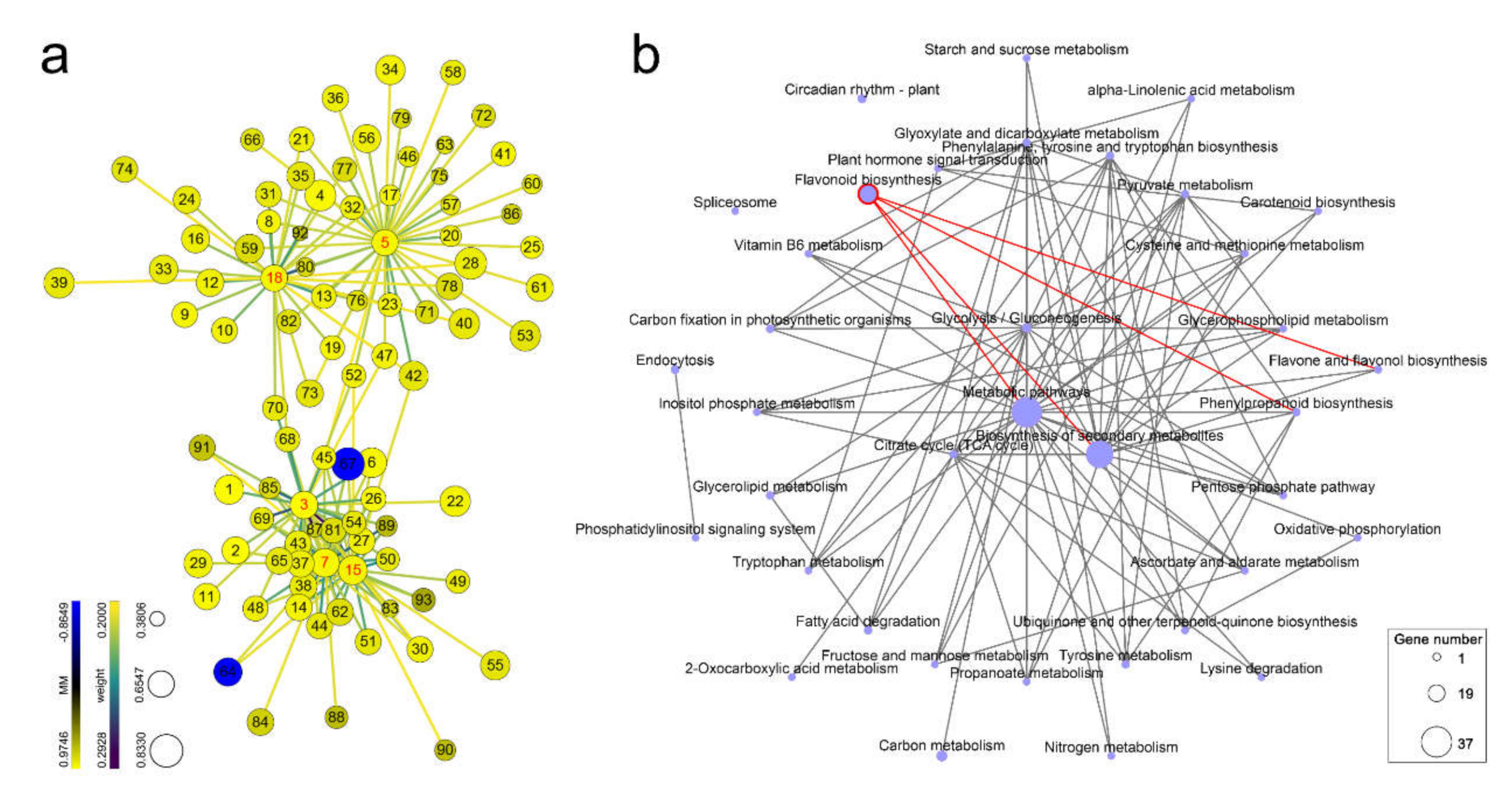
2.4. Functional and Pathway Enrichment Analyses of Genes in a Nematode Population-Related Module
2.5. Finding Genes with High Gene Significance and High Intramodular Connectivity in Interesting Modules
2.6. Expression Profiles and Coexpression Network of Candidate Genes
3. Discussion
4. Materials and Methods
4.1. Biological Material and Pine Wood Nematode Inoculation
4.2. RNA Extraction, cDNA Synthesis, Library Preparation and Sequencing
4.3. De Novo Assembly, Unigene Annotation and Functional Classification
4.4. Weighted Gene Coexpression Network Analysis
4.5. Real-Time Quantitative PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Futai, K. Pine wood nematode, Bursaphelenchus xylophilus. Annu. Rev. Phytopathol. 2013, 51, 61–83. [Google Scholar] [CrossRef] [PubMed]
- Linit, M.J. Nemtaode-vector relationships in the pine wilt disease system. J. Nematol. 1988, 20, 227–235. [Google Scholar]
- Naves, P.M.; Camacho, S.; Sousa, E.; Quartau, J.A. Transmission of the pine wood nematode Bursaphelenchus xylophilus through feeding activity of Monochamus galloprovincialis (Col., Cerambycidae). J. Appl. Èntomol. 2007, 131, 21–25. [Google Scholar] [CrossRef]
- Linit, M.J. Transmission of pinewood nematode through feeding wounds of Monochamus carolinensis (Coleoptera: Cerambycidae). J. Nematol. 1990, 22, 231–236. [Google Scholar]
- Naves, P.; Camacho, S.; Sousa, E.; Quartau, J.A. Transmission of the pine wood nematode Bursaphelenchus xylophilus through oviposition activity of Monochamus galloprovincialis (Coleoptera: Cerambycidae). Èntomol. Fenn. 2007, 18, 193–198. [Google Scholar] [CrossRef]
- Edwards, O.R.; Linit, M.J. Transmission of Bursaphelenchus xylophilus through oviposition wounds of Monochamm carolinensis (Coleoptera: Cerambycidae). J. Nematol. 1992, 24, 133–139. [Google Scholar]
- Ichihara, Y.; Fukuda, K.; Suzuki, K. Early symptom development and histological changes associated with migration of Bursaphelenchus xylophilus in seedling tissues of Pinus thunbergii. Plant. Dis. 2000, 84, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, T. Xylem dysfunction in Bursaphelenchus xylophilus-infected Pinus thunbergii in relation to xylem cavitation and water status. Jpn. J. Phytopathol. 1996, 62, 554–558. [Google Scholar] [CrossRef]
- Baojun, Y.; Qouli, W. Distribution of the pinewood nematode in China and susceptibility of some Chinese and exotic pines to the nematode. Can. J. For. Res. 1989, 19, 1527–1530. [Google Scholar] [CrossRef]
- Mamiya, Y.; Kiyohara, T. Description of Bursaphelenchus lignicolus, N. Sp. (Nematoda: Aphelenchoididae) from pine wood and histopathology of nematode-infested trees. Nematologica 1972, 18, 120–124. [Google Scholar] [CrossRef]
- Mamiya, Y. History of pine wilt disease in Japan. J. Nematol. 1988, 20, 219–226. [Google Scholar]
- Yi, C.K.; Byun, B.H.; Park, J.D.; Yang, S.I.; Chang, K.H. First finding of the pine wood nematode, Bursaphelenchus xylophilus (Steiner et Buhrer) nickle and its insect vector in Korea. Res. Rep. For. Res. Inst. 1989, 38, 141–149. [Google Scholar]
- Mamiya, Y. Pathology of the pine wilt disease caused by Bursaphelenchus xylophilus. Annu. Rev. Phytopathol. 1983, 21, 201–220. [Google Scholar] [CrossRef] [PubMed]
- Dwinell, L.D. First Report of pinewood Nematode (Bursaphelenchus xylophilus) in Mexico. Plant Dis. 1993, 77, 846A. [Google Scholar] [CrossRef]
- Mota, M.M.; Braasch, H.; Bravo, M.A.; Penas, A.C.; Burgermeister, W.; Metge, K.; Sousa, E. First report of Bursaphelenchus xylophilus in Portugal and in Europe. Nematology 1999, 1, 727–734. [Google Scholar] [CrossRef]
- Robertson, L.; Arcos, S.C.; Escuer, M.; Merino, R.S.; Esparrago, G.; Abelleira, A.; Navas, A. Incidence of the pinewood Nematode Bursaphelenchus xylophlius Steiner & Buhrer, 1934 (Nickle, 1970) in Spain. Nematology 2011, 13, 755–757. [Google Scholar] [CrossRef]
- Abelleira, A.; Picoaga, A.; Mansilla, J.P.; Aguin, O. Detection of Bursaphelenchus xylophilus, causal agent of pine wilt disease on Pinus pinaster in northwestern Spain. Plant Dis. 2011, 95, 776. [Google Scholar] [CrossRef]
- Adams, J.C.; Morehart, A.L. Decline and death of Pinus spp. in Delaware caused by Bursaphelenchus xylophilus. J. Nematol. 1982, 14, 382–385. [Google Scholar]
- Sutherland, J.R.; Peterson, M.J. The pinewood nematode in Canada: History, distribution, hosts, potential vectors and research. In Proceedings of the International Symposium on Sustainability of Pine Forests in Relation to Pine Wilt and Decline, Tokyo, Japan, 27–28 October 1999; Shokado Shoten: Kyoto, Japan, 1999; pp. 247–253. [Google Scholar]
- Kikuchi, T.; Cotton, J.; Dalzell, J.; Hasegawa, K.; Kanzaki, N.; McVeigh, P.; Takanashi, T.; Tsai, I.J.; Assefa, S.A.; Cock, P.; et al. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 2011, 7, e1002219. [Google Scholar] [CrossRef]
- Gaspar, D.; Trindade, C.; Usié, A.; Meireles, B.; Fortes, A.M.; Guimarães, J.B.; Simões, F.; Costa, R.L.; Ramos, A.M. Comparative transcriptomic response of two Pinus species to infection with the pine wood nematode Bursaphelenchus xylophilus. Forests 2020, 11, 204. [Google Scholar] [CrossRef]
- Gaspar, D.; Trindade, C.; Usié, A.; Meireles, B.; Barbosa, P.; Fortes, A.M.; Pesquita, C.; Costa, R.L.; Ramos, A.M. Expression profiling in Pinus pinaster in response to infection with the pine wood nematode Bursaphelenchus xylophilus. Forests 2017, 8, 279. [Google Scholar] [CrossRef]
- Santos, C.S.; Pinheiro, M.; Silva, A.I.; Egas, C.; Vasconcelos, M.W. Searching for resistance genes to Bursaphelenchus xylophilus using high throughput screening. BMC Genom. 2012, 13, 599. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wei, Y.; Xuelian, C.; Hao, Y.; Yongcheng, W.; Zhou, Z. Transcriptomic profiling reveals differentially expressed genes associated with pine wood nematode resistance in masson pine (Pinus massoniana Lamb.). Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhou, Z.; Wei, Y.; Shen, D.; Feng, Z.; Hong, S. Genome-wide identification of differentially expressed genes associated with the high yielding of oleoresin in secondary xylem of masson pine (Pinus massoniana Lamb) by transcriptomic analysis. PLoS ONE 2015, 10, e0132624. [Google Scholar] [CrossRef]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, R.; Li, D.; Wang, F. Integrating transcriptome and coexpression network analyses to characterize salicylic acid- and jasmonic acid-related genes in tolerant poplars infected with rust. Int. J. Mol. Sci. 2021, 22, 5001. [Google Scholar] [CrossRef]
- Martin, C. Structure, function, and regulation of the chalcone synthase. Int. Rev. Cytol. 1993, 147, 233–284. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.T.H.; Linthorst, H.J.M.; Verpoorte, R. Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 2011, 10, 397–412. [Google Scholar] [CrossRef]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell 1995, 7, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Gläßgen, W.E.; Rose, A.; Madlung, J.; Koch, W.; Gleitz, J.; Seitz, H.U. Regulation of enzymes involved in anthocyanin biosynthesis in carrot cell cultures in response to treatment with ultraviolet light and fungal elicitors. Planta 1998, 204, 490–498. [Google Scholar] [CrossRef]
- VanEtten, H.D.; Matthews, E.D.; Matthews, P.S. Phytoalexin detoxification: Importance for pathogenicity and practical implications. Annu. Rev. Phytopathol. 1989, 27, 143–164. [Google Scholar] [CrossRef] [PubMed]
- Ryder, T.B.; Cramer, C.L.; Bell, J.N.; Robbins, M.P.; Dixon, R.A.; Lamb, C.J. Elicitor rapidly induces chalcone synthase mRNA in Phaseolus vulgaris cells at the onset of the phytoalexin defense response. Proc. Natl. Acad. Sci. USA 1984, 81, 5724–5728. [Google Scholar] [CrossRef]
- Brignolas, F.; Lacroix, B.; Lieutier, F.; Sauvard, D.; Drouet, A.; Claudot, A.C.; Yart, A.; Berryman, A.A.; Christiansen, E. Induced responses in phenolic metabolism in two Norway spruce clones after wounding and inoculations with Ophiostoma polonicum, a bark beetle-associated fungus. Plant Physiol. 1995, 109, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Morkunas, I.; Marczak, Ł.; Stachowiak, J.; Stobiecki, M. Sucrose-induced lupine defense against Fusarium oxysporum: Sucrose-stimulated accumulation of isoflavonoids as a defense response of lupine to Fusarium oxysporum. Plant Physiol. Biochem. 2005, 43, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Mamiya, Y. Movement of the pinewood nematode, Bursaphelenchus xylophilus, through tracheids in diseased pine trees. Jpn. J. Nematol. 2008, 38, 41–44. [Google Scholar] [CrossRef][Green Version]
- Wang, F.; Chen, Q.; Zhang, R.; Li, D.; Ling, Y.; Song, R. The anti-phytoalexin gene Bx-cathepsin W supports the survival of Bursaphelenchus xylophilus under Pinus massoniana phytoalexin stress. BMC Genom. 2019, 20, 779. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Q.; Zhang, R.; Li, D.; Wang, F. Transcriptomic and Coexpression Network Analyses Revealed Pine Chalcone Synthase Genes Associated with Pine Wood Nematode Infection. Int. J. Mol. Sci. 2021, 22, 11195. https://doi.org/10.3390/ijms222011195
Chen Q, Zhang R, Li D, Wang F. Transcriptomic and Coexpression Network Analyses Revealed Pine Chalcone Synthase Genes Associated with Pine Wood Nematode Infection. International Journal of Molecular Sciences. 2021; 22(20):11195. https://doi.org/10.3390/ijms222011195
Chicago/Turabian StyleChen, Qiaoli, Ruizhi Zhang, Danlei Li, and Feng Wang. 2021. "Transcriptomic and Coexpression Network Analyses Revealed Pine Chalcone Synthase Genes Associated with Pine Wood Nematode Infection" International Journal of Molecular Sciences 22, no. 20: 11195. https://doi.org/10.3390/ijms222011195
APA StyleChen, Q., Zhang, R., Li, D., & Wang, F. (2021). Transcriptomic and Coexpression Network Analyses Revealed Pine Chalcone Synthase Genes Associated with Pine Wood Nematode Infection. International Journal of Molecular Sciences, 22(20), 11195. https://doi.org/10.3390/ijms222011195





