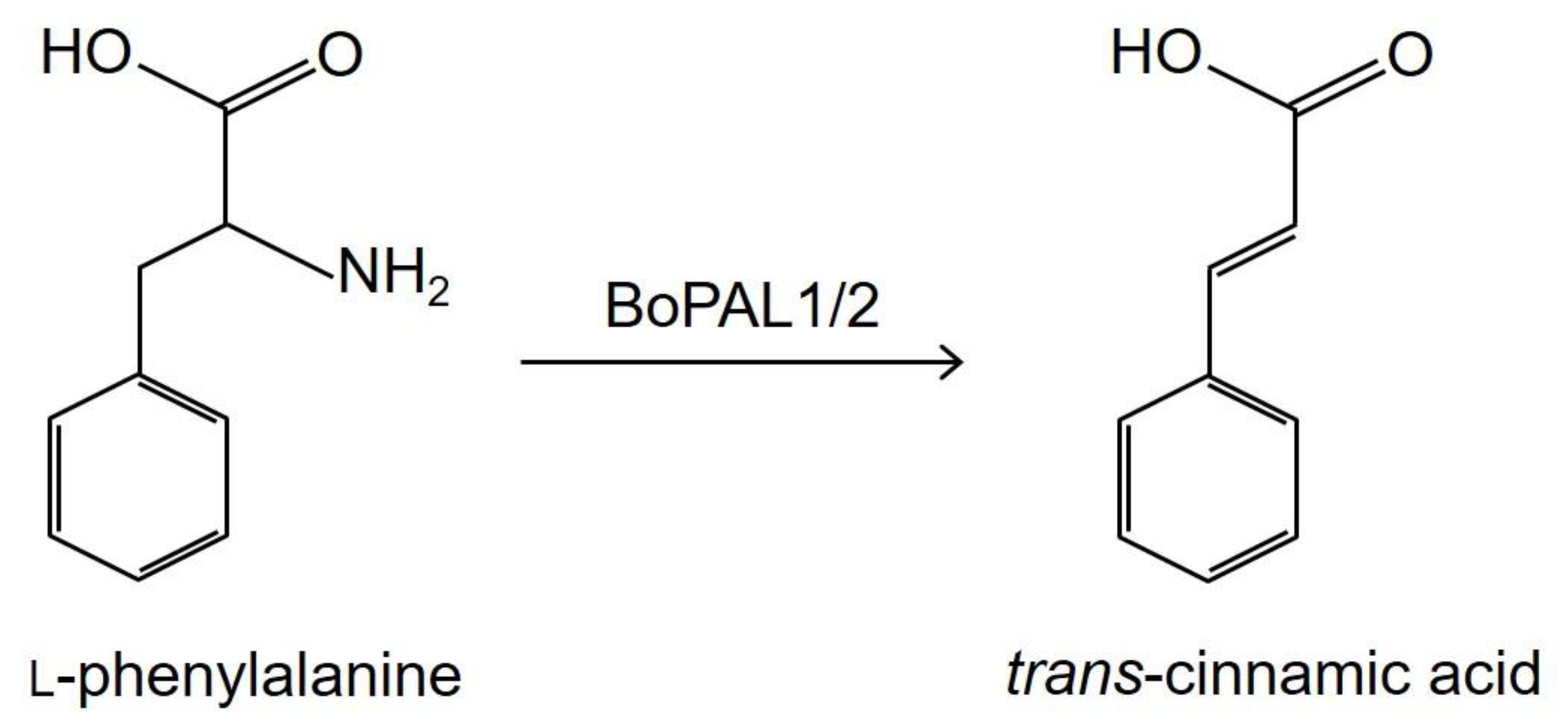
Production of Trans-Cinnamic Acid by Immobilization of the Bambusa oldhamii BoPAL1 and BoPAL2 Phenylalanine Ammonia-Lyases on Electrospun Nanofibers
Abstract
:1. Introduction
2. Results
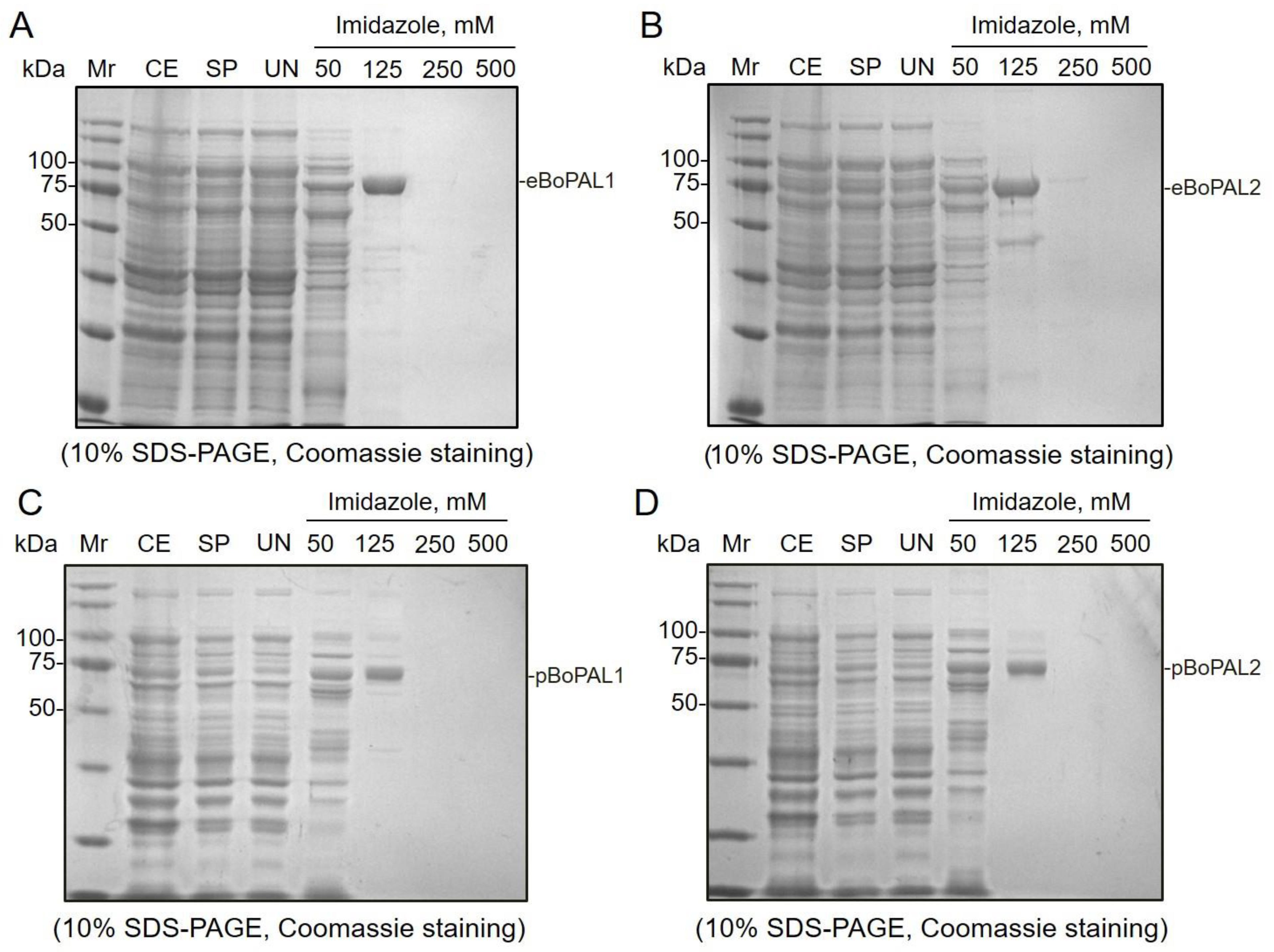
2.1. Purification of Recombinant Proteins by Affinity Chromatography
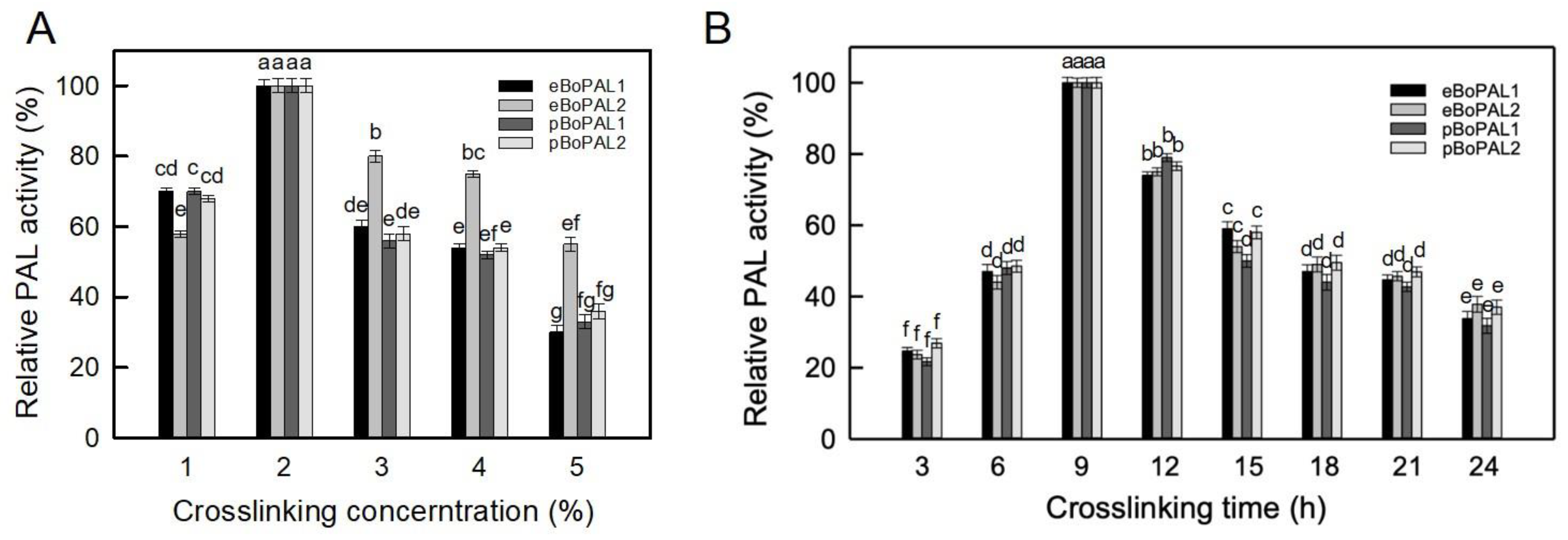
2.2. Optimum Crosslinking Conditions
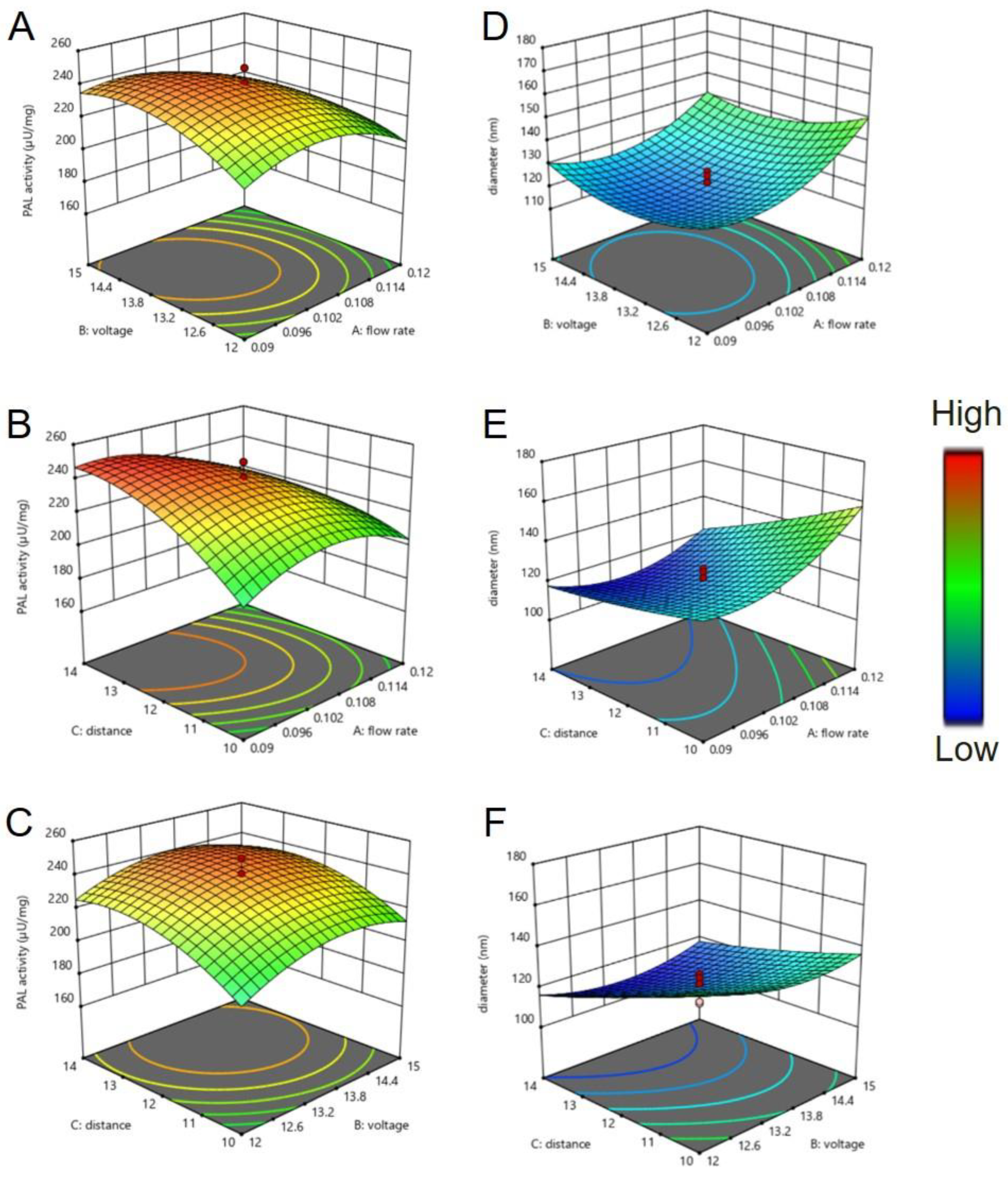
2.3. Central Composite Design (CCD)-Response Surface Methodology (RSM)
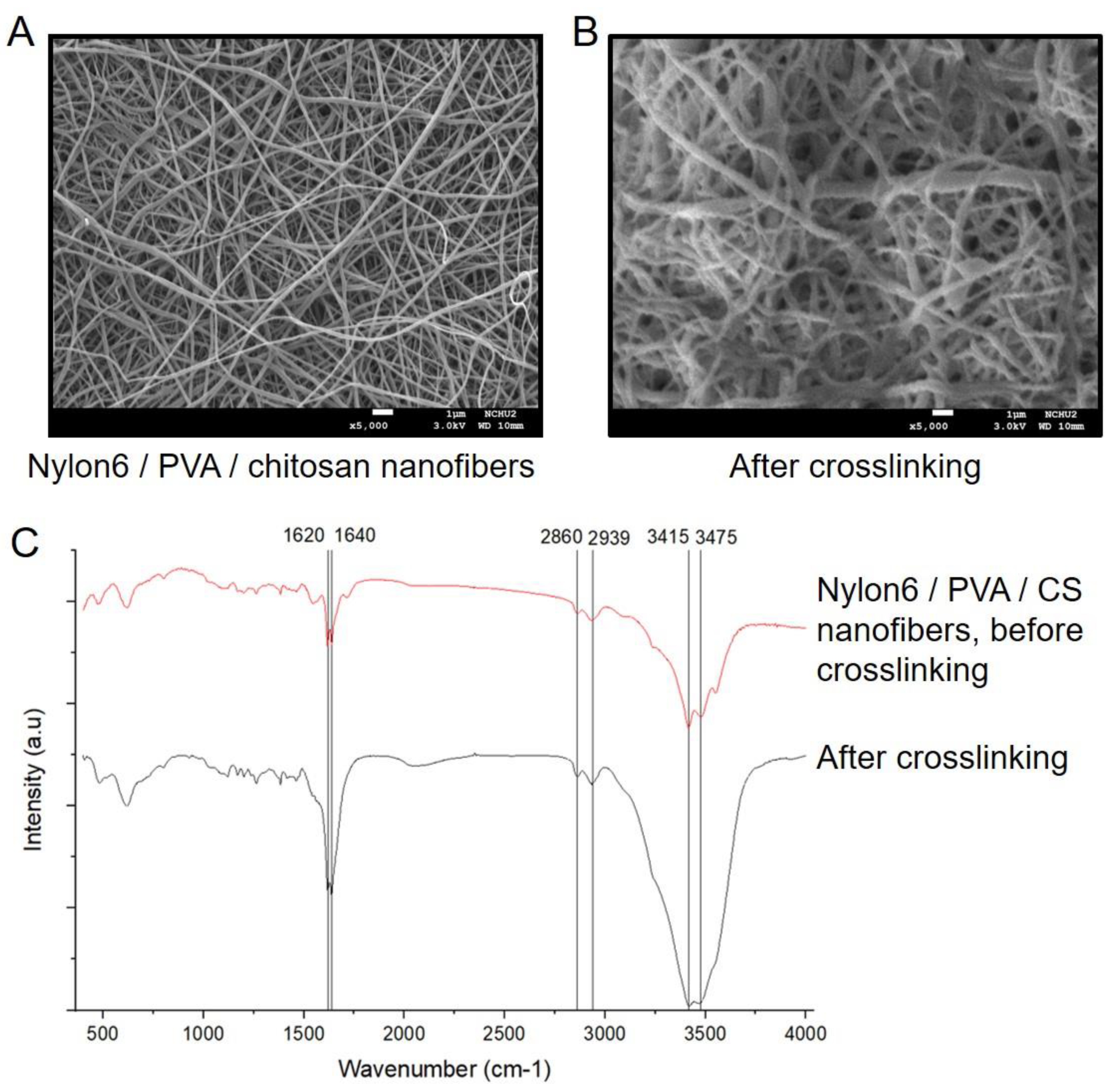
2.4. Surface Morphology
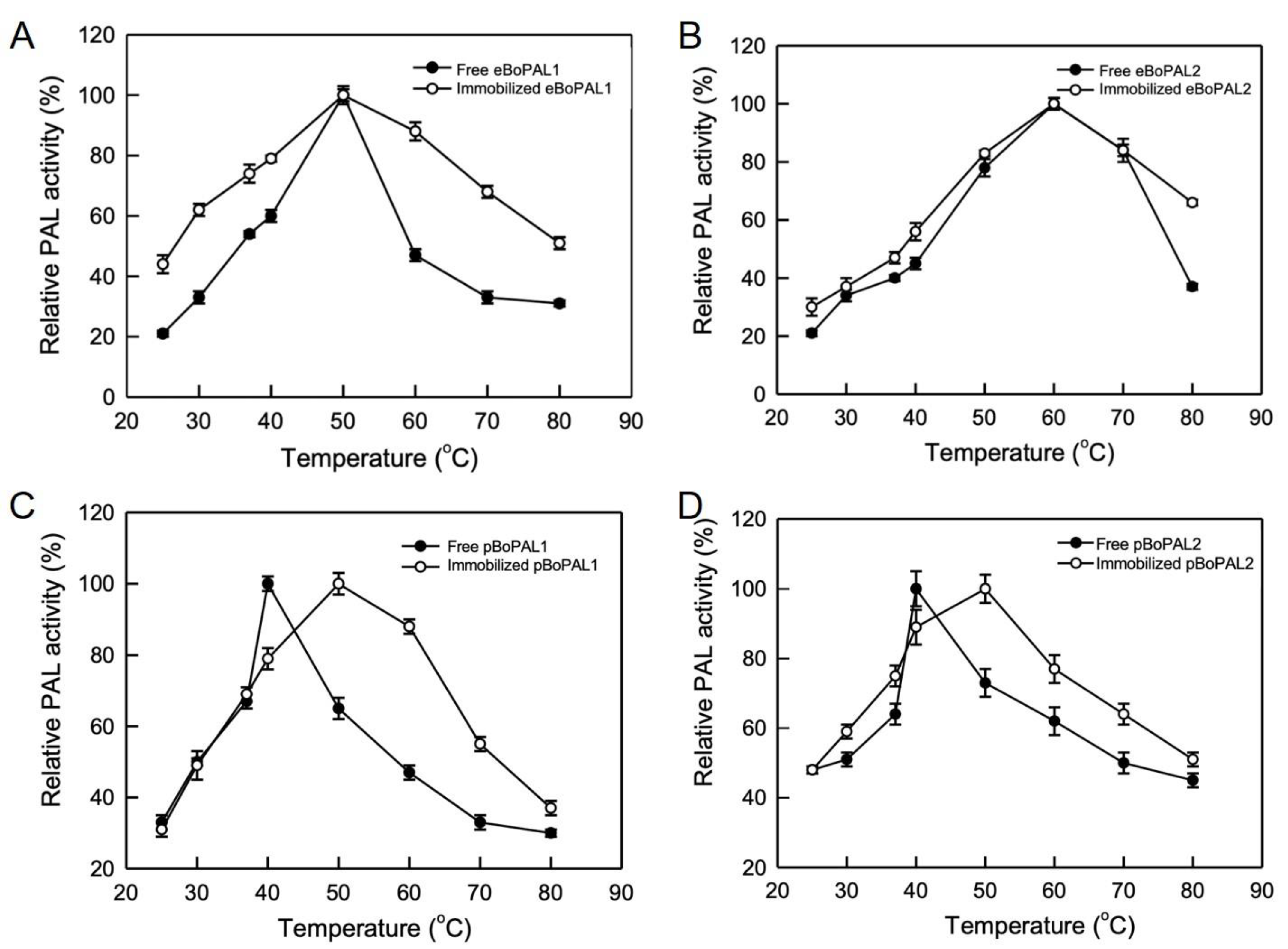
2.5. Temperature and pH Stability of Free and Immobilized BoPAL Proteins
2.6. Kinetic Parameters of Free and Immobilized BoPAL Proteins
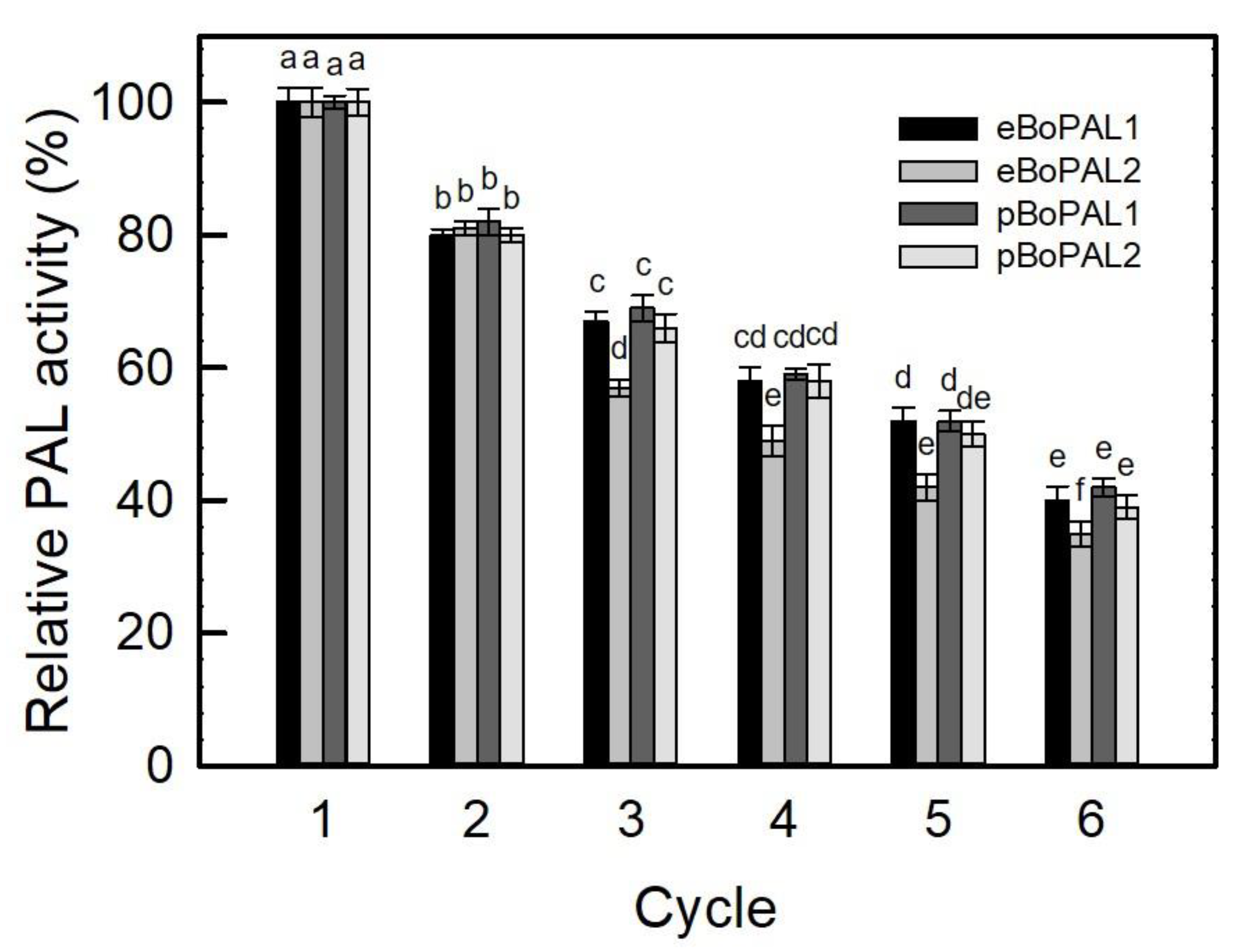
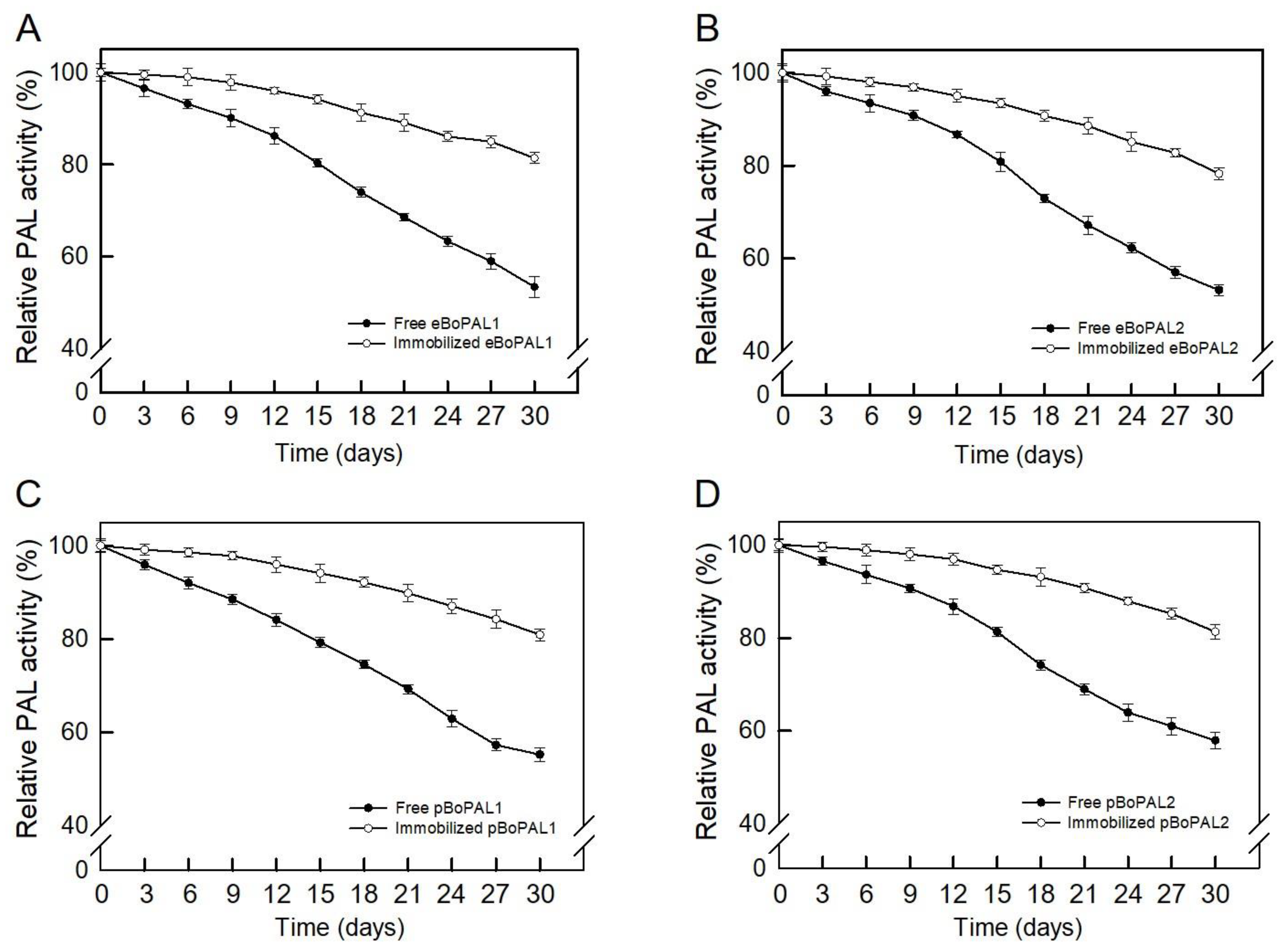
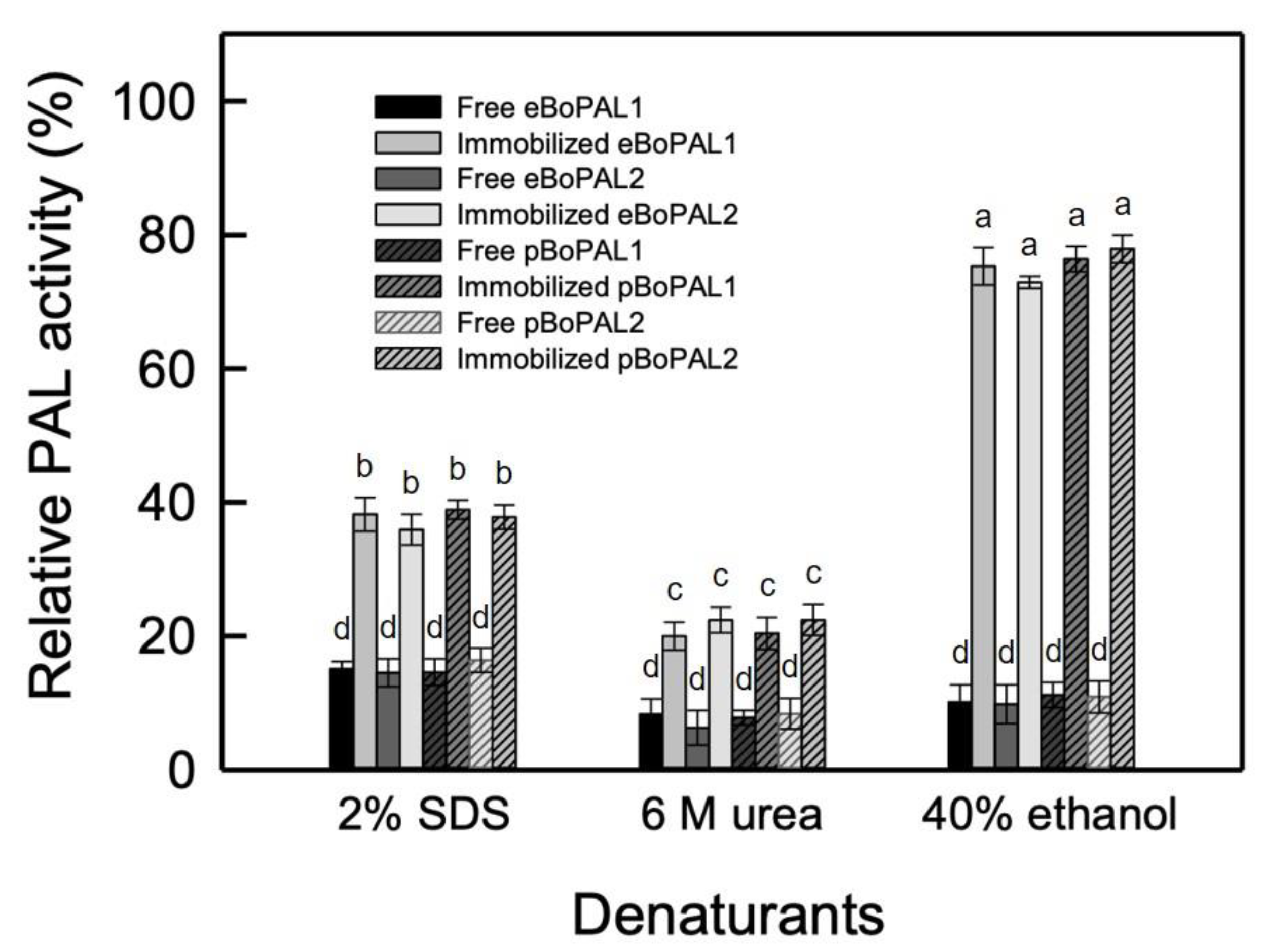
2.7. Recyclability, Storage Stability, and Denaturant Tolerance of Free and immobiLized BoPAL Proteins
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Expressions of BoPAL1 and BoPAL2 Proteins in Escherichia coli and Pichia pastoris
4.3. Purification of Recombinant BoPAL1 and BoPAL2 Proteins
4.4. Preparation of Nanofibers by Electrospinning Method
4.5. Crosslinking
4.6. Central Composite Design (CCD) Response Surface Methodology (RSM)
4.7. Membrane Surface morphOlogy Characterization Using Scanning Electron Microscope (SEM) and Fourier Transform Infrared Spectroscopy (FT-IR)
4.8. PAL Activity Assay
4.9. PAL Enzyme Kinetic
4.10. Biochemical Properties Analysis
4.11. Recyclability and Denaturant Treatment
4.12. Data Analysis and Statistic
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| ANOVA | analysis of variances |
| BMGY | buffered glycerol-complex medium |
| BMMY | buffered methanol-complex medium |
| CBR | Coomassie Brilliant Blue R-25 |
| CCD | central composite design |
| CS | chitosan |
| FT-IR | Fourier transform infrared spectroscopy |
| IPTG | β-D-thiogalactopyranoside |
| MD | minimal dextrose |
| MM | minimal methanol |
| PAL | phenylalanine ammonia-lyase |
| PVA | polyvinyl alcohol |
| RSM | response surface methodology |
| SDS-PAGE | sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| TFE-SEM | thermal field emission scanning electron microscope |
References
- Valletta, A.; Iozia, L.M.; Leonelli, F. Impact of environmental factors on stilbene biosynthesis. Plants 2021, 10, 90. [Google Scholar] [CrossRef]
- Marchica, A.; Cotrozzi, L.; Detti, R.; Lorenzini, G.; Pellegrini, E.; Petersen, M.; Nali, C. The biosynthesis of phenolic compounds is an integrated defence mechanism to prevent ozone injury in Salvia officinalis. Antioxidants 2020, 9, 1274. [Google Scholar] [CrossRef] [PubMed]
- Attridge, T.; Stewart, G.; Smith, H. End-product inhibition of Pisum phenylalanine ammonia-lyase by the Pisum flavonoids. FEBS Lett. 1971, 17, 84–86. [Google Scholar] [CrossRef] [Green Version]
- Ilahy, R.; Tlili, I.; Siddiqui, M.W.; Hdider, C.; Lenucci, M.S. Inside and beyond color: Comparative overview of functional quality of tomato and watermelon fruits. Front. Plant Sci. 2019, 10, 769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, R.A.; Broadhurst, C.L.; Polansky, M.M.; Schmidt, W.F.; Khan, A.; Flanagan, V.P.; Graves, D.J. Isolation and characterization of polyphenol type-A polymers from cinnamon with insulin-like biological activity. J. Agric. Food Chem. 2004, 52, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Soliman, M.M.; Attia, H.F.; El-Shazly, S.A.; Saleh, O.M. Biomedical effects of cinnamon extract on obesity and diabetes relevance in Wistar rats. Am. J. Biochem. Mol. Biol. 2012, 2, 133–135. [Google Scholar] [CrossRef]
- Yilmaz, S.; Ergün, S. Trans-cinnamic acid application for rainbow trout (Oncorhynchus mykiss): I. Effects on haematological, serum biochemical, non-specific immune and head kidney gene expression responses. Fish Shellfish. Immunol. 2018, 78, 140–157. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Y.; Wang, J.; Zhou, M.; Wang, M.; Feng, J. Antifungal activity and action mechanism of the natural product cinnamic acid against Sclerotinina sclerotiorum. Plant Dis. 2019, 103, 944–950. [Google Scholar] [CrossRef]
- Silva, A.T.; Bento, C.M.; Pena, A.C.; Figueiredo, L.M.; Prudêncio, C.; Aguiar, L.; Teixeira, C. Cinnamic acid conjugates in the rescuing and repurposing of classical antimalarial drugs. Molecules 2019, 25, 66. [Google Scholar] [CrossRef] [Green Version]
- Kang, N.H.; Mukherjee, S.; Yun, J.W. Trans-cinnamic acid stimulates white fat browning and activates brown adipocytes. Nutrients 2019, 11, 577. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Ge, S.; Li, S.; Lin, H.; Lin, S. Anti-obesity effect of trans-cinnamic acid on HepG2 cells and HFD-fed mice. Food Chem. Toxicol. 2020, 137, 111148. [Google Scholar] [CrossRef]
- Gunia-Krzyżak, A.; Słoczyńska, K.; Popiół, J.; Koczurkiewicz, P.; Marona, H.; Pękala, E. Cinnamic acid derivatives in cosmetics: Current use and future prospects. Int. J. Cosmet. Sci. 2018, 40, 356–366. [Google Scholar] [CrossRef] [Green Version]
- De, P.; Baltas, M.; Bedos-Belval, F. Cinnamic acid derivatives as anticancer agents-a review. Curr. Med. Chem. 2011, 18, 1672–1703. [Google Scholar] [CrossRef]
- Sova, M. Antioxidant and antimicrobial activities of cinnamic acid derivatives. Mini. Rev. Med. Chem. 2012, 12, 749–767. [Google Scholar] [CrossRef]
- Ruwizhi, N.; Aderibigbe, B.A. Cinnamic acid derivatives and their biological efficacy. Int. J. Mol. Sci. 2020, 21, 5712. [Google Scholar] [CrossRef]
- Ge, J.; Lu, D.; Liu, Z.; Liu, Z. Recent advances in nanostructured biocatalysts. Biochem. Eng. J. 2009, 44, 53–59. [Google Scholar] [CrossRef]
- Datta, S.; Christena, L.; Rajaram, Y. Enzyme immobilization: An overview on techniques and support materials. 3 Biotech. 2013, 3, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Galan, C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of different enzyme immobilization strategies to improve enzyme performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Berenguer-Murcia, Á.; Carballares, D.; Morellon-Sterling, R.; Fernandez-Lafuente, R. Stabilization of enzymes via immobilization: Multipoint covalent attachment and other stabilization strategies. Biotechnol. Adv. 2021, 52, 107821. [Google Scholar] [CrossRef]
- Misson, M.; Dai, S.; Jin, B.; Chen, B.H.; Zhang, H. Manipulation of nanofiber-based β-galactosidase nanoenvironment for enhancement of galacto-oligosaccharide production. J. Biotechnol. 2016, 222, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Virly, C.-H.C.; Tsai, T.-Y.; Yeh, Y.-C.; Wang, R. Encapsulation of β-glucosidase within PVA fibers by CCD-RSM-guided coelectrospinning: A novel approach for specific mogroside sweetener production. J. Agric. Food Chem. 2020, 68, 11790–11801. [Google Scholar] [CrossRef]
- Kamoun, E.A.; Loutfy, S.A.; Hussein, Y.; Kenawy, E.-R.S. Recent advances in PVA-polysaccharide based hydrogels and electrospun nanofibers in biomedical applications: A review. Int. J. Biol. Macromol. 2021, 187, 755–768. [Google Scholar] [CrossRef]
- Huang, Z.-M.; Zhang, Y.-Z.; Kotaki, M.; Ramakrishna, S. A review on polymer nanofibers by electrospinning and their applications in nanocomposites. Compos. Sci. Technol. 2003, 63, 2223–2253. [Google Scholar] [CrossRef]
- Li, Z.; Wang, C. One-Dimensional Nanostructures: Electrospinning Technique and Unique Nanofibers; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Haider, A.; Haider, S.; Kang, I.-K. A comprehensive review summarizing the effect of electrospinning parameters and potential applications of nanofibers in biomedical and biotechnology. Arab. J. Chem. 2018, 11, 1165–1188. [Google Scholar] [CrossRef]
- Misson, M.; Saallah, S.; Zhang, H. Nanofiber-Immobilized β-Galactosidase for Dairy Waste Conversion into Galacto-Oligosaccharides. Advances in Waste Processing Technology; Springer: Singapore, 2020; pp. 37–48. [Google Scholar]
- Wu, L.; Yuan, X.; Sheng, J. Immobilization of cellulase in nanofibrous PVA membranes by electrospinning. J. Membr. Sci. 2005, 250, 167–173. [Google Scholar] [CrossRef]
- Dai, M.; Jin, S.; Nugen, S. Water-soluble electrospun nanofibers as a method for on-chip reagent storage. Biosensors 2012, 2, 388–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsieh, L.S.; Yeh, C.S.; Pan, H.C.; Cheng, C.Y.; Yang, C.C.; Lee, P.D. Cloning and expression of a phenylalanine ammonia-lyase gene (BoPAL2) from Bambusa oldhamii in Escherichia coli and Pichia pastoris. Protein Expr. Purif. 2010, 71, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, L.S.; Ma, G.J.; Yang, C.C.; Lee, P.D. Cloning, expression, site-directed mutagenesis and immunolocalization of phenylalanine ammonia-lyase in Bambusa oldhamii. Phytochemistry 2010, 71, 1999–2009. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, L.S.; Hsieh, Y.L.; Yeh, C.S.; Cheng, C.Y.; Yang, C.C.; Lee, P.D. Molecular characterization of a phenylalanine ammonia-lyase gene (BoPAL1) from Bambusa oldhamii. Mol. Biol. Rep. 2011, 38, 283–290. [Google Scholar] [CrossRef]
- Feng, Q.; Zhao, Y.; Wei, A.; Li, C.; Wei, Q.; Fong, H. Immobilization of catalase on electrospun PVA/PA6–Cu (II) nanofibrous membrane for the development of efficient and reusable enzyme membrane reactor. Environ. Sci. Technol. 2014, 48, 10390–10397. [Google Scholar] [CrossRef]
- Hsieh, C.Y.; Huang, Y.H.; Lin, Z.Y.; Hsieh, L.S. Insights into the substrate selectivity of Bambusa oldhamii phenylalanine ammonia-lyase 1 and 2 through mutational analysis. Phytochem. Lett. 2020, 38, 140–143. [Google Scholar] [CrossRef]
- Sojitra, U.V.; Nadar, S.S.; Rathod, V.K. Immobilization of pectinase onto chitosan magnetic nanoparticles by macromolecular cross-linker. Carbohydr. Polym. 2017, 157, 677–685. [Google Scholar] [CrossRef]
- Wang, B.; Cheng, F.; Lu, Y.; Ge, W.; Zhang, M.; Yue, B. Immobilization of pectinase from Penicillium oxalicum F67 onto magnetic cornstarch microspheres: Characterization and application in juice production. J. Mol. Catal. B Enzym. 2013, 97, 137–143. [Google Scholar] [CrossRef]
- Chen, J.-P.; Ho, K.-H.; Chiang, Y.-P.; Wu, K.-W. Fabrication of electrospun poly(methyl methacrylate) nanofibrous membranes by statistical approach for application in enzyme immobilization. J. Membr. Sci. 2009, 340, 9–15. [Google Scholar] [CrossRef]
- Fatarella, E.; Spinelli, D.; Ruzzante, M.; Pogni, R. Nylon 6 film and nanofiber carriers: Preparation and laccase immobilization performance. J. Mol. Catal. B Enzym. 2014, 102, 41–47. [Google Scholar] [CrossRef]
- Nadar, S.S.; Rathod, V.K. Magnetic macromolecular cross linked enzyme aggregates (CLEAs) of glucoamylase. Enzym. Microb. Technol. 2016, 83, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Zhao, Y.; Feng, Y.; Lin, T.; Zhong, C.; Tan, Z.; Jia, S. Encapsulation of spherical cross-linked phenylalanine ammonia lyase aggregates in mesoporous biosilica. J. Agric. Food Chem. 2017, 65, 618–625. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Zhao, Y.; Tan, Z.; Zhong, C.; Han, P.; Jia, S. Mesoporous phenylalanine ammonia lyase microspheres with improved stability through calcium carbonate templating. Int. J. Biol. Macromol. 2017, 98, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Porto, M.D.A.; Santos, J.P.D.; Hackbart, H.; Bruni, G.P.; Fonseca, L.M.; da Rosa Zavareze, E.; Dias, A.R.G. Immobilization of—Amylase in ultrafine polyvinyl alcohol (PVA) fibers via electrospinning and their stability on different substrates. Int. J. Biol. Macromol. 2019, 126, 834–841. [Google Scholar] [CrossRef] [PubMed]
- El-Aassar, M.R.; Shibraen, M.H.M.A.; Abdel-Fattah, Y.R.; Elzain, A.A. Functionalization of electrospun poly(acrylonitrile-co-styrene/pyrrole) copolymer nanofibers for using as a high-performance carrier for laccase immobilization. Fibers Polym. 2019, 20, 2268–2279. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye-binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Hsieh, L.S.; Su, W.M.; Han, G.S.; Carman, G.M. Phosphorylation regulates the ubiquitin-independent degradation of yeast Pah1 phosphatidate phosphatase by the 20S proteasome. J. Biol. Chem. 2015, 290, 11467–11478. [Google Scholar] [CrossRef] [Green Version]
- Hsiao, C.J.; Hsieh, C.Y.; Hsieh, L.S. Cloning and characterization of the Bambusa oldhamii BoMDH-encoded malate dehydrogenase. Protein Expr. Purif. 2020, 174, 105665. [Google Scholar] [CrossRef] [PubMed]
- Michaelis, L.; Menten, M.L. Die kinetik der invertinwirkung. Biochem. Z. 1913, 49, 333–369. [Google Scholar]
- Lineweaver, H.; Burk, D. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 1934, 56, 658–666. [Google Scholar] [CrossRef]
- Davis, B.J. Disc electrophoresis. II. Method and application to human serum proteins. Ann. N. Y. Acad. Sci. 1964, 121, 404–427. [Google Scholar] [CrossRef] [PubMed]
- Hsu, W.H.; Huang, Y.H.; Chen, P.R.; Hsieh, L.-S. NLIP and HAD-like domains of Pah1 and Lipin 1 phosphatidate phosphatases are essential for their catalytic activities. Molecules 2021, 26, 5470. [Google Scholar] [CrossRef] [PubMed]

| Enzyme | kcat (s−1) | Km (µM) | kcat/Km (s−1 µM−1) | |||
|---|---|---|---|---|---|---|
| Free | Immobilized | Free | Immobilized | Free | Immobilized | |
| eBoPAL1 | 1.01 | 1.21 | 518 | 534 | 1.91 × 10−3 | 2.2 × 10−3 |
| eBoPAL2 | 4.02 | 3.99 | 329 | 379 | 1.23 × 10−2 | 1.05 × 10−2 |
| pBoPAL1 | 1.03 | 1.25 | 376 | 453 | 2.72 × 10−3 | 3.32 × 10−3 |
| pBoPAL2 | 3.81 | 4.01 | 294 | 364 | 1.29 × 10−2 | 1.1 × 10−2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, P.-Y.; Huang, Y.-H.; Lim, G.C.W.; Chen, Y.-P.; Hsiao, C.-J.; Chen, L.-H.; Ciou, J.-Y.; Hsieh, L.-S. Production of Trans-Cinnamic Acid by Immobilization of the Bambusa oldhamii BoPAL1 and BoPAL2 Phenylalanine Ammonia-Lyases on Electrospun Nanofibers. Int. J. Mol. Sci. 2021, 22, 11184. https://doi.org/10.3390/ijms222011184
Hong P-Y, Huang Y-H, Lim GCW, Chen Y-P, Hsiao C-J, Chen L-H, Ciou J-Y, Hsieh L-S. Production of Trans-Cinnamic Acid by Immobilization of the Bambusa oldhamii BoPAL1 and BoPAL2 Phenylalanine Ammonia-Lyases on Electrospun Nanofibers. International Journal of Molecular Sciences. 2021; 22(20):11184. https://doi.org/10.3390/ijms222011184
Chicago/Turabian StyleHong, Pei-Yu, Yi-Hao Huang, GiGi Chin Wen Lim, Yen-Po Chen, Che-Jen Hsiao, Li-Hsien Chen, Jhih-Ying Ciou, and Lu-Sheng Hsieh. 2021. "Production of Trans-Cinnamic Acid by Immobilization of the Bambusa oldhamii BoPAL1 and BoPAL2 Phenylalanine Ammonia-Lyases on Electrospun Nanofibers" International Journal of Molecular Sciences 22, no. 20: 11184. https://doi.org/10.3390/ijms222011184
APA StyleHong, P.-Y., Huang, Y.-H., Lim, G. C. W., Chen, Y.-P., Hsiao, C.-J., Chen, L.-H., Ciou, J.-Y., & Hsieh, L.-S. (2021). Production of Trans-Cinnamic Acid by Immobilization of the Bambusa oldhamii BoPAL1 and BoPAL2 Phenylalanine Ammonia-Lyases on Electrospun Nanofibers. International Journal of Molecular Sciences, 22(20), 11184. https://doi.org/10.3390/ijms222011184








