The Amyloid Forming Peptides Islet Amyloid Polypeptide and Amyloid β Interact at the Molecular Level
Abstract
:1. Introduction
2. Results
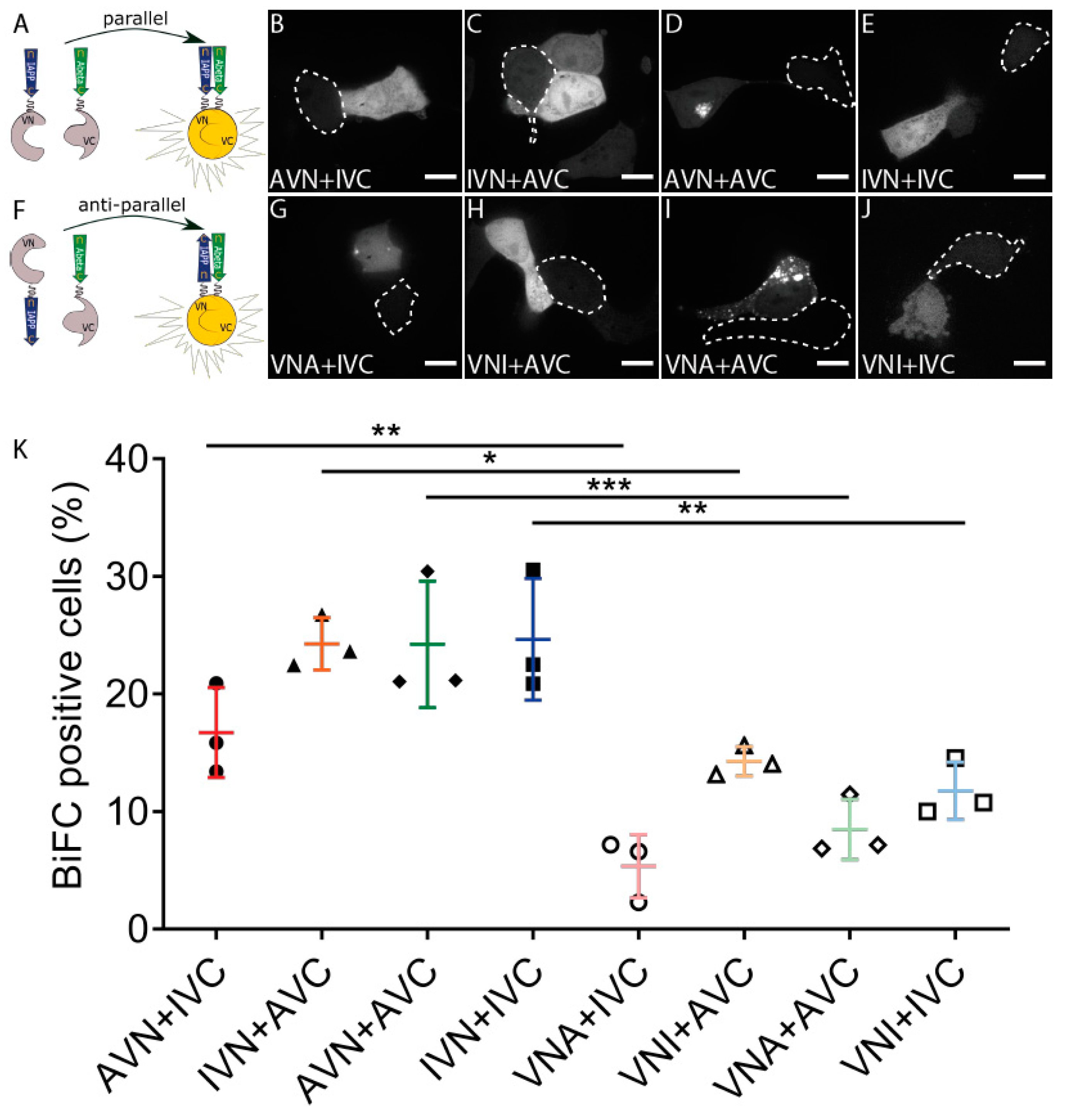
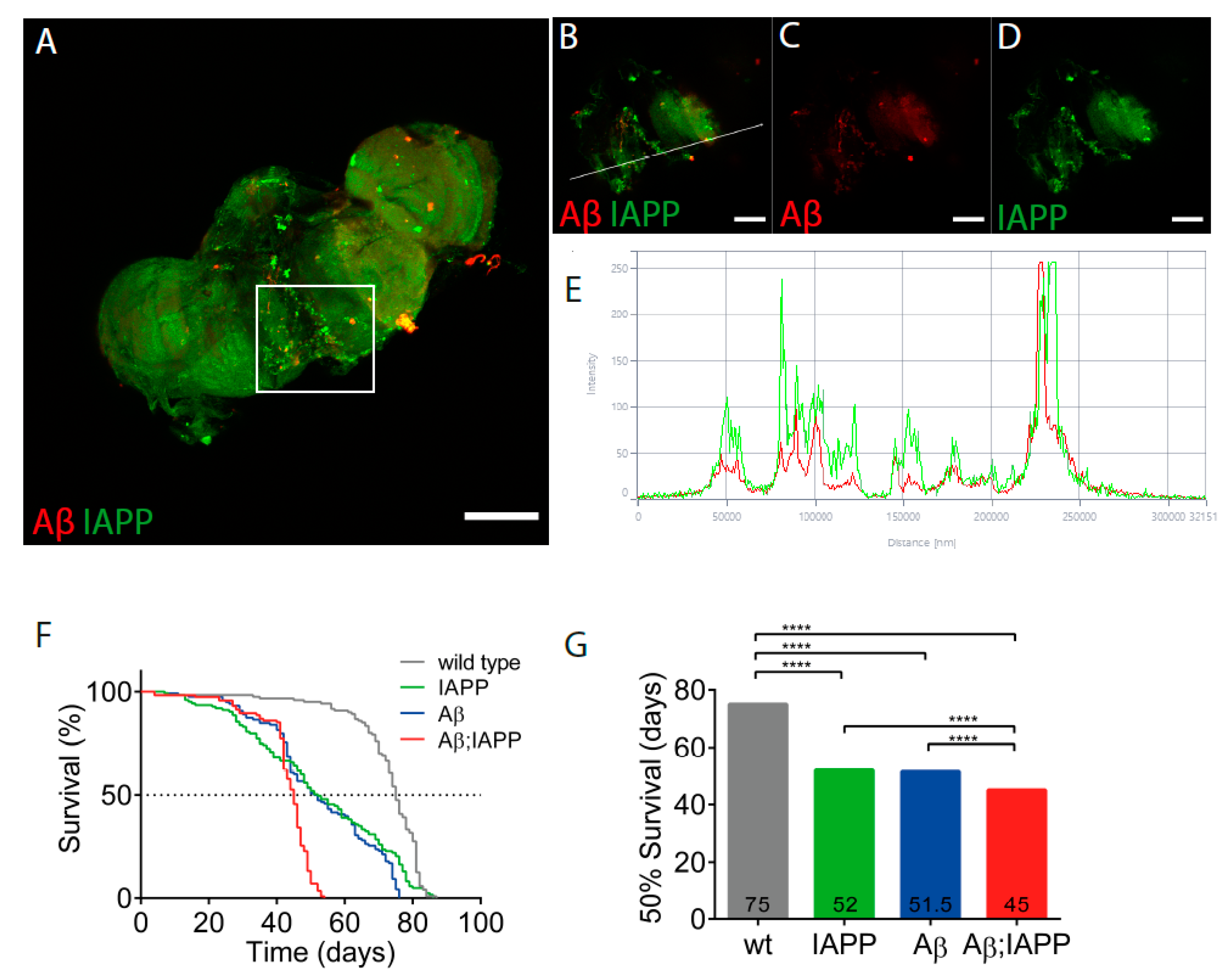
2.1. Aβ and IAPP Interact on a Molecular Level
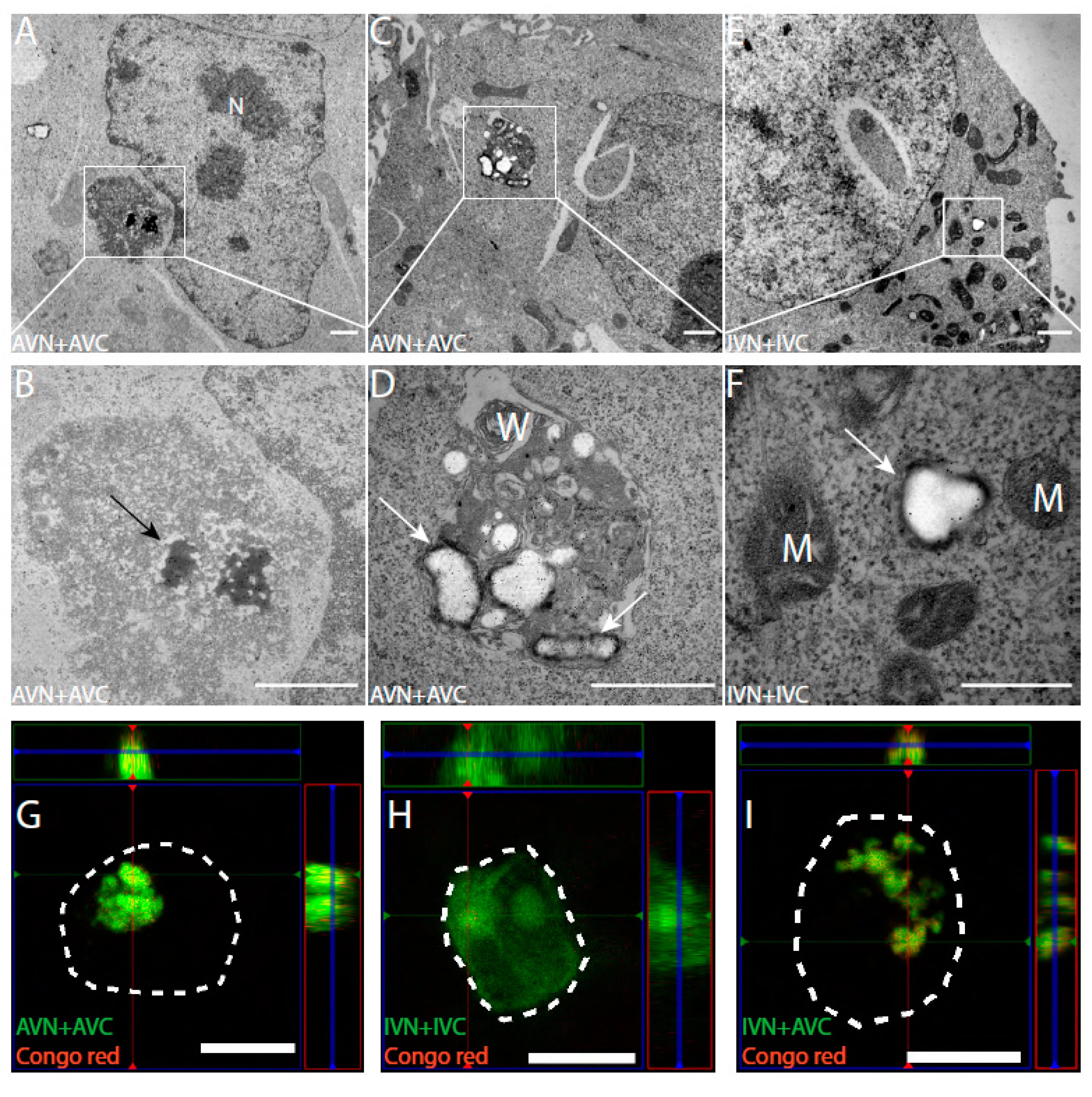
2.2. In TEM, Dot-Like Inclusion Bodies Contain a Mixture of Amorphous and Fibrillar Material
2.3. Amyloid Develops in Cells Expressing Aβ-Containing BiFC Constructs
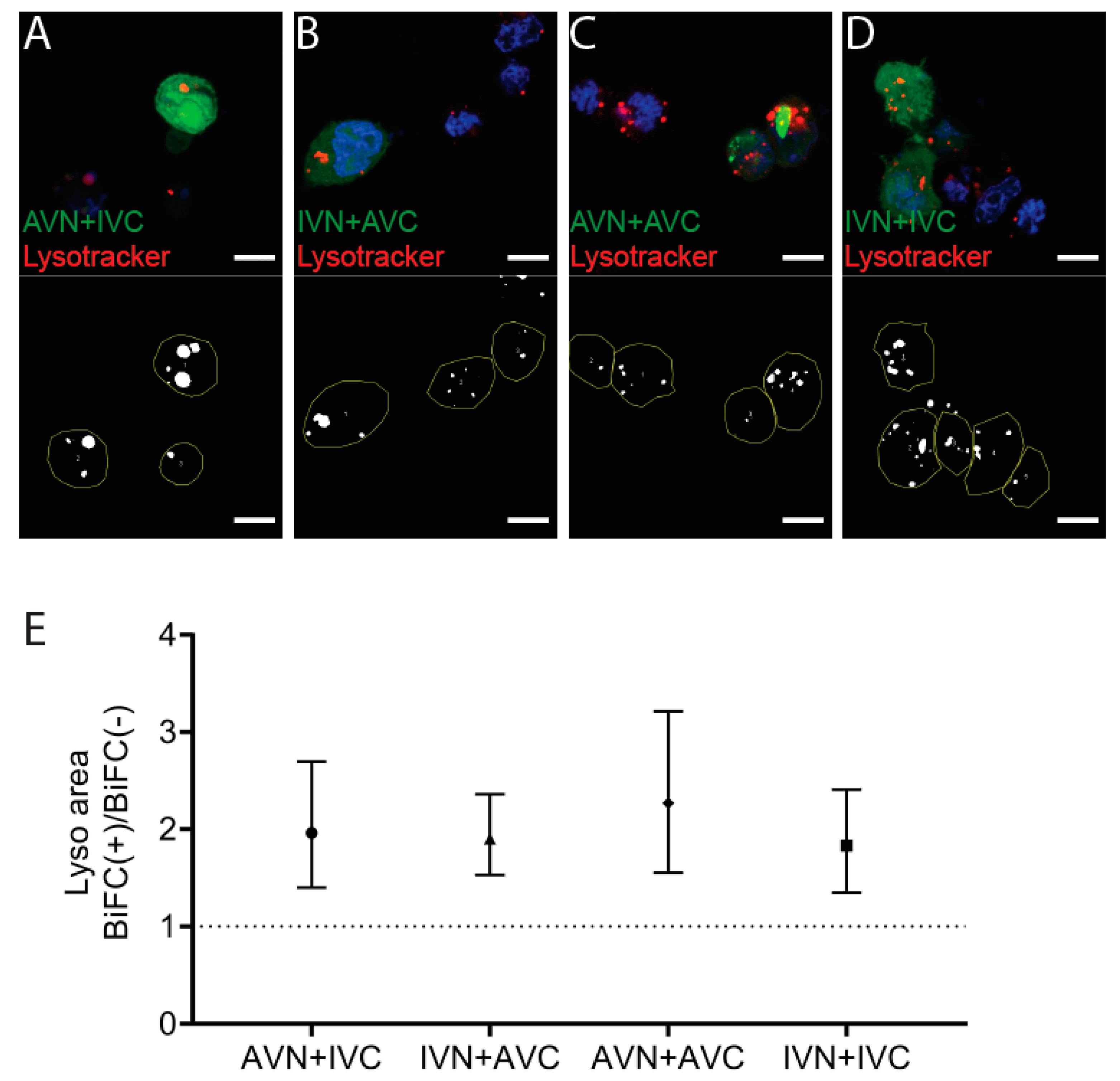
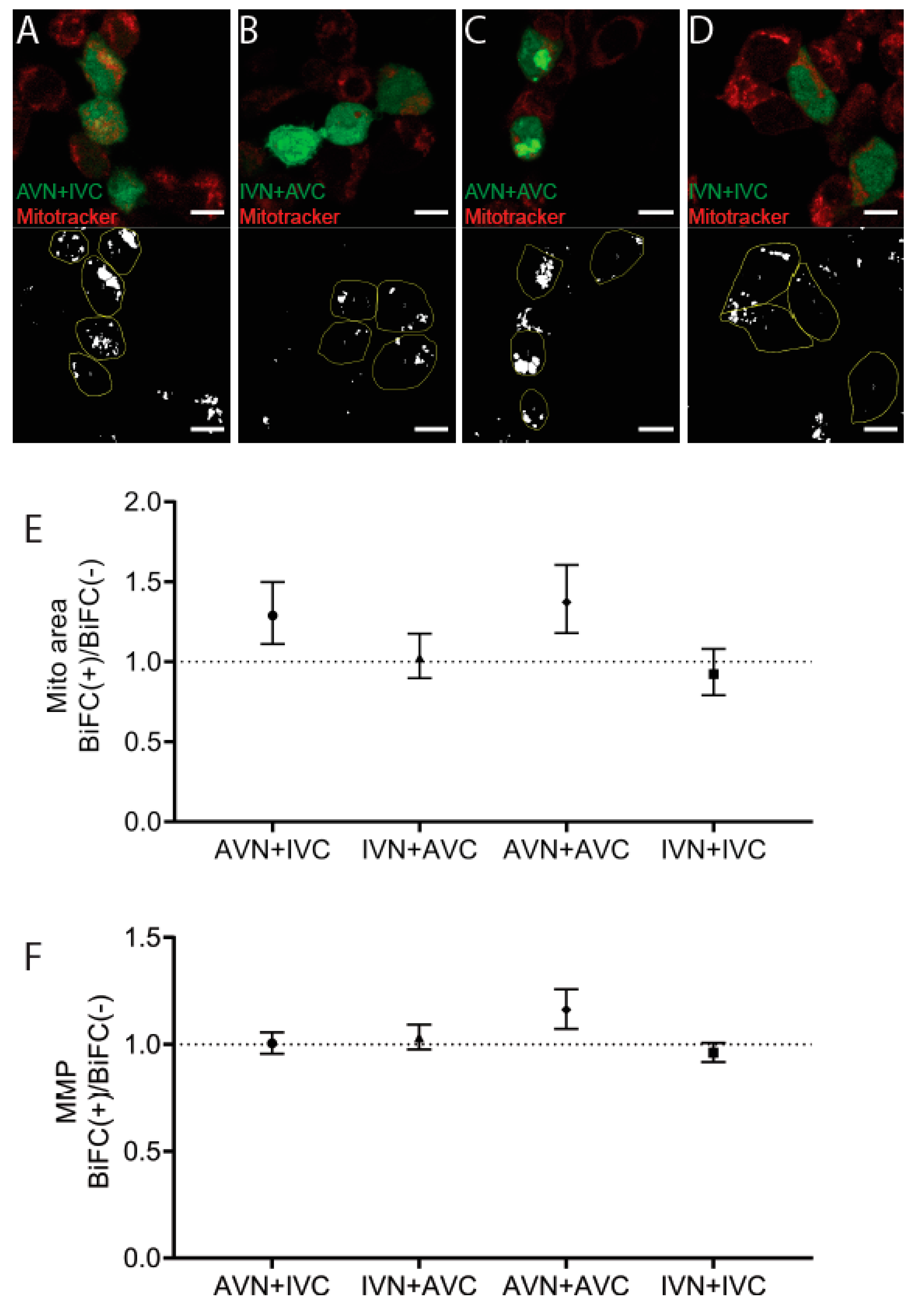
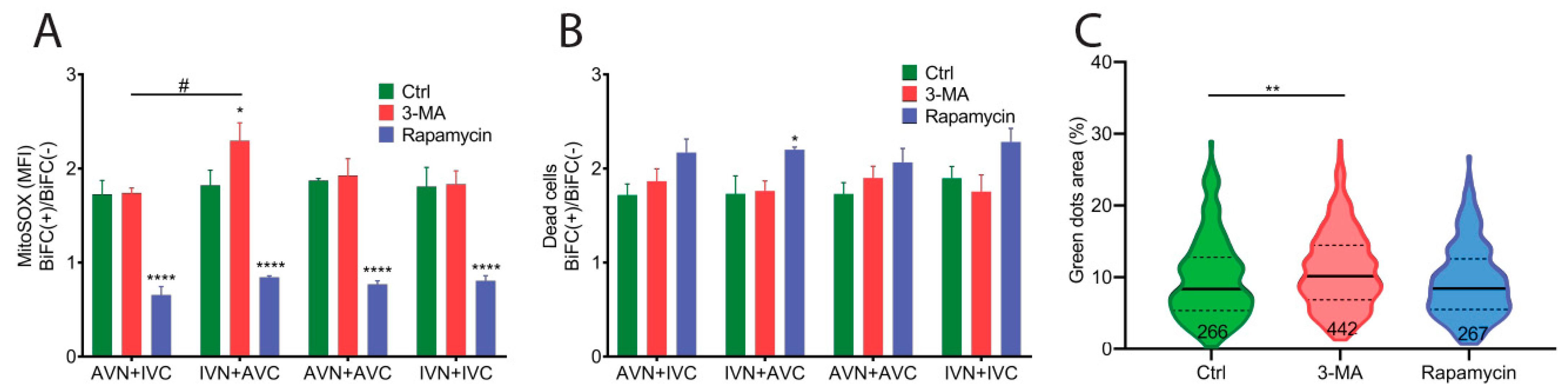
2.4. Aβ/IAPP, Aβ/Aβ, and IAPP/IAPP Expression Increase Lysosomal and Mitochondrial Area, but Only Aβ/Aβ Expression Elevates MMP
2.5. Expression of Aβ, IAPP, and Aβ/IAPP Increase Superoxide Production and Susceptibility to Cell Death
2.6. Co-Expression of Aβ and IAPP Shortens the Lifespan in Drosophila melanogaster
3. Discussion
4. Materials and Methods
4.1. Material Used for the Study
4.2. BiFC Vectors
4.3. Cell Culture and Transfection
4.4. Congo Red Staining
4.5. Transmission Electron Microscopy
4.6. Detection of Lysosomes
4.7. Detection of Mitochondria
4.8. Detection of Mitochondrial Superoxide
4.9. Modulation of Autophagy
4.10. Detection of Dead Cells
4.11. Confocal Imaging
4.12. Drosophila melanogaster
4.13. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Benson, M.D.; Buxbaum, J.N.; Eisenberg, D.S.; Merlini, G.; Saraiva, M.J.M.; Sekijima, Y. Amyloid nomenclature 2020: Update and recommendations by the International Society of Amyloidosis (ISA) nomenclature committee. Amyloid 2020, 27, 217–222. [Google Scholar] [CrossRef]
- Westermark, P. Quantitative studies on amyloid in the islets of Langerhans. Ups. J. Med. Sci. 1972, 77, 91–94. [Google Scholar] [CrossRef] [Green Version]
- Clark, A.; Wells, C.A.; Buley, I.D.; Cruickshank, J.K.; Vanhegan, R.I.; Matthews, D.R. Islet amyloid, increased A-cells, reduced B-cells and exocrine fibrosis: Quantitative changes in the pancreas in type 2 diabetes. Diabetes Res. 1988, 9, 151–159. [Google Scholar]
- Marzban, L.; Trigo-Gonzalez, G.; Zhu, X.; Rhodes, C.J.; Halban, P.A.; Steiner, D.F. Role of beta-cell prohormone convertase (PC)1/3 in processing of pro-islet amyloid polypeptide. Diabetes 2004, 53, 141–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Xu, J.; Finnerty, J.; Furuta, M.; Steiner, D.F.; Verchere, C.B. The prohormone convertase enzyme 2 (PC2) is essential for processing pro-islet amyloid polypeptide at the NH2-terminal cleavage site. Diabetes 2001, 50, 534–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westermark, P.; Andersson, A.; Westermark, G.T. Islet amyloid polypeptide, islet amyloid, and diabetes mellitus. Physiol. Rev. 2011, 91, 795–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akter, R.; Cao, P.; Noor, H.; Ridgway, Z.; Tu, L.H.; Wang, H. Islet Amyloid Polypeptide: Structure, Function, and Pathophysiology. J. Diabetes Res. 2016, 2016, 2798269. [Google Scholar] [CrossRef] [Green Version]
- Milardi, D.; Gazit, E.; Radford, S.E.; Xu, Y.; Gallardo, R.U.; Caflisch, A. Proteostasis of Islet Amyloid Polypeptide: A Molecular Perspective of Risk Factors and Protective Strategies for Type II Diabetes. Chem. Rev. 2021, 121, 1845–1893. [Google Scholar] [CrossRef]
- Taylor, R. Insulin resistance and type 2 diabetes. Diabetes 2012, 61, 778–779. [Google Scholar] [CrossRef] [Green Version]
- Kahn, S.E.; Halban, P.A. Release of incompletely processed proinsulin is the cause of the disproportionate proinsulinemia of NIDDM. Diabetes 1997, 46, 1725–1732. [Google Scholar] [CrossRef]
- Paulsson, J.F.; Westermark, G.T. Aberrant processing of human proislet amyloid polypeptide results in increased amyloid formation. Diabetes 2005, 54, 2117–2125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paulsson, J.F.; Andersson, A.; Westermark, P.; Westermark, G.T. Intracellular amyloid-like deposits contain unprocessed pro-islet amyloid polypeptide (proIAPP) in beta cells of transgenic mice overexpressing the gene for human IAPP and transplanted human islets. Diabetologia 2006, 49, 1237–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glenner, G.G.; Wong, C.W. Alzheimer’s disease: Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 1984, 120, 885–890. [Google Scholar] [CrossRef]
- Masters, C.L.; Simms, G.; Weinman, N.A.; Multhaup, G.; McDonald, B.L.; Beyreuther, K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc. Natl. Acad. Sci. USA 1985, 82, 4245–4249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfe, M.S.; Xia, W.; Moore, C.L.; Leatherwood, D.D.; Ostaszewski, B.; Rahmati, T. Peptidomimetic probes and molecular modeling suggest that Alzheimer’s gamma-secretase is an intramembrane-cleaving aspartyl protease. Biochemistry 1999, 38, 4720–4727. [Google Scholar] [CrossRef] [PubMed]
- Mori, H.; Takio, K.; Ogawara, M.; Selkoe, D.J. Mass spectrometry of purified amyloid beta protein in Alzheimer’s disease. J. Biol. Chem. 1992, 267, 17082–17086. [Google Scholar] [CrossRef]
- Naslund, J.; Schierhorn, A.; Hellman, U.; Lannfelt, L.; Roses, A.D.; Tjernberg, L.O. Relative abundance of Alzheimer A beta amyloid peptide variants in Alzheimer disease and normal aging. Proc. Natl. Acad. Sci. USA 1994, 91, 8378–8382. [Google Scholar] [CrossRef] [Green Version]
- Allinquant, B.; Hantraye, P.; Mailleux, P.; Moya, K.; Bouillot, C.; Prochiantz, A. Downregulation of amyloid precursor protein inhibits neurite outgrowth in vitro. J. Cell. Biol. 1995, 128, 919–927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabo, S.L.; Ikin, A.F.; Buxbaum, J.D.; Greengard, P. The amyloid precursor protein and its regulatory protein, FE65, in growth cones and synapses in vitro and in vivo. J. Neurosci. 2003, 23, 5407–5415. [Google Scholar] [CrossRef] [Green Version]
- Grundke-Iqbal, I.; Iqbal, K.; George, L.; Tung, Y.C.; Kim, K.S.; Wisniewski, H.M. Amyloid protein and neurofibrillary tangles coexist in the same neuron in Alzheimer disease. Proc. Natl. Acad. Sci. USA 1989, 86, 2853–2857. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N. Amyloidogenesis of natively unfolded proteins. Curr. Alzheimer Res. 2008, 5, 260–287. [Google Scholar] [CrossRef]
- Westermark, P. Amyloid and polypeptide hormones: What is their interrelationship? Amyloid 1994, 1, 47–60. [Google Scholar] [CrossRef]
- O’Nuallain, B.; Williams, A.D.; Westermark, P.; Wetzel, R. Seeding specificity in amyloid growth induced by heterologous fibrils. J. Biol. Chem. 2004, 279, 17490–17499. [Google Scholar] [CrossRef] [Green Version]
- Andreetto, E.; Yan, L.M.; Tatarek-Nossol, M.; Velkova, A.; Frank, R.; Kapurniotu, A. Identification of hot regions of the Abeta-IAPP interaction interface as high-affinity binding sites in both cross- and self-association. Angew Chem. Int. Ed. Engl. 2010, 49, 3081–3085. [Google Scholar] [CrossRef]
- Oskarsson, M.E.; Paulsson, J.F.; Schultz, S.W.; Ingelsson, M.; Westermark, P.; Westermark, G.T. In vivo seeding and cross-seeding of localized amyloidosis: A molecular link between type 2 diabetes and Alzheimer disease. Am. J. Pathol. 2015, 185, 834–846. [Google Scholar] [CrossRef]
- Jackson, K.; Barisone, G.A.; Diaz, E.; Jin, L.W.; DeCarli, C.; Despa, F. Amylin deposition in the brain: A second amyloid in Alzheimer disease? Ann. Neurol. 2013, 74, 517–526. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.D.; Chinenov, Y.; Kerppola, T.K. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002, 9, 789–798. [Google Scholar] [CrossRef]
- Harmon, M.; Larkman, P.; Hardingham, G.; Jackson, M.; Skehel, P. A Bi-fluorescence complementation system to detect associations between the Endoplasmic reticulum and mitochondria. Sci. Rep. 2017, 7, 17467. [Google Scholar] [CrossRef] [Green Version]
- Boland, B.; Kumar, A.; Lee, S.; Platt, F.M.; Wegiel, J.; Yu, W.H. Autophagy induction and autophagosome clearance in neurons: Relationship to autophagic pathology in Alzheimer’s disease. J. Neurosci. 2008, 28, 6926–6937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Park, K.; Kim, M.J.; Lim, H.; Kim, K.H.; Kim, S.W. An autophagy enhancer ameliorates diabetes of human IAPP-transgenic mice through clearance of amyloidogenic oligomer. Nat. Commun. 2021, 12, 183. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.T.; Blaser, G.; Behrens, C.; Cabrita, L.D.; Dobson, C.M.; Jackson, S.E. Folding study of Venus reveals a strong ion dependence of its yellow fluorescence under mildly acidic conditions. J. Biol. Chem. 2010, 285, 4859–4869. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.D.; Kerppola, T.K. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 2003, 21, 539–545. [Google Scholar] [CrossRef]
- Nilsberth, C.; Westlind-Danielsson, A.; Eckman, C.B.; Condron, M.M.; Axelman, K.; Forsell, C. The ‘Arctic’ APP mutation (E693G) causes Alzheimer’s disease by enhanced Abeta protofibril formation. Nat. Neurosci. 2001, 4, 887–893. [Google Scholar] [CrossRef]
- Kerppola, T.K. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 2006, 1, 1278–1286. [Google Scholar] [CrossRef] [Green Version]
- Outeiro, T.F.; Putcha, P.; Tetzlaff, J.E.; Spoelgen, R.; Koker, M.; Carvalho, F. Formation of toxic oligomeric alpha-synuclein species in living cells. PLoS ONE 2008, 3, e1867. [Google Scholar] [CrossRef]
- Herrera, F.; Goncalves, S.; Outeiro, T.F. Imaging protein oligomerization in neurodegeneration using bimolecular fluorescence complementation. Methods Enzymol. 2012, 506, 157–174. [Google Scholar]
- Kim, J.; Lee, H.; Lee, J.H.; Kwon, D.Y.; Genovesio, A.; Fenistein, D. Dimerization, oligomerization, and aggregation of human amyotrophic lateral sclerosis copper/zinc superoxide dismutase 1 protein mutant forms in live cells. J. Biol. Chem. 2014, 289, 15094–15103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frey, B.; AlOkda, A.; Jackson, M.P.; Riguet, N.; Duce, J.A.; Lashuel, H.A. Monitoring alpha-synuclein oligomerization and aggregation using bimolecular fluorescence complementation assays: What you see is not always what you get. J. Neurochem. 2021, 157, 872–888. [Google Scholar] [CrossRef] [PubMed]
- Buckig, A.; Tikkanen, R.; Herzog, V.; Schmitz, A. Cytosolic and nuclear aggregation of the amyloid beta-peptide following its expression in the endoplasmic reticulum. Histochem. Cell Biol. 2002, 118, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Johnston, J.A.; Ward, C.L.; Kopito, R.R. Aggresomes: A cellular response to misfolded proteins. J. Cell. Biol. 1998, 143, 1883–1898. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, M.; Kim, Y.M.; Lee, G.; Junn, E.; Iwatsubo, T.; Mouradian, M.M. Aggresomes formed by alpha-synuclein and synphilin-1 are cytoprotective. J. Biol. Chem. 2004, 279, 4625–4631. [Google Scholar] [CrossRef] [Green Version]
- Wong, E.S.; Tan, J.M.; Soong, W.E.; Hussein, K.; Nukina, N.; Dawson, V.L. Autophagy-mediated clearance of aggresomes is not a universal phenomenon. Hum. Mol. Genet. 2008, 17, 2570–2582. [Google Scholar] [CrossRef]
- Schultz, S.W.; Nilsson, K.P.; Westermark, G.T. Drosophila melanogaster as a model system for studies of islet amyloid polypeptide aggregation. PLoS ONE 2011, 6, e20221. [Google Scholar] [CrossRef] [Green Version]
- Crowther, D.C.; Page, R.; Chandraratna, D.; Lomas, D.A. A Drosophila model of Alzheimer’s disease. Methods Enzymol. 2006, 412, 234–255. [Google Scholar]
- Bharadwaj, P.; Solomon, T.; Sahoo, B.R.; Ignasiak, K.; Gaskin, S.; Rowles, J. Amylin and beta amyloid proteins interact to form amorphous heterocomplexes with enhanced toxicity in neuronal cells. Sci. Rep. 2020, 10, 10356. [Google Scholar] [CrossRef]
- Bharadwaj, P.; Solomon, T.; Malajczuk, C.J.; Mancera, R.L.; Howard, M.; Arrigan, D.W.M. Role of the cell membrane interface in modulating production and uptake of Alzheimer’s beta amyloid protein. Biochim. Biophys Acta Biomembr. 2018, 1860, 1639–1651. [Google Scholar] [CrossRef]
- Hu, C.D.; Grinberg, A.V.; Kerppola, T.K. Visualization of protein interactions in living cells using bimolecular fluorescence complementation (BiFC) analysis. Curr. Protoc. Protein Sci. 2005, 19, 19. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fieller, E.C. Some problems in interval estimation. J. R. Stat. Soc. Ser. B 1954, 16, 175–185. [Google Scholar] [CrossRef]
| IAPP | Forward: 5′-TATTAAGCTTATGAAATGCAACACT-3′ Reverse: 5′-AAGGGAATTCATATGTATTGGATCC-3′ |
| Aβ | Forward: 5′-TATTAAGCTTATGGATGCGGAATTT-3′ Reverse: 5′-AAGGGAATTCCGCAATCACCAC-3′ |
| Venus 1-173 (VN): | Forward: 5′-TTAATCTAGAGTGAGCAAGGGC-3′ Reverse: 5′-AAACGGGCCCCTATTACTCGATGTTGTG-3′ |
| Venus 155-239 (VC) | Forward: 5′-TTAATCTAGAGCCGACAAGCAG-3′ Reverse: 5′-AAACGGGCCCCTATTACTTGTACAGCTC-3′ |
| Venus 1-173 (VN) Antiparallel | Forward: 5′-TTAAAAGCTTATGGTGAGCAAGGGC-3′ Reverse: 5′-AAACGAATTCCTCGATGTTGTG-3′ |
| IAPP Antiparallel | Forward: 5′-TATTTCTAGAAAATGCAACACT-3′ Reverse: 5′-AAGGGGGCCCCTATTAATATGTATTGGA-3′ |
| Aβ Antiparallel | Forward: 5′-TATTTCTAGAGATGCGGAATTT-3′ Reverse: 5′-AAGGGGGCCCCTATTACGCAATCACCAC-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Westermark, G.T. The Amyloid Forming Peptides Islet Amyloid Polypeptide and Amyloid β Interact at the Molecular Level. Int. J. Mol. Sci. 2021, 22, 11153. https://doi.org/10.3390/ijms222011153
Wang Y, Westermark GT. The Amyloid Forming Peptides Islet Amyloid Polypeptide and Amyloid β Interact at the Molecular Level. International Journal of Molecular Sciences. 2021; 22(20):11153. https://doi.org/10.3390/ijms222011153
Chicago/Turabian StyleWang, Ye, and Gunilla T. Westermark. 2021. "The Amyloid Forming Peptides Islet Amyloid Polypeptide and Amyloid β Interact at the Molecular Level" International Journal of Molecular Sciences 22, no. 20: 11153. https://doi.org/10.3390/ijms222011153
APA StyleWang, Y., & Westermark, G. T. (2021). The Amyloid Forming Peptides Islet Amyloid Polypeptide and Amyloid β Interact at the Molecular Level. International Journal of Molecular Sciences, 22(20), 11153. https://doi.org/10.3390/ijms222011153







