Transcriptional Regulation of GDF15 by EGR1 Promotes Head and Neck Cancer Progression through a Positive Feedback Loop
Abstract
:1. Introduction
2. Results
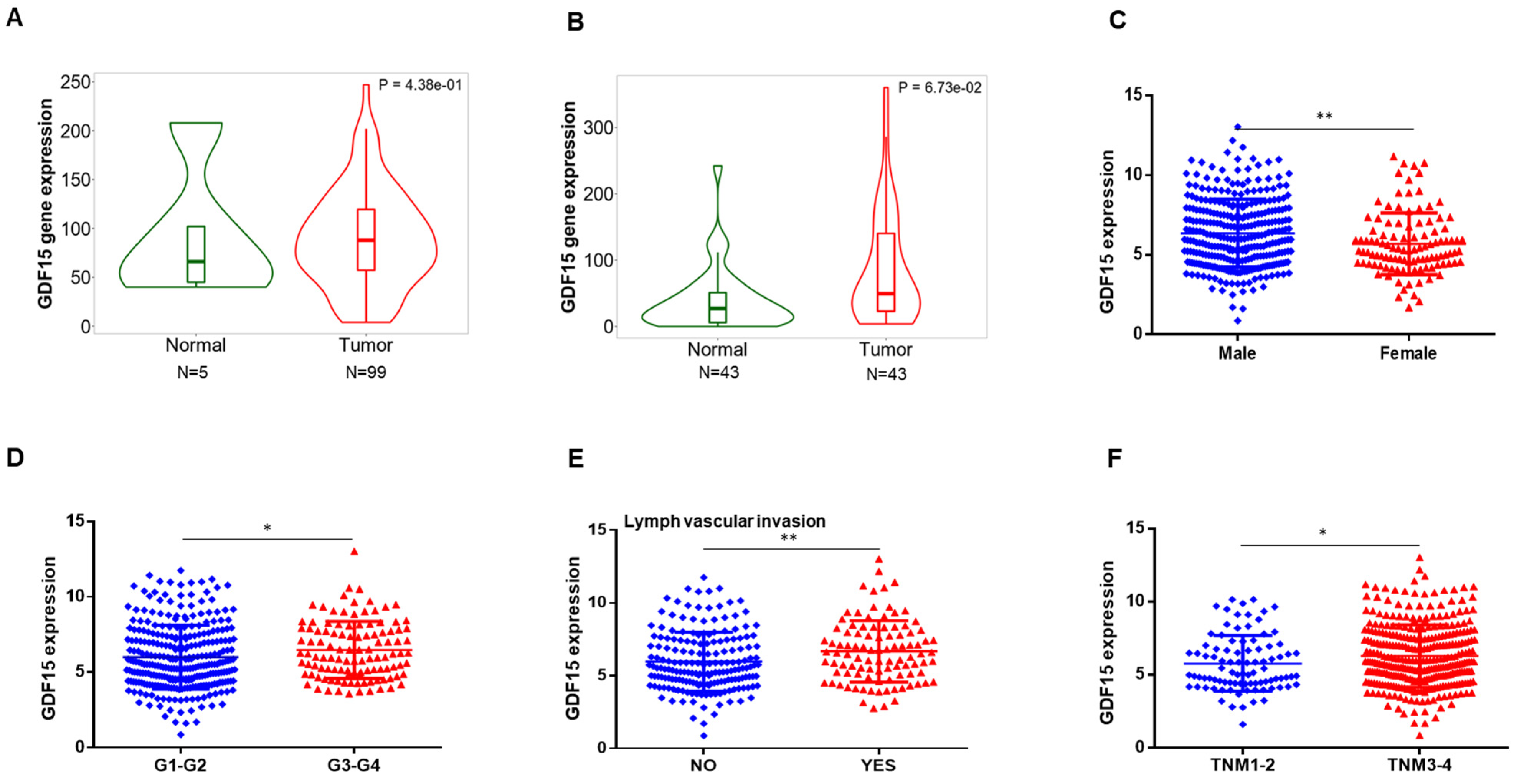
2.1. GDF15 Expression in the Public Database
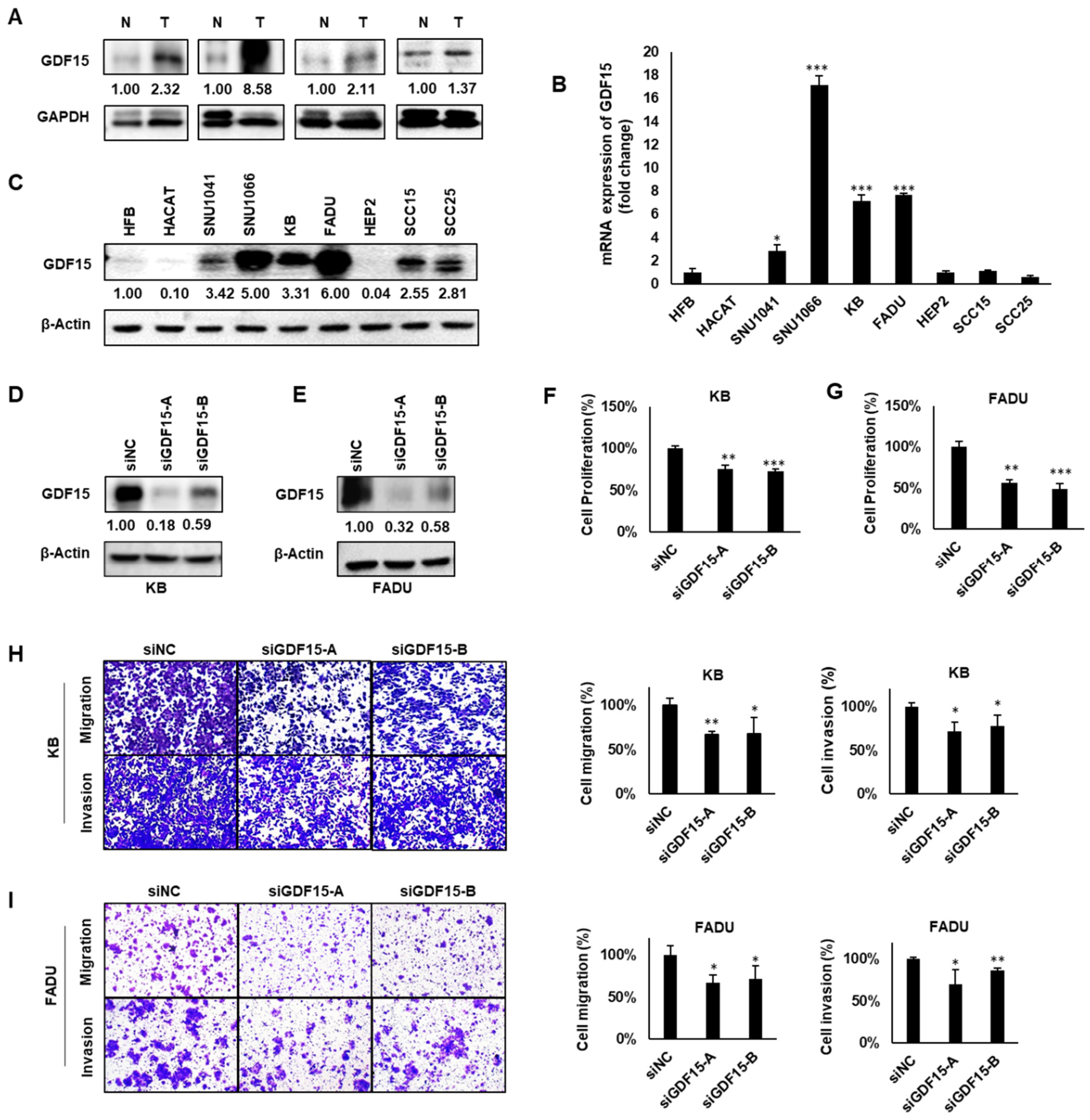
2.2. GDF15 Promotes Cancer Cell Proliferation, Migration and Invasion
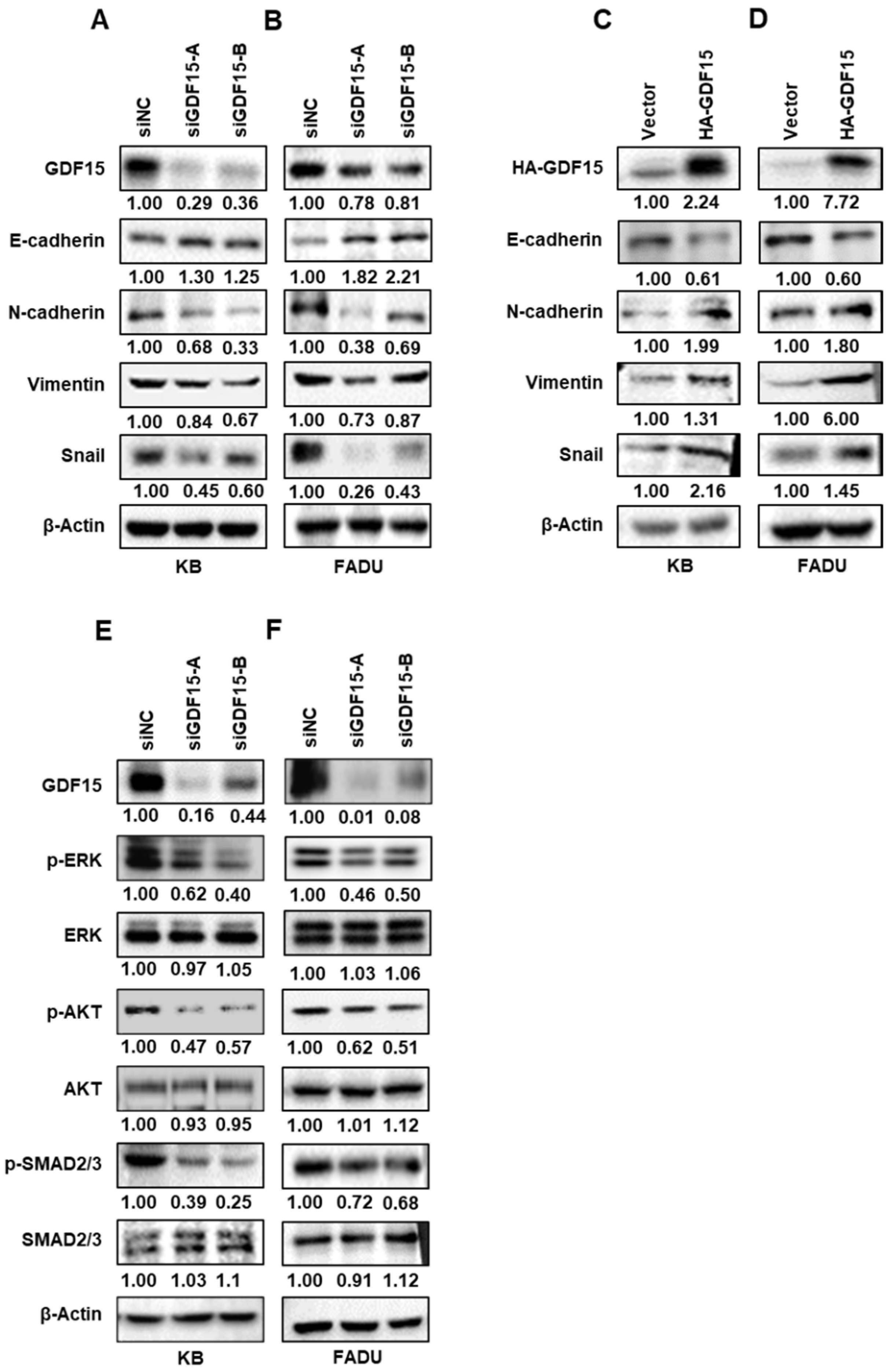
2.3. GDF15 Regulates Epithelial–Mesenchymal Transition (EMT)-Related Proteins via Phosphorylating SMAD, ERK and AKT in HNC
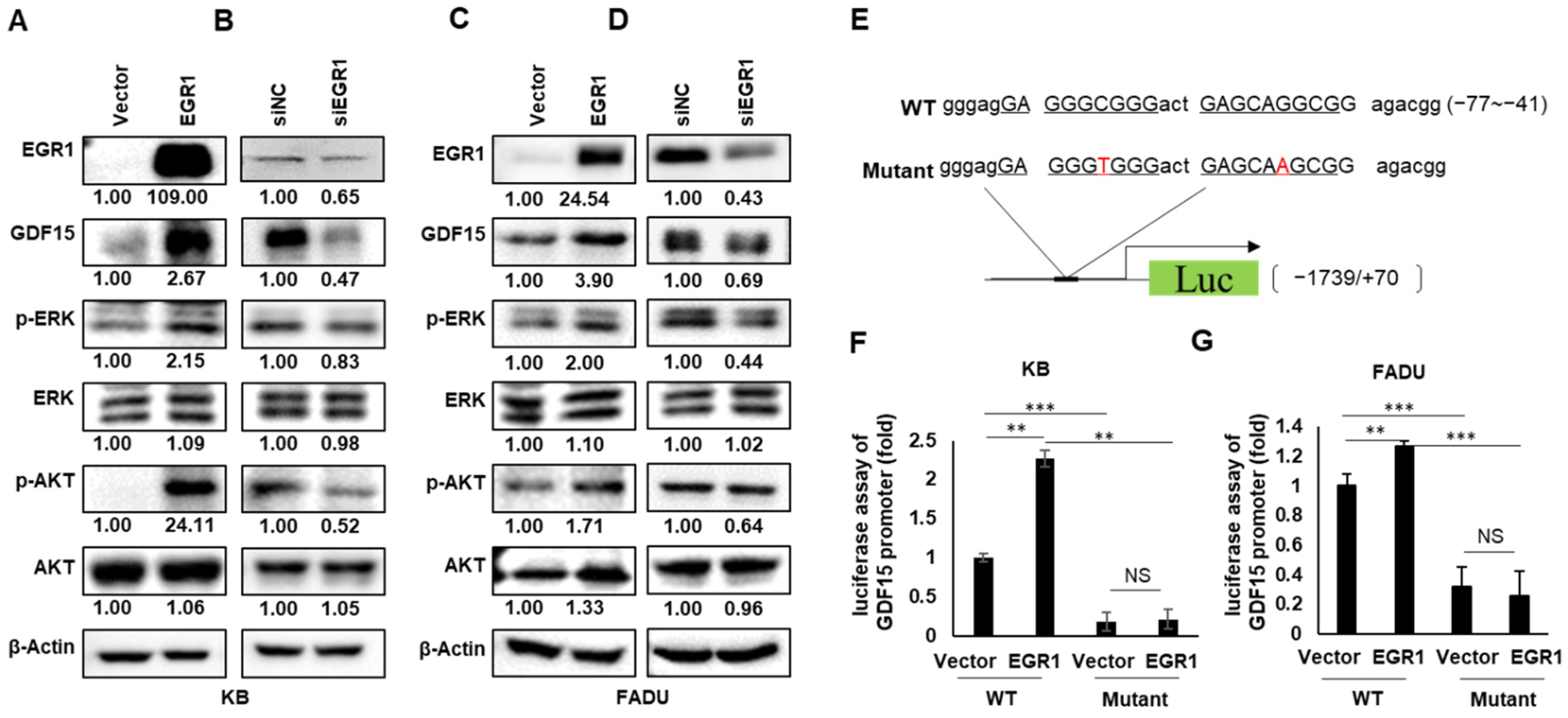
2.4. EGR1 Is a Transcriptional Regulator of GDF15
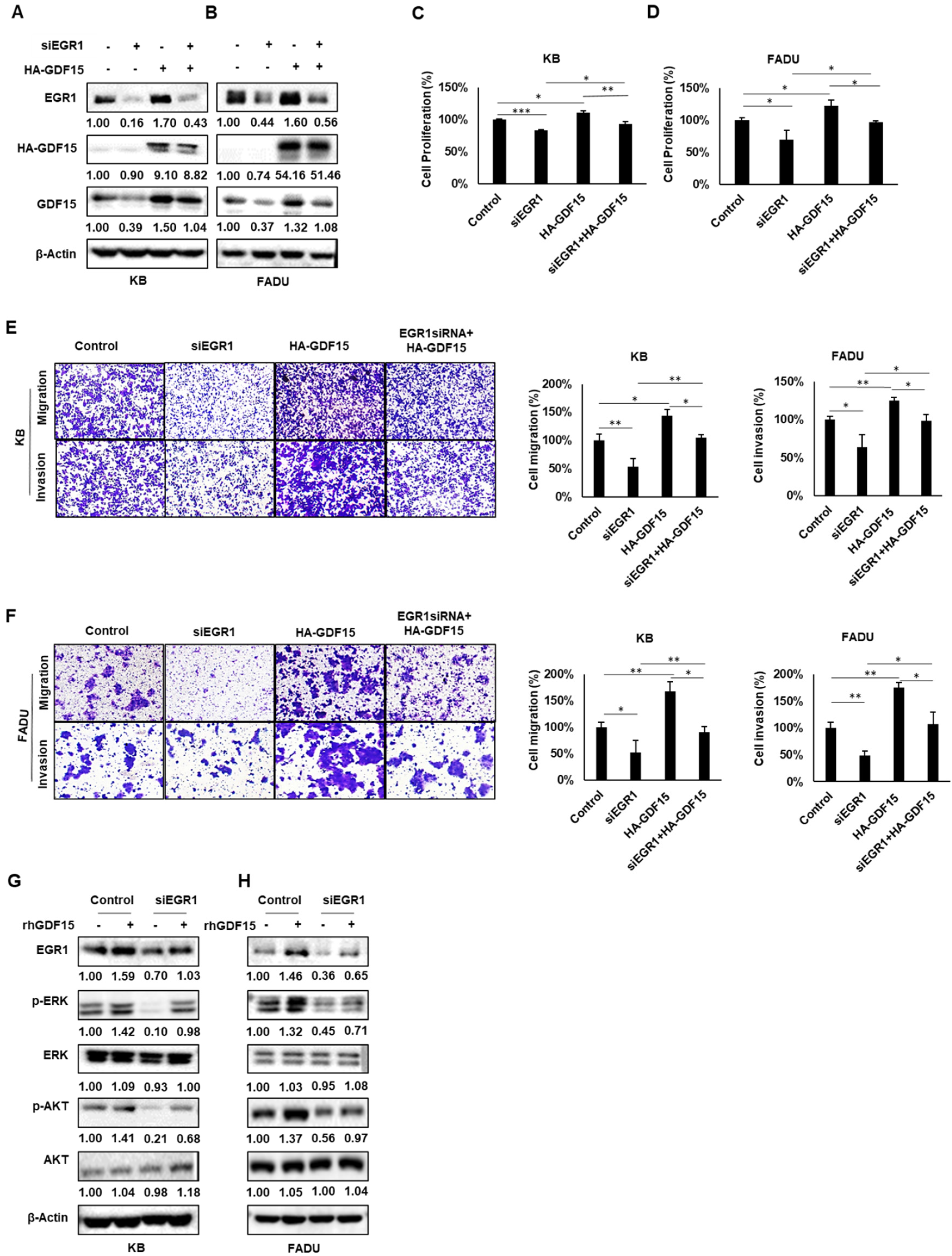
2.5. A Positive Feedback Loop of EGR1 and GDF15 Facilitates Proliferation, Migration and Invasion in HNC
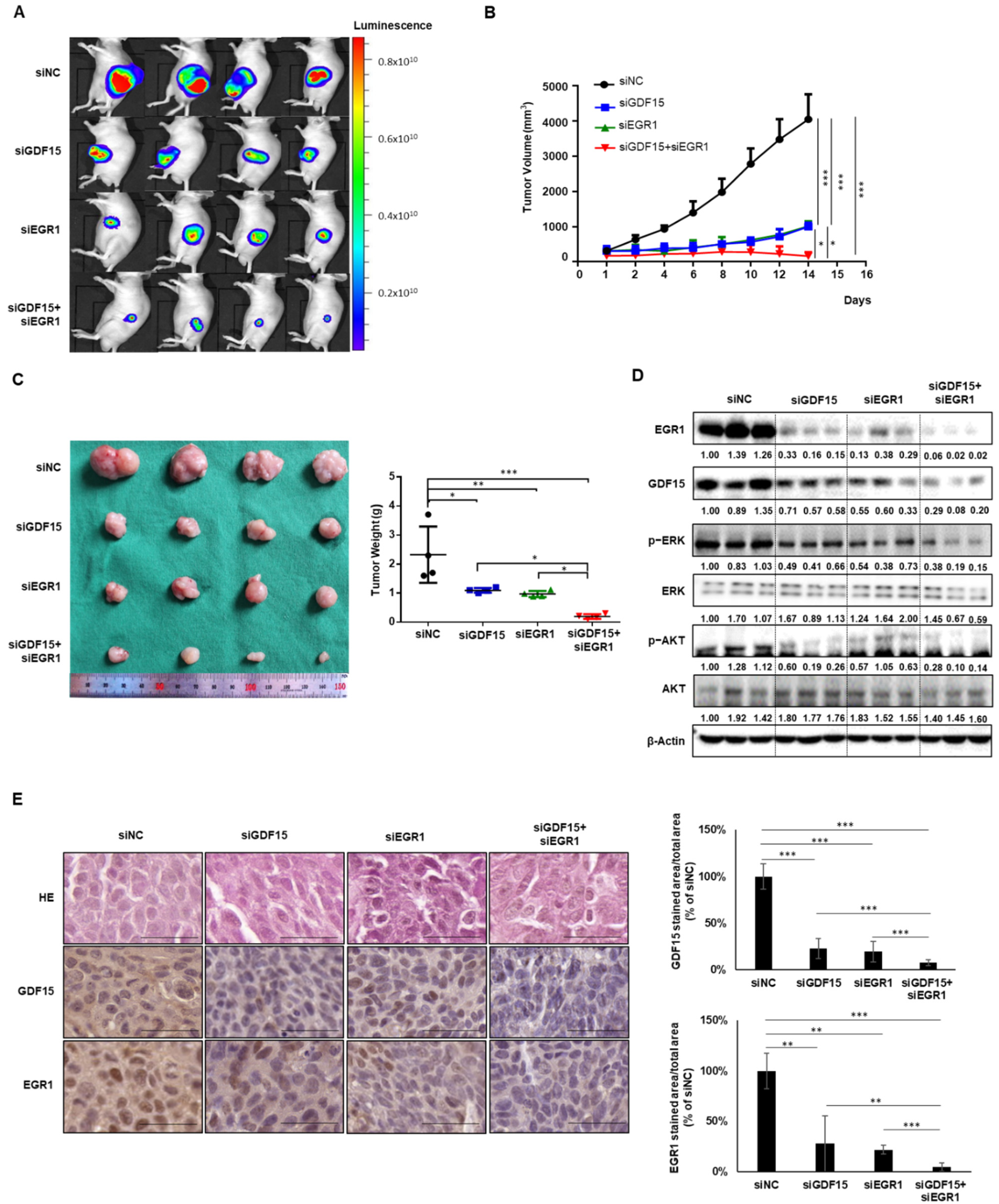
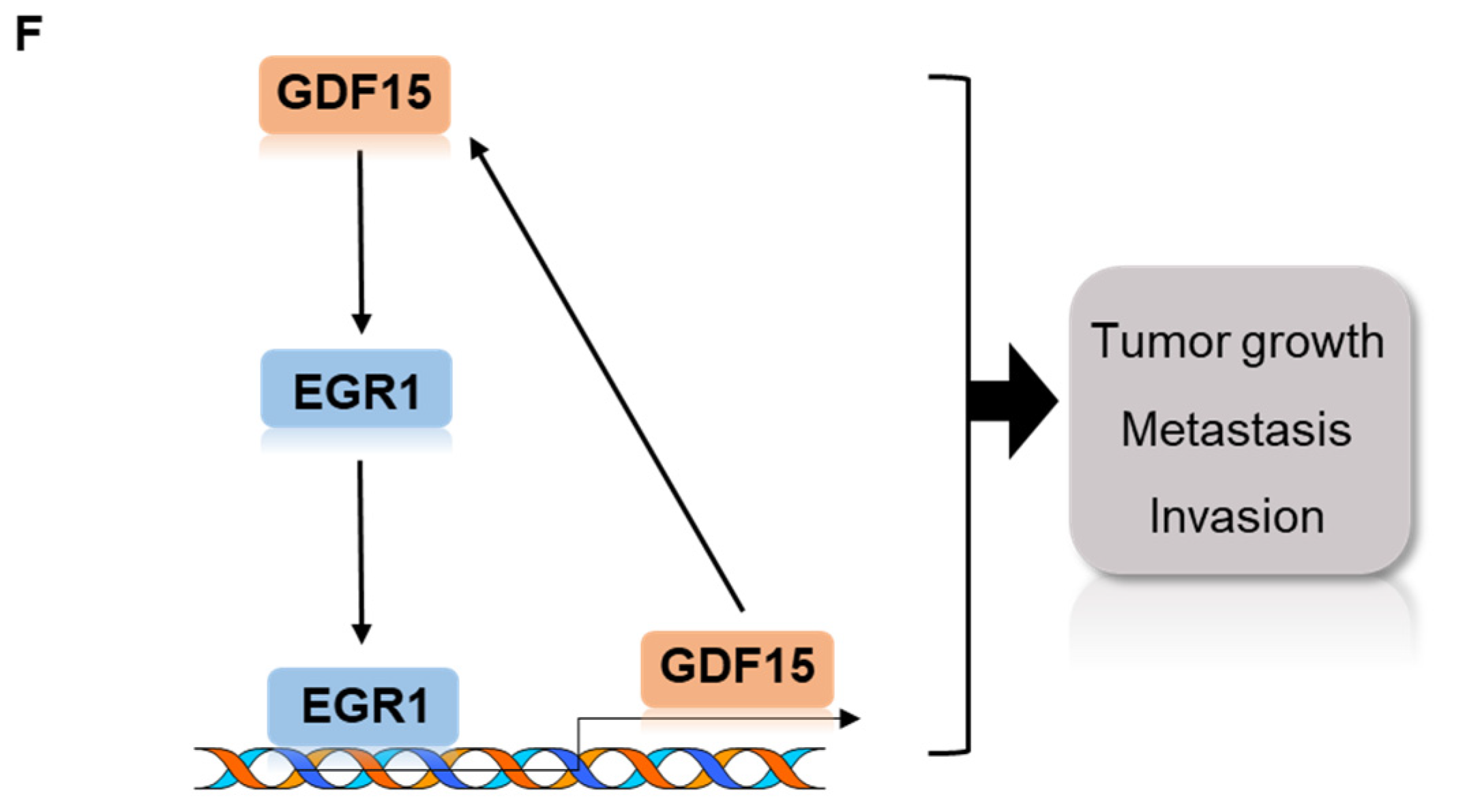
2.6. Combined Treatment Targeting Both GDF15 and EGR1 Synergistically Reduced the Tumor Growth In Vivo
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Materials
4.2. RNA Isolation and Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
4.3. Western Blot Analysis
4.4. Small-Interfering RNA (siRNA) Transfection
4.5. Cell Proliferation Assay
4.6. Cell Migration and Invasion Assay
4.7. Plasmid Constructs and Site-Directed Mutagenesis
4.8. Luciferase Assay
4.9. Animal Experiments
4.10. In Vivo Imaging
4.11. Histological and Immunohistochemical Analysis
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GDF15 | Growth and differentiation factor 15 |
| EGR1 | Early growth response 1 |
| TGF-β1 | Transforming growth factor β1 |
| p53 | Tumor protein p53 |
| HE | Hematoxylin and eosin |
| RT-PCR | Quantitative real-time reverse transcriptase polymerase chain reaction |
| cDNA | Complementary DNA |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
References
- Li, Y.-L.; Chang, J.T.; Lee, L.-Y.; Fan, K.-H.K.-H.; Lu, Y.-C.; Li, Y.-C.; Chiang, C.-H.; You, G.-R.; Chen, H.-Y.; Cheng, A.-J. GDF15 contributes to radioresistance and cancer stemness of head and neck cancer by regulating cellular reactive oxygen species via a SMAD-associated signaling pathway. Oncotarget 2017, 8, 1508–1528. [Google Scholar] [CrossRef] [Green Version]
- Miyauchi, S.; Kim, S.S.; Pang, J.; Gold, K.A.; Gutkind, J.S.; Califano, J.A.; Mell, L.K.; Cohen, E.E.W.; Sharabi, A.B. Immune Modulation of Head and Neck Squamous Cell Carcinoma and the Tumor Microenvironment by Conventional Therapeutics. Clin. Cancer Res. 2019, 25, 4211–4223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wittekindt, C.; Wagner, S.; Sharma, S.J.; Würdemann, N.; Knuth, J.; Reder, H.; Klußmann, J.P. HPV—A different view on Head and Neck Cancer. Laryngorhinootologie 2018, 97, S48–S113. [Google Scholar] [PubMed] [Green Version]
- Kaidar-Person, O.; Gil, Z.; Billan, S. Precision medicine in head and neck cancer. Drug Resist. Updat. 2018, 40, 13–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, M.; Tang, Y.; Sun, B.; Wang, K.; Zhu, F. Perioperative mortality of head and neck cancers. BMC Cancer 2021, 21, 256. [Google Scholar] [CrossRef]
- Coll, A.; Chen, M.; Taskar, P.; Rimmington, D.; Patel, S.; Tadross, J.A.; Cimino, I.; Yang, M.; Welsh, P.; Virtue, S.; et al. GDF15 mediates the effects of metformin on body weight and energy balance. Nature 2020, 578, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Barton, H.; Zechendorf, E.; Ostareck, D.; Ostareck-Lederer, A.; Stoppe, C.; Zayat, R.; Simon-Philipp, T.; Marx, G.; Bickenbach, J. Prognostic Value of GDF-15 in Predicting Prolonged Intensive Care Stay following Cardiac Surgery: A Pilot Study. Dis. Markers 2021, 2021, 5564334. [Google Scholar] [CrossRef]
- Wollert, K.C.; Kempf, T.; Wallentin, L. Growth Differentiation Factor 15 as a Biomarker in Cardiovascular Disease. Clin. Chem. 2017, 63, 140–151. [Google Scholar] [CrossRef]
- Lee, J.; Kim, I.; Yoo, E.; Baek, S.J. Competitive inhibition by NAG-1/GDF-15 NLS peptide enhances its anti-cancer activity. Biochem. Biophys. Res. Commun. 2019, 519, 29–34. [Google Scholar] [CrossRef]
- Wang, X.; Baek, S.J.; Eling, T.E. The diverse roles of nonsteroidal anti-inflammatory drug activated gene (NAG-1/GDF15) in cancer. Biochem. Pharmacol. 2013, 85, 597–606. [Google Scholar] [CrossRef] [Green Version]
- Baek, S.J.; Eling, T. Growth differentiation factor 15 (GDF15): A survival protein with therapeutic potential in metabolic diseases. Pharmacol. Ther. 2019, 198, 46–58. [Google Scholar] [CrossRef]
- Wang, B.; Guo, H.; Yu, H.; Chen, Y.; Xu, H.; Zhao, G. The Role of the Transcription Factor EGR1 in Cancer. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef]
- Gitenay, D.; Baron, V.T. Is EGR1 a potential target for prostate cancer therapy? Future Oncol. 2009, 5, 993–1003. [Google Scholar] [CrossRef] [Green Version]
- Baron, V.; Adamson, E.D.; Calogero, A.; Ragona, G.; Mercola, D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFbeta1, PTEN, p53, and fibronectin. Cancer Gene Ther. 2006, 13, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Xia, Q.; Liu, Y.; Bai, W.; Yao, Y.; Ding, J.; Lin, L.; Xu, Z.; Cai, Z.; Wang, S.; et al. TCF7L2 and EGR1 synergistic activation of transcription of LCN2 via an ERK1/2-dependent pathway in esophageal squamous cell carcinoma cells. Cell. Signal. 2019, 55, 8–16. [Google Scholar] [CrossRef]
- Rochette, L.; Méloux, A.; Zeller, M.; Cottin, Y.; Vergely, C. Functional roles of GDF15 in modulating microenvironment to promote carcinogenesis. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165798. [Google Scholar] [CrossRef]
- Tsai, V.W.W.; Husaini, Y.; Sainsbury, A.; Brown, D.A.; Breit, S.N. The MIC-1/GDF15-GFRAL Pathway in Energy Homeostasis: Implications for Obesity, Cachexia, and Other Associated Diseases. Cell Metab. 2018, 28, 353–368. [Google Scholar] [CrossRef] [Green Version]
- Kadowaki, M.; Yoshioka, H.; Kamitani, H.; Watanabe, T.; Wade, P.; Eling, T.E. DNA methylation-mediated silencing of nonsteroidal anti-inflammatory drug-activated gene (NAG-1/GDF15) in glioma cell lines. Int. J. Cancer 2012, 130, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Li, T.-T.; Liu, M.-R.; Pei, D.-S. Friend or foe, the role of EGR-1 in cancer. Med Oncol. 2019, 37, 7. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-C.; Gajdošik, M.; Josic, D.; Clifton, J.G.; Logothetis, C.; Yu-Lee, L.-Y.; Gallick, G.E.; Maity, S.N.; Lin, S.-H. Secretome Analysis of an Osteogenic Prostate Tumor Identifies Complex Signaling Networks Mediating Cross-talk of Cancer and Stromal Cells Within the Tumor Microenvironment. Mol. Cell. Proteom. 2015, 14, 471–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urakawa, N.; Utsunomiya, S.; Nishio, M.; Shigeoka, M.; Takase, N.; Arai, N.; Kakeji, Y.; Koma, Y.-I.; Yokozaki, H. GDF15 derived from both tumor-associated macrophages and esophageal squamous cell carcinomas contributes to tumor progression via Akt and Erk pathways. Lab. Investig. 2015, 95, 491–503. [Google Scholar] [CrossRef] [Green Version]
- Bruzzese, F.; Hägglöf, C.; Leone, A.; Sjöberg, E.; Roca, M.S.; Kiflemariam, S.; Sjöblom, T.; Hammarsten, P.; Egevad, L.; Bergh, A.; et al. Local and Systemic Protumorigenic Effects of Cancer-Associated Fibroblast-Derived GDF15. Cancer Res. 2014, 74, 3408–3417. [Google Scholar] [CrossRef] [Green Version]
- Pisani, E.B.P.; Airoldi, M.; Allais, A.; Valletti, P.A.; Battista, M.; Benazzo, M.; Briatore, R.; Cacciola, S.; Cocuzza, S.; Colombo, A.; et al. Metastatic disease in head & neck oncology. Acta Otorhinolaryngol. Ital. 2020, 40, S1–S86. [Google Scholar] [CrossRef]
- Liu, Y.-Q.; Zou, H.-Y.; Xie, J.-J.; Fang, W.-K. Paradoxical Roles of Desmosomal Components in Head and Neck Cancer. Biomolecules 2021, 11, 914. [Google Scholar] [CrossRef]
- Cassell, A.; Grandis, J.R. Investigational EGFR-targeted therapy in head and neck squamous cell carcinoma. Expert Opin. Investig. Drugs 2010, 19, 709–722. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Ma, Y.M.; Zheng, P.S.; Zhang, P. GDF15 promotes the proliferation of cervical cancer cells by phosphorylating AKT1 and Erk1/2 through the receptor ErbB2. J. Exp. Clin. Cancer Res. 2018, 37, 80. [Google Scholar] [CrossRef]
- Li, C.; Wang, J.; Kong, J.; Tang, J.; Wu, Y.; Xu, E.; Zhang, H.; Lai, M. GDF15 promotes EMT and metastasis in colorectal cancer. Oncotarget 2016, 7, 860–872. [Google Scholar] [CrossRef]
- Welsh, J.B.; Sapinoso, L.M.; Kern, S.G.; Brown, D.A.; Liu, T.; Bauskin, A.R.; Ward, R.; Hawkins, N.; Quinn, D.; Russell, P.; et al. Large-scale delineation of secreted protein biomarkers overexpressed in cancer tissue and serum. Proc. Natl. Acad. Sci. USA 2003, 100, 3410–3415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wischhusen, J.; Melero, I.; Fridman, W.H. Growth/Differentiation Factor-15 (GDF-15): From Biomarker to Novel Targetable Immune Checkpoint. Front. Immunol. 2020, 11, 951. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Zhang, M.; Zhang, Z.; Jiang, L.; Li, L. GDF15 promotes epithelial-to-mesenchymal transition in colorectal. Artif. Cells Nanomed. Biotechnol. 2018, 46, 652–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corre, J.; Labat, E.; Espagnolle, N.; Hébraud, B.; Avet-Loiseau, H.; Roussel, M.; Huynh, A.; Gadelorge, M.; Cordelier, P.; Klein, B.; et al. Bioactivity and Prognostic Significance of Growth Differentiation Factor GDF15 Secreted by Bone Marrow Mesenchymal Stem Cells in Multiple Myeloma. Cancer Res. 2012, 72, 1395–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, N.H.; Yang, Y.; Lee, S.J.; Kim, K.-Y.; Koo, T.H.; Shin, S.M.; Song, K.S.; Lee, Y.H.; Kim, Y.-J.; Lee, J.J.; et al. Macrophage inhibitory cytokine-1 induces the invasiveness of gastric cancer cells by up-regulating the urokinase-type plasminogen activator system. Cancer Res. 2003, 63, 4648–4655. [Google Scholar] [PubMed]
- Si, Y.; Liu, X.; Cheng, M.; Wang, M.; Gong, Q.; Yang, Y.; Wang, T.; Yang, W. Growth Differentiation Factor 15 Is Induced by Hepatitis C Virus Infection and Regulates Hepatocellular Carcinoma-Related Genes. PLoS ONE 2011, 6, e19967. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.K.; Lee, J.J.; Yang, Y.; You, K.H.; Lee, J.H. Macrophage inhibitory cytokine-1 activates AKT and ERK-1/2 via the transactivation of ErbB2 in human breast and gastric cancer cells. Carcinogenesis 2008, 29, 704–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.; Wang, M.; Liu, X. GDF15 promotes osteosarcoma cell migration and invasion by regulating the TGF-β signaling pathway. Mol. Med. Rep. 2019, 20, 4262–4270. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Ma, J.; Li, Y.; Huang, J.; Zhang, S.; Yin, Z.; Ren, J.; Huang, K.; Wu, G.; Yang, K.; et al. CDP138 silencing inhibits TGF-β/Smad signaling to impair radioresistance and metastasis via GDF15 in lung cancer. Cell Death Dis. 2017, 8, e3036. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kang, S.-M.; Oh, S.Y.; Kang, S.H.; Lee, I.; Hwang, J.C.; Lee, H.-J.; Choi, S.-Y.; Hong, S.-H. Early Growth Response 1-Dependent Downregulation of Matrix Metalloproteinase 9 and Mouse Double Minute 2 Attenuates Head and Neck Squamous Cell Carcinoma Metastasis. Cell. Physiol. Biochem. 2018, 50, 1869–1881. [Google Scholar] [CrossRef]
- Yoon, T.; Kim, S.-A.; Lee, N.; Lee, J.; Park, Y.-L.; Lee, K.-H.; Chung, I.-J.; Joo, Y.-E.; Lim, S. EGR1 regulates radiation-induced apoptosis in head and neck squamous cell carcinoma. Oncol. Rep. 2015, 33. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.; Na, K.; Chen, D.; Wang, Y.; Pan, H.; Wang, X. Effects of non-steroidal anti-inflammatory drug-activated gene-1 on Ganoderma lucidum polysaccharides-induced apoptosis of human prostate cancer PC-3 cells. Int. J. Oncol. 2018, 53, 2356–2368. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.H.; Kim, J.; Khan, I.A.; Walker, L.A.; Khan, S.I. Nonsteroidal anti-inflammatory drug activated gene-1 (NAG-1) modulators from natural products as anti-cancer agents. Life Sci. 2014, 100, 75–84. [Google Scholar] [CrossRef]
- Yu, C.Y.; Su, K.-Y.; Lee, P.-L.; Jhan, J.-Y.; Tsao, P.-H.; Chan, D.-C.; Chen, Y.-L.S. Potential Therapeutic Role of Hispidulin in Gastric Cancer through Induction of Apoptosis via NAG-1 Signaling. Evid.-Based Complement. Altern. Med. 2013, 2013, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, S.; Kadowaki, M.; Yoshioka, H.; Kambe, A.; Watanabe, T.; Kinyamu, H.K.; Eling, T.E. Proteasome inhibitor MG132 induces NAG-1/GDF15 expression through the p38 MAPK pathway in glioblastoma cells. Biochem. Biophys. Res. Commun. 2013, 430, 1277–1282. [Google Scholar] [CrossRef] [Green Version]
- Gabriel, K.N.; Jones, A.C.; Nguyen, J.P.; Antillon, K.S.; Janos, S.N.; Overton, H.N.; Jenkins, S.M.; Frisch, E.H.; Trujillo, K.A.; Bisoffi, M. Association and regulation of protein factors of field effect in prostate tissues. Int. J. Oncol. 2016, 49, 1541–1552. [Google Scholar] [CrossRef] [Green Version]
- Stangelberger, A.; Schally, A.V.; Varga, J.L.; Zarandi, M.; Cai, R.-Z.; Baker, B.; Hammann, B.D.; Armatis, P.; Kanashiro, C.A. Inhibition of human androgen-independent PC-3 and DU-145 prostate cancers by antagonists of bombesin and growth hormone releasing hormone is linked to PKC, MAPK and c-jun intracellular signalling. Eur. J. Cancer 2005, 41, 2735–2744. [Google Scholar] [CrossRef]
- Brower, V. Capecitabine and lapatinib treatment in head and neck cancer. Lancet Oncol. 2016, 17, e271. [Google Scholar] [CrossRef]
- Wang, W.; Yang, X.; Dai, J.; Lu, Y.; Zhang, J.; Keller, E.T. Prostate cancer promotes a vicious cycle of bone metastasis progression through inducing osteocytes to secrete GDF15 that stimulates prostate cancer growth and invasion. Oncogene 2019, 38, 4540–4559. [Google Scholar] [CrossRef]
- Yuan, J.S.; Reed, A.; Chen, F.; Stewartjr, C.N. Statistical analysis of real-time PCR data. BMC Bioinform. 2006, 7, 85. [Google Scholar] [CrossRef] [Green Version]
- Jung, S.-N.; Shin, D.-S.; Kim, H.-N.; Jeon, Y.J.; Yun, J.; Lee, Y.-J.; Kang, J.S.; Han, D.C.; Kwon, B.-M. Sugiol inhibits STAT3 activity via regulation of transketolase and ROS-mediated ERK activation in DU145 prostate carcinoma cells. Biochem. Pharmacol. 2015, 97, 38–50. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, Y.; Jung, S.-N.; Lim, M.A.; Oh, C.; Piao, Y.; Kim, H.J.; Liu, L.; Kang, Y.E.; Chang, J.W.; Won, H.-R.; et al. Transcriptional Regulation of GDF15 by EGR1 Promotes Head and Neck Cancer Progression through a Positive Feedback Loop. Int. J. Mol. Sci. 2021, 22, 11151. https://doi.org/10.3390/ijms222011151
Jin Y, Jung S-N, Lim MA, Oh C, Piao Y, Kim HJ, Liu L, Kang YE, Chang JW, Won H-R, et al. Transcriptional Regulation of GDF15 by EGR1 Promotes Head and Neck Cancer Progression through a Positive Feedback Loop. International Journal of Molecular Sciences. 2021; 22(20):11151. https://doi.org/10.3390/ijms222011151
Chicago/Turabian StyleJin, Yanli, Seung-Nam Jung, Mi Ae Lim, Chan Oh, Yudan Piao, Hae Jong Kim, Lihua Liu, Yea Eun Kang, Jae Won Chang, Ho-Ryun Won, and et al. 2021. "Transcriptional Regulation of GDF15 by EGR1 Promotes Head and Neck Cancer Progression through a Positive Feedback Loop" International Journal of Molecular Sciences 22, no. 20: 11151. https://doi.org/10.3390/ijms222011151
APA StyleJin, Y., Jung, S.-N., Lim, M. A., Oh, C., Piao, Y., Kim, H. J., Liu, L., Kang, Y. E., Chang, J. W., Won, H.-R., Song, K., & Koo, B. S. (2021). Transcriptional Regulation of GDF15 by EGR1 Promotes Head and Neck Cancer Progression through a Positive Feedback Loop. International Journal of Molecular Sciences, 22(20), 11151. https://doi.org/10.3390/ijms222011151




