The Heparanase Regulatory Network in Health and Disease
Abstract
1. Introduction
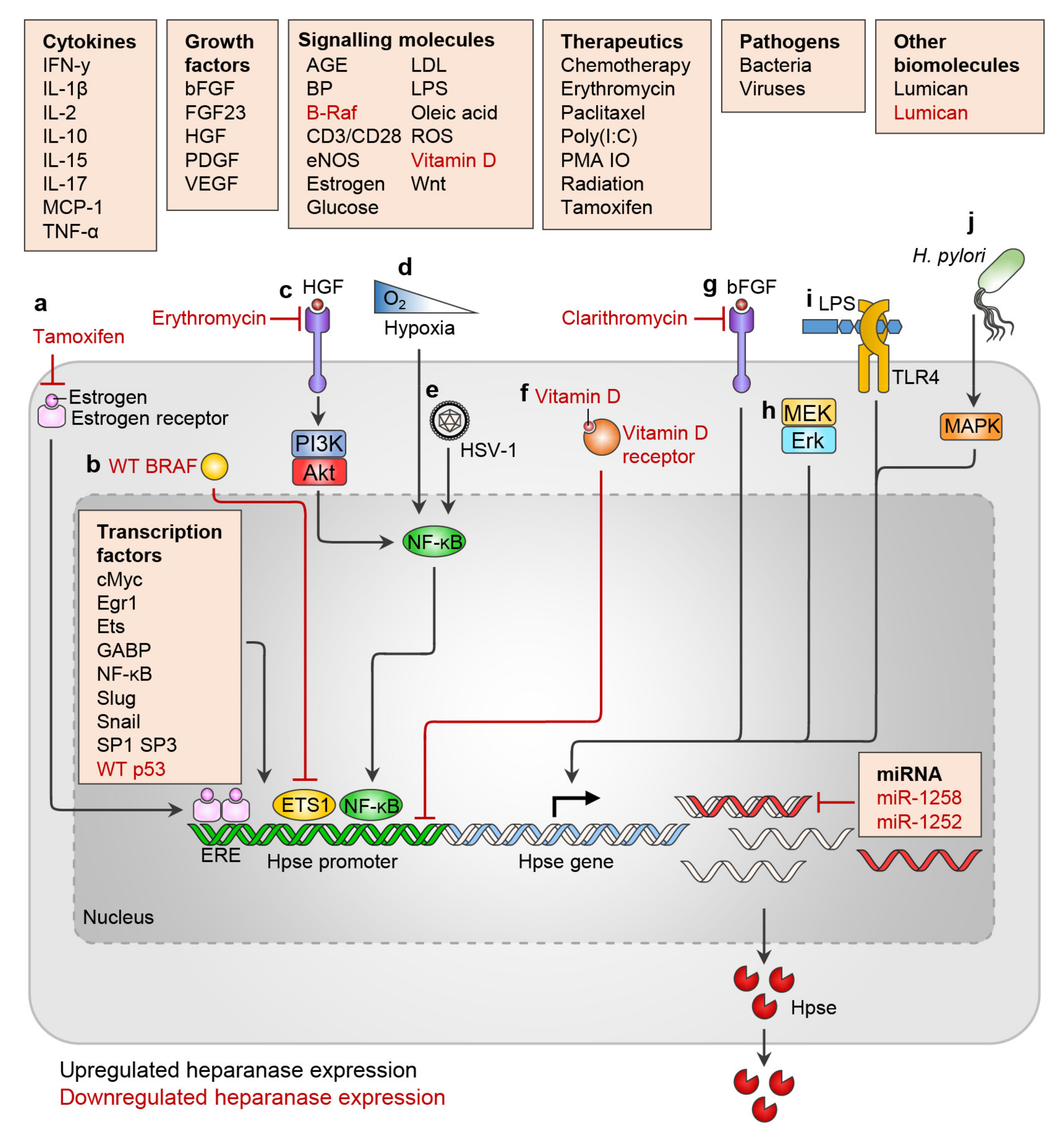
2. Regulation of Heparanase Expression
2.1. Heparanase Expression and Tissue Distribution
2.2. Transcription Factors
2.3. miRNA
2.4. Cytokines
2.5. Growth Factors
2.6. Hormones and Metabolites
2.7. Pathogens
2.8. Therapies
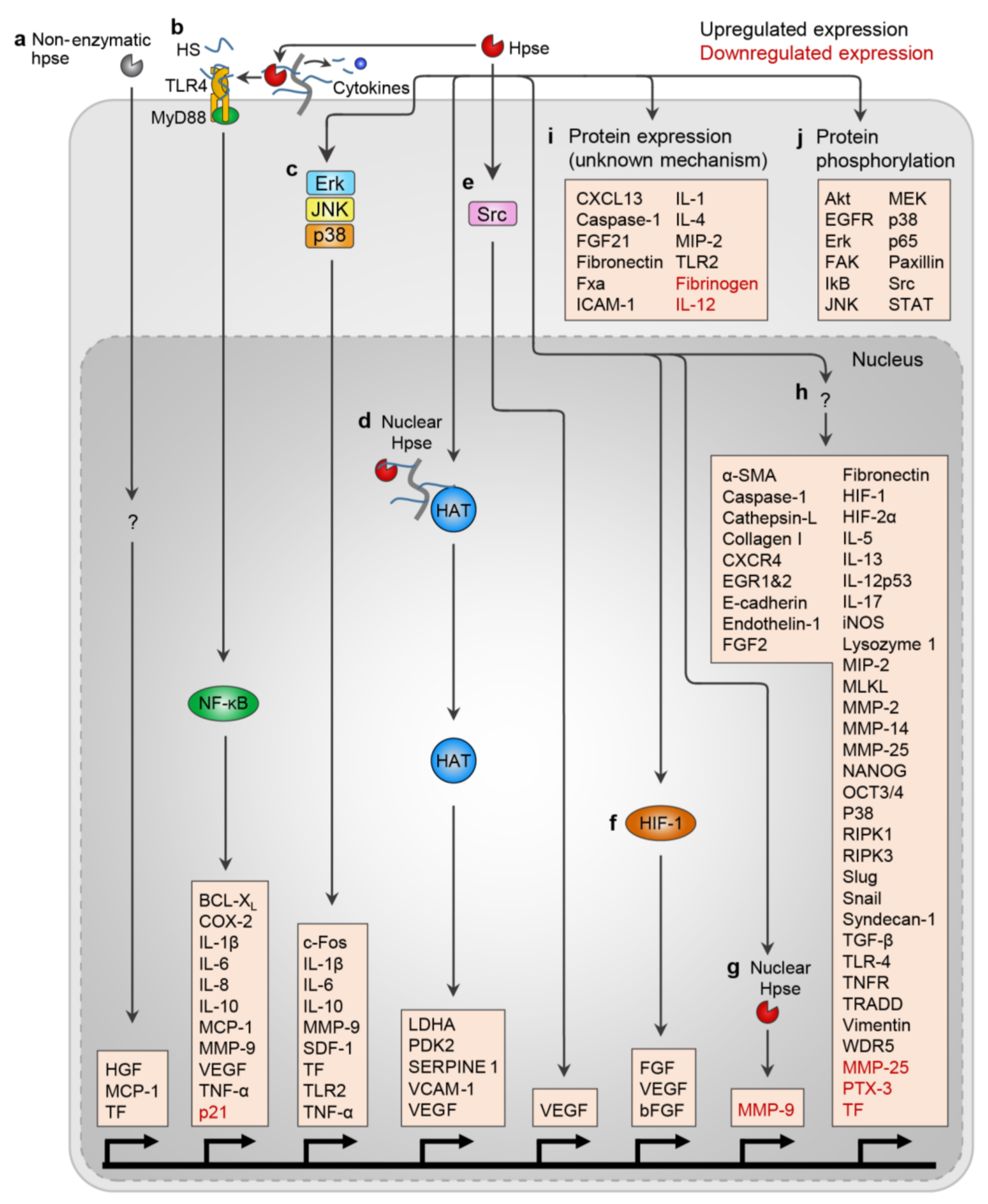
3. Regulation of Heparanase Enzymatic Activity: Proteolytic Activation and Natural Inhibitors
4. Heparanase in Regulating Gene Expression, Protein Expression, and Protein Phosphorylation
4.1. Nuclear Heparanase Regulates Gene Transcription
4.2. Heparanase Regulates Gene and Protein Expression and Protein Activation
4.3. Protein Phosphorylation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef]
- Sarrazin, S.; Lamanna, W.C.; Esko, J.D. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 2011, 3, a004952. [Google Scholar] [CrossRef] [PubMed]
- Bernfield, M.; Götte, M.; Park, P.W.; Reizes, O.; Fitzgerald, M.L.; Lincecum, J.; Zako, M. Functions of Cell Surface Heparan Sulfate Proteoglycans. Annu. Rev. Biochem. 1999, 68, 729–777. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.D.; Sanderson, R.D. Heparan sulfate in the nucleus and its control of cellular functions. Matrix Biol. 2014, 35, 56–59. [Google Scholar] [CrossRef] [PubMed]
- Ori, A.; Wilkinson, M.C.; Fernig, D.G. A systems biology approach for the investigation of the heparin/heparan sulfate interactome. J. Biol. Chem. 2011, 286, 19892–19904. [Google Scholar] [CrossRef] [PubMed]
- Kreuger, J.; Jemth, P.; Sanders-Lindberg, E.; Eliahu, L.; Ron, D.; Basilico, C.; Salmivirta, M.; Lindahl, U. Fibroblast growth factors share binding sites in heparan sulphate. Biochem. J. 2005, 389, 145–150. [Google Scholar] [CrossRef]
- Sekiguchi, K.; Hakomori, S.; Funahashi, M.; Matsumoto, I.; Seno, N. Binding of fibronectin and its proteolytic fragments to glycosaminoglycans. Exposure of cryptic glycosaminoglycan-binding domains upon limited proteolysis. J. Biol. Chem. 1983, 258, 14359–14365. [Google Scholar] [CrossRef]
- Ogamo, O.; Nagai, A.; Nagasawa, N. Binding of heparin fractions and other polysulfated polysaccharides to plasma fibronectin: Effects of molecular size and degree of sulfation of polysaccharides. BBA Gen. Subj. 1985, 841, 30–41. [Google Scholar] [CrossRef]
- Dyer, D.P.; Salanga, C.L.; Volkman, B.F.; Kawamura, T.; Handel, T.M. The dependence of chemokine-glycosaminoglycan interactions on chemokine oligomerization. Glycobiology 2015, 26, 312–326. [Google Scholar] [CrossRef]
- Wu, Z.L.; Zhang, L.; Yabe, T.; Kuberan, B.; Beeler, D.L.; Love, A.; Rosenberg, R.D. The involvement of heparan sulfate (HS) in FGF1/HS/FGFR1 signaling complex. J. Biol. Chem. 2003, 278, 17121–17129. [Google Scholar] [CrossRef]
- Jakobsson, L.; Kreuger, J.; Holmborn, K.; Lundin, L.; Eriksson, I.; Kjellén, L.; Claesson-Welsh, L. Heparan Sulfate in trans Potentiates VEGFR-Mediated Angiogenesis. Dev. Cell 2006, 10, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.V.; Katakam, S.K.; Urbanowitz, A.-K.; Gotte, M. Heparan sulphate as a regulator of leukocyte recruitment in inflammation. Curr. Protein Pept. Sci. 2015, 16, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Faye, C.; Chautard, E.; Olsen, B.R.; Ricard-Blum, S. The first draft of the endostatin interaction network. J. Biol. Chem. 2009, 284, 22041–22047. [Google Scholar] [CrossRef]
- Gralle, M.; Botelho, M.G.; Wouters, F.S. Neuroprotective secreted amyloid precursor protein acts by disrupting amyloid precursor protein dimers. J. Biol. Chem. 2009, 284, 15016–15025. [Google Scholar] [CrossRef] [PubMed]
- Ishitsuka, R.; Kojima, K.; Utsumi, H.; Ogawa, H.; Matsumoto, I. Glycosaminoglycan Binding Properties of Annexin IV, V, and VI. J. Biol. Chem. 1998, 273, 9935–9941. [Google Scholar] [CrossRef]
- Moscatelli, D. High and low affinity binding sites for basic fibroblast growth factor on cultured cells: Absence of a role for low affinity binding in the stimulation of plasminogen activator production by bovine capillary endothelial cells. J. Cell. Physiol. 1987, 131, 123–130. [Google Scholar] [CrossRef]
- Bashkin, P.; Doctrow, S.; Klagsbrun, M.; Svahn, C.M.; Folkman, J.; Vlodavsky, I. Basic Fibroblast Growth Factor Binds to Subendothelial Extracellular Matrix and Is Released by Heparitinase and Heparin-like Molecules. Biochemistry 1989, 28, 1737–1743. [Google Scholar] [CrossRef]
- Rapraeger, A.C.; Krufka, A.; Olwin, B.B. Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science 1991, 252, 1705–1708. [Google Scholar] [CrossRef]
- Andres, J.L.; DeFalcis, D.; Noda, M.; Massague, J. Binding of two growth factor families to separate domains of the proteoglycan betaglycan. J. Biol. Chem. 1992, 267, 5927–5930. [Google Scholar] [CrossRef]
- Stamatoglou, S.C.; Keller, J.M. Correlation between cell substrate attachment in vitro and cell surface heparan sulfate affinity for fibronectin and collagen. J. Cell Biol. 1983, 96, 1820–1823. [Google Scholar] [CrossRef]
- LeBaron, R.G.; Hook, A.; Esko, J.D.; Gay, S.; Hook, M. Binding of heparan sulfate to type V collagen. A mechanism of cell-substrate adhesion. J. Biol. Chem. 1989, 264, 7950–7956. [Google Scholar] [CrossRef]
- Ricard-Blum, S.; Beraud, M.; Raynal, N.; Farndale, R.W.; Ruggiero, F. Structural requirements for heparin/heparan sulfate binding to type V collagen. J. Biol. Chem. 2006, 281, 25195–25204. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, C.; Mayer, U.; Aumailley, M.; Timpl, R. Basement-membrane heparan sulfate proteoglycan binds to laminin by its heparan sulfate chains and to nidogen by sites in the protein core. Eur. J. Biochem. 1992, 208, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Pettersson, U.S.; Hoorelbeke, B.; Kolaczkowska, E.; Schelfhout, K.; Martens, E.; Kubes, P.; van Damme, J.; Phillipson, M.; Opdenakker, G. Interference with Glycosaminoglycan-Chemokine Interactions with a Probe to Alter Leukocyte Recruitment and Inflammation In Vivo. PLoS ONE 2014, 9, e104107. [Google Scholar] [CrossRef] [PubMed]
- Luster, A.D.; Greenberg, S.M.; Leder, P. The IP-10 chemokine binds to a specific cell surface heparan sulfate site shared with platelet factor 4 and inhibits endothelial cell proliferation. J. Exp. Med. 1995, 182, 219–231. [Google Scholar] [CrossRef]
- Monneau, Y.R.; Luo, L.; Sankaranarayanan, N.V.; Nagarajan, B.; Vivès, R.R.; Baleux, F.; Desai, U.R.; Arenzana-Seidedos, F.; Lortat-Jacob, H. Solution structure of CXCL13 and heparan sulfate binding show that GAG binding site and cellular signalling rely on distinct domains. Open Biol. 2017, 7, 170133. [Google Scholar] [CrossRef] [PubMed]
- Karumanchi, S.A.; Jha, V.; Ramchandran, R.; Karihaloo, A.; Tsiokas, L.; Chan, B.; Dhanabal, M.; Hanai, J.I.; Venkataraman, G.; Shriver, Z.; et al. Cell surface glypicans are low-affinity endostatin receptors. Mol. Cell 2001, 7, 811–822. [Google Scholar] [CrossRef]
- Blackhall, F.H.; Merry, C.L.R.; Lyon, M.; Jayson, G.C.; Folkman, J.; Javaherian, K.; Gallagher, J.T. Binding of endostatin to endothelial heparan sulphate shows a differential requirement for specific sulphates. Biochem. J. 2003, 375, 131–139. [Google Scholar] [CrossRef]
- Loo, B.-M.; Kreuger, J.; Jalkanen, M.; Lindahl, U.; Salmivirta, M. Binding of Heparin/Heparan Sulfate to Fibroblast Growth Factor Receptor 4. J. Biol. Chem. 2001, 276, 16868–16876. [Google Scholar] [CrossRef]
- Lyon, M.; Deakin, J.A.; Mizuno, K.; Nakamura, T.; Gallagher, J.T. Interaction of hepatocyte growth factor with heparan sulfate. Elucidation of the major heparan sulfate structural determinants. J. Biol. Chem. 1994, 269, 11216–11223. [Google Scholar] [CrossRef]
- Jones, A.L.; Hulett, M.D.; Parish, C.R. Histidine-rich glycoprotein binds to cell-surface heparan sulfate via its N-terminal domain following Zn2+ chelation. J. Biol. Chem. 2004, 279, 30114–30122. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Young, J.; Song, D.; Esko, J.D. Heparan sulfate is essential for high mobility group protein 1 (HMGB1) signaling by the receptor for advanced glycation end products (RAGE). J. Biol. Chem. 2011, 286, 41736–41744. [Google Scholar] [CrossRef] [PubMed]
- Gordts, P.L.S.M.; Foley, E.M.; Lawrence, R.; Sinha, R.; Lameda-Diaz, C.; Deng, L.; Nock, R.; Glass, C.K.; Erbilgin, A.; Lusis, A.J.; et al. Reducing Macrophage Proteoglycan Sulfation Increases Atherosclerosis and Obesity through Enhanced Type I Interferon Signaling. Cell Metab. 2014, 20, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Witt, D.P.; Lander, A.D. Differential binding of chemokines to glycosaminoglycan subpopulations. Curr. Biol. 1994, 4, 394–400. [Google Scholar] [CrossRef]
- Norgard-Sumnicht, K.E.; Varki, N.M.; Varki, A. Calcium-dependent heparin-like ligands for L-selectin in nonlymphoid endothelial cells. Science 1993, 261, 480–483. [Google Scholar] [CrossRef]
- Koenig, A.; Norgard-Sumnicht, K.; Linhardt, R.; Varki, A. Differential interactions of heparin and heparan sulfate glycosaminoglycans with the selectins: Implications for the use of unfractionated and low molecular weight heparins as therapeutic agents. J. Clin. Investig. 1998, 101, 877–889. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, L.; Rogers, L.; Quach, T.; Breckenridge, S.; Kolattukudy, P.E. Lysine 58 and histidine 66 at the C-terminal α-helix of monocyte chemoattractant protein-1 are essential for glycosaminoglycan binding. J. Biol. Chem. 1998, 273, 29641–29647. [Google Scholar] [CrossRef]
- Pasqualon, T.; Lue, H.; Groening, S.; Pruessmeyer, J.; Jahr, H.; Denecke, B.; Bernhagen, J.; Ludwig, A. Cell surface syndecan-contributes to binding and function of macrophage migration inhibitory factor (MIF) on epithelial tumor cells. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 717–726. [Google Scholar] [CrossRef]
- Stringer, S.E.; Forster, M.J.; Mulloy, B.; Bishop, C.R.; Graham, G.J.; John, T.; Gallagher, J.T. Characterization of the binding site on heparan sulfate for macrophage inflammatory protein 1α. Blood 2002, 100, 1543–1550. [Google Scholar] [CrossRef]
- Zilka, A.; Landau, G.; Hershkovitz, O.; Bloushtain, N.; Bar-Ilan, A.; Benchetrit, F.; Fima, E.; van Kuppevelt, T.H.; Gallagher, J.T.; Elgavish, S.; et al. Characterization of the heparin/heparan sulfate binding site of the natural cytotoxicity receptor NKp46. Biochemistry 2005, 44, 14477–14485. [Google Scholar] [CrossRef]
- Abramsson, A.; Kurup, S.; Busse, M.; Yamada, S.; Lindblom, P.; Schallmeiner, E.; Stenzel, D.; Sauvaget, D.; Ledin, J.; Ringvall, M.; et al. Defective N-sulfation of heparan sulfate proteoglycans limits PDGF-BB binding and pericyte recruitment in vascular development. Genes Dev. 2007, 21, 316–331. [Google Scholar] [CrossRef] [PubMed]
- Lustig, F.; Hoebeke, J.; Östergren-Lundèn, G.; Velge-Roussel, F.; Bondjers, G.; Olsson, U.; Rüetschi, U.; Fager, G. Alternative splicing determines the binding of platelet-derived growth factor (PDGF-AA) to glycosaminoglycans. Biochemistry 1996, 35, 12077–12085. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Young, J.H.; Krahn, J.M.; Song, D.; Corbett, K.D.; Chazin, W.J.; Pedersen, L.C.; Esko, J.D. Stable RAGE-Heparan Sulfate Complexes Are Essential for Signal Transduction. ACS Chem. Biol. 2013, 8, 1611–1620. [Google Scholar] [CrossRef] [PubMed]
- Øynebråten, I.; Barois, N.; Bergeland, T.; Küchler, A.M.; Bakke, O.; Haraldsen, G. Oligomerized, filamentous surface presentation of RANTES/CCL5 on vascular endothelial cells. Sci. Rep. 2015, 5, 9261. [Google Scholar] [CrossRef]
- Aricescu, A.R.; McKinnell, I.W.; Halfter, W.; Stoker, A.W. Heparan Sulfate Proteoglycans Are Ligands for Receptor Protein Tyrosine Phosphatase. Mol. Cell. Biol. 2002, 22, 1881–1892. [Google Scholar] [CrossRef]
- Netelenbos, T.; van den Born, J.; Kessler, F.L.; Zweegman, S.; Merle, P.A.; van Oostveen, J.W.; Zwaginga, J.J.; Huijgens, P.C.; Dräger, A.M. Proteoglycans on bone marrow endothelial cells bind and present SDF-1 towards hematopoietic progenitor cells. Leukemia 2003, 17, 175–184. [Google Scholar] [CrossRef]
- Houck, K.A.; Leung, D.W.; Rowland, A.M.; Winer, J.; Ferrara, N. Dual regulation of vascular endothelial growth factor bioavailability by genetic and proteolytic mechanisms. J. Biol. Chem. 1992, 267, 26031–26037. [Google Scholar] [CrossRef]
- Wilson, J.C.; Laloo, A.E.; Singh, S.; Ferro, V. 1H NMR spectroscopic studies establish that heparanase is a retaining glycosidase. Biochem. Biophys. Res. Commun. 2014, 443, 185–188. [Google Scholar] [CrossRef]
- Zcharia, E.; Zilka, R.; Yaar, A.; Yacoby-Zeevi, O.; Zetser, A.; Metzger, S.; Sarid, R.; Naggi, A.; Casu, B.; Ilan, N.; et al. Heparanase accelerates wound angiogenesis and wound healing in mouse and rat models. FASEB J. 2005, 19, 211–221. [Google Scholar] [CrossRef]
- Putz, E.M.; Mayfosh, A.J.; Kos, K.; Barkauskas, D.S.; Nakamura, K.; Town, L.; Goodall, K.J.; Yee, D.Y.; Poon, I.K.H.; Baschuk, N.; et al. NK cell heparanase controls tumor invasion and immune surveillance. J. Clin. Investig. 2017, 127, 2777–2788. [Google Scholar] [CrossRef]
- Poon, I.K.H.; Goodall, K.J.; Phipps, S.; Chow, J.D.Y.; Pagler, E.B.; Andrews, D.M.; Conlan, C.L.; Ryan, G.F.; White, J.A.; Wong, M.K.L.; et al. Mice deficient in heparanase exhibit impaired dendritic cell migration and reduced airway inflammation. Eur. J. Immunol. 2014, 44, 1016–1030. [Google Scholar] [CrossRef] [PubMed]
- Lider, O.; Mekori, Y.A.; Miller, T.; Bar-Tana, R.; Vlodavsky, I.; Baharav, E.; Cohen, I.R.; Naparstek, Y. Inhibition of T lymphocyte heparanase by heparin prevents T cell migration and T cell-mediated immunity. Eur. J. Immunol. 1990, 20, 493–499. [Google Scholar] [CrossRef]
- Sasaki, N.; Higashi, N.; Taka, T.; Nakajima, M.; Irimura, T. Cell Surface Localization of Heparanase on Macrophages Regulates Degradation of Extracellular Matrix Heparan Sulfate. J. Immunol. 2004, 172, 3830–3835. [Google Scholar] [CrossRef]
- Shteper, P.J.; Zcharia, E.; Ashhab, Y.; Peretz, T.; Vlodavsky, I.; Ben-Yehuda, D. Role of promoter methylation in regulation of the mammalian heparanase gene. Oncogene 2003, 22, 7737–7749. [Google Scholar] [CrossRef]
- Jiao, F.; Bai, S.; Ma, Y.; Yan, Z.; Yue, Z.; Yu, Y.; Wang, X.; Wang, J. DNA Methylation of Heparanase Promoter Influences Its Expression and Associated with the Progression of Human Breast Cancer. PLoS ONE 2014, 9, e92190. [Google Scholar] [CrossRef]
- Ostrovsky, O.; Korostishevsky, M.; Shafat, I.; Mayorov, M.; Ilan, N.; Vlodavsky, I.; Nagler, A. Inverse correlation between HPSE gene single nucleotide polymorphisms and heparanase expression: Possibility of multiple levels of heparanase regulation. J. Leukoc. Biol. 2009, 86, 445–455. [Google Scholar] [CrossRef]
- Ostrovsky, O.; Korostishevsky, M.; Levite, I.; Leiba, M.; Galski, H.; Vlodavsky, I.; Nagler, A. Association of heparanase gene (HPSE) single nucleotide polymorphisms with hematological malignancies. Leukemia 2007, 21, 2296–2303. [Google Scholar] [CrossRef] [PubMed]
- Hulett, M.D.; Freeman, C.; Hamdorf, B.J.; Baker, R.T.; Harris, M.J.; Parish, C.R. Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis. Nat. Med. 1999, 5, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Rabelink, T.J.; van den Berg, B.M.; Garsen, M.; Wang, G.; Elkin, M.; van der Vlag, J. Heparanase: Roles in cell survival, extracellular matrix remodelling and the development of kidney disease. Nat. Rev. Nephrol. 2017, 13, 201–212. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- De Mestre, A.M.; Soe-Htwe, T.; Sutcliffe, E.L.; Rao, S.; Pagler, E.B.; Hornby, J.R.; Hulett, M.D. Regulation of mouse Heparanase gene expression in T lymphocytes and tumor cells. Immunol. Cell Biol. 2007, 85, 205–214. [Google Scholar] [CrossRef] [PubMed]
- De Mestre, A.M.; Staykova, M.A.; Hornby, J.R.; Willenborg, D.O.; Hulett, M.D. Expression of the heparan sulfate-degrading enzyme heparanase is induced in infiltrating CD4 + T cells in experimental autoimmune encephalomyelitis and regulated at the level of transcription by early growth response gene. J. Leukoc. Biol. 2007, 82, 1289–1300. [Google Scholar] [CrossRef]
- Ostrovsky, O.; Shimoni, A.; Baryakh, P.; Morgulis, Y.; Mayorov, M.; Beider, K.; Shteingauz, A.; Ilan, N.; Vlodavsky, I.; Nagler, A. Modification of heparanase gene expression in response to conditioning and LPS treatment: Strong correlation to rs4693608 SNP. J. Leukoc. Biol. 2014, 95, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Jayatilleke, K.M.; Hulett, M.D. Heparanase and the hallmarks of cancer. J. Transl. Med. 2020, 18, 1–25. [Google Scholar] [CrossRef]
- Tang, B.; Xie, R.; Qin, Y.; Xiao, Y.; Yong, X.; Zheng, L.; Dong, H.; Yang, S.-M. Human telomerase reverse transcriptase (hTERT) promotes gastric cancer invasion through cooperating with c-Myc to upregulate heparanase expression. Oncotarget 2015, 7, 11364–11379. [Google Scholar] [CrossRef] [PubMed]
- De Mestre, A.M.; Khachigian, L.M.; Santiago, F.S.; Staykova, M.A.; Hulett, M.D. Regulation of Inducible Heparanase Gene Transcription in Activated T Cells by Early Growth Response 1. J. Biol. Chem. 2003, 278, 50377–50385. [Google Scholar] [CrossRef]
- De Mestre, A.M.; Rao, S.; Hornby, J.R.; Soe-Htwe, T.; Khachigian, L.M.; Hulett, M.D. Early growth response gene 1 (EGR1) regulates heparanase gene transcription in tumor cells. J. Biol. Chem. 2005, 280, 35136–35147. [Google Scholar] [CrossRef]
- Gil, N.; Goldberg, R.; Neuman, T.; Garsen, M.; Zcharia, E.; Rubinstein, A.M.; van Kuppevelt, T.; Meirovitz, A.; Pisano, C.; Li, J.P.; et al. Heparanase is essential for the development of diabetic nephropathy in mice. Diabetes 2012, 61, 208–216. [Google Scholar] [CrossRef]
- Lu, W.C.; Liu, Y.N.; Kang, B.B.; Chen, J.H. Trans-activation of heparanase promoter by ETS transcription factors. Oncogene 2003, 22, 919–923. [Google Scholar] [CrossRef]
- Jiang, P.; Kumar, A.; Parrillo, J.E.; Dempsey, L.A.; Platt, J.L.; Prinz, R.A.; Xu, X. Cloning and characterization of the human heparanase-1 (HPR1) gene promoter. Role of GA-binding protein and Sp1 in regulating HPR1 basal promoter activity. J. Biol. Chem. 2002, 277, 8989–8998. [Google Scholar] [CrossRef] [PubMed]
- Rao, G.; Liu, D.; Xing, M.; Tauler, J.; Prinz, R.A.; Xu, X. Induction of Heparanase-1 Expression by Mutant B-Raf Kinase: Role of GA Binding Protein in Heparanase-1 Promoter Activation. Neoplasia 2010, 12, 946–956. [Google Scholar] [CrossRef]
- Andela, V.B.; Schwarz, E.M.; Puzas, J.E.; O’Keefe, R.J.; Rosier, R.N. Tumor metastasis and the reciprocal regulation of prometastatic and antimetastatic factors by nuclear factor κB. Cancer Res. 2000, 60, 6557–6562. [Google Scholar] [PubMed]
- Rops, A.L.; van den Hoven, M.J.; Baselmans, M.M.; Lensen, J.F.; Wijnhoven, T.J.; van den Heuvel, L.P.; van Kuppevelt, T.H.; Berden, J.H.; van der Vlag, J. Heparan sulfate domains on cultured activated glomerular endothelial cells mediate leukocyte trafficking. Kidney Int. 2008, 73, 52–62. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, W.; Pan, C.; Meng, K.; Zhao, L.; Du, L.; Liu, Q.; Lin, R. Hypoxia activates heparanase expression in an NF-kappaB dependent manner. Oncol. Rep. 2010, 23, 255–261. [Google Scholar] [CrossRef]
- Hadigal, S.R.; Agelidis, A.M.; Karasneh, G.A.; Antoine, T.E.; Yakoub, A.M.; Ramani, V.C.; Djalilian, A.R.; Sanderson, R.D.; Shukla, D. Heparanase is a host enzyme required for herpes simplex virus-1 release from cells. Nat. Commun. 2015, 6, 6985. [Google Scholar] [CrossRef]
- Ramani, V.C.; Vlodavsky, I.; Ng, M.; Zhang, Y.; Barbieri, P.; Noseda, A.; Sanderson, R.D. Chemotherapy induces expression and release of heparanase leading to changes associated with an aggressive tumor phenotype. Matrix Biol. 2016, 55, 22–34. [Google Scholar] [CrossRef]
- Baraz, L.; Haupt, Y.; Elkin, M.; Peretz, T.; Vlodavsky, I. Tumor suppressor p53 regulates heparanase gene expression. Oncogene 2006, 25, 3939–3947. [Google Scholar] [CrossRef]
- Karamanou, K.; Franchi, M.; Proult, I.; Rivet, R.; Vynios, D.; Brézillon, S. Lumican Inhibits In Vivo Melanoma Metastasis by Altering Matrix-Effectors and Invadopodia Markers. Cells 2021, 10, 841. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sullivan, P.S.; Goodman, J.C.; Gunaratne, P.H.; Marchetti, D. MicroRNA-1258 suppresses breast cancer brain metastasis by targeting heparanase. Cancer Res. 2011, 71, 645–654. [Google Scholar] [CrossRef]
- Rodrigues, D.M., Jr.; Pelarin, M.F.d.A.; Nader, H.B.; Vettore, A.L.; Pinhal, M.A.S. MicroRNA-1252-5p associated with extracellular vesicles enhances bortezomib sensitivity in multiple myeloma cells by targeting heparanase. OncoTargets Ther. 2021, 14, 455–467. [Google Scholar] [CrossRef]
- Edovitsky, E.; Lerner, I.; Zcharia, E.; Peretz, T.; Vlodavsky, I.; Elkin, M. Role of endothelial heparanase in delayed-type hypersensitivity. Blood 2006, 107, 3609–3616. [Google Scholar] [CrossRef]
- Chen, G.; Wang, D.; Vikramadithyan, R.; Yagyu, H.; Saxena, U.; Pillarisetti, S.; Goldberg, I.J. Inflammatory Cytokines and Fatty Acids Regulate Endothelial Cell Heparanase Expression. Biochemistry 2004, 43, 4971–4977. [Google Scholar] [CrossRef]
- El-Nadi, M.; Hassan, H.; Saleh, M.E.; Nassar, E.; Ismail, Y.M.; Amer, M.; Greve, B.; Götte, M.; El-Shinawi, M.; Ibrahim, S.A. Induction of heparanase via IL-10 correlates with a high infiltration of CD163+ M2-type tumor-associated macrophages in inflammatory breast carcinomas. Matrix Biol. Plus 2020, 6–7, 6–7. [Google Scholar] [CrossRef]
- Lv, Q.; Wu, K.; Liu, F.; Wu, W.; Chen, Y.; Zhang, W. Interleukin-17A and heparanase promote angiogenesis and cell proliferation and invasion in cervical cancer. Int. J. Oncol. 2018, 53, 1809–1817. [Google Scholar] [CrossRef]
- Boels, M.G.S.; Koudijs, A.; Avramut, M.C.; Sol, W.M.P.J.; Wang, G.; van Oeveren-Rietdijk, A.M.; van Zonneveld, A.J.; de Boer, H.C.; van der Vlag, J.; van Kooten, C.; et al. Systemic Monocyte Chemotactic Protein-1 Inhibition Modifies Renal Macrophages and Restores Glomerular Endothelial Glycocalyx and Barrier Function in Diabetic Nephropathy. Am. J. Pathol. 2017, 187, 2430–2440. [Google Scholar] [CrossRef]
- Tang, X.-D.; Liang, G.-P.; Li, C.; Wan, Y.; Chen, T.; Chen, L.; Yu, S.-T.; Xiong, Z.; Fang, D.-C.; Wang, G.-Z.; et al. Cytotoxic T lymphocyte epitopes from human heparanase can elicit a potent anti-tumor immune response in mice. Cancer Immunol. Immunother. 2010, 59, 1041–1047. [Google Scholar] [CrossRef]
- Schmidt, E.P.; Yang, Y.; Janssen, W.J.; Gandjeva, A.; Perez, M.J.; Barthel, L.; Zemans, R.L.; Bowman, J.C.; Koyanagi, D.E.; Yunt, Z.X.; et al. The pulmonary endothelial glycocalyx regulates neutrophil adhesion and lung injury during experimental sepsis. Nat. Med. 2012, 18, 1217–1223. [Google Scholar] [CrossRef]
- Secchi, M.F.; Crescenzi, M.; Masola, V.; Russo, F.P.; Floreani, A.; Onisto, M. Heparanase and macrophage interplay in the onset of liver fibrosis. Sci. Rep. 2017, 7, 14956. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Ito, T.; Kashima, M.; Fukui, S.; Izumiyama, N.; Watanabe, A.; Sano, M.; Fujiwara, Y.; Miura, M. Erythromycin and clarithromycin modulation of growth factor-induced expression of heparanase mRNA on human lung cancer cells in vitro. Mediat. Inflamm. 2001, 10, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Hao, N.B.; Tang, B.; Wang, G.Z.; Xie, R.; Hu, C.J.; Wang, S.M.; Wu, Y.Y.; Liu, E.; Xie, X.; Yang, S.M. Hepatocyte growth factor (HGF) upregulates heparanase expression via the PI3K/Akt/NF-κB signaling pathway for gastric cancer metastasis. Cancer Lett. 2015, 361, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Suvannasankha, A.; Tompkins, D.R.; Edwards, D.F.; Petyaykina, K.V.; Crean, C.D.; Fournier, P.G.; Parker, J.M.; Sandusky, G.E.; Ichikawa, S.; Imel, E.A.; et al. FGF23 is elevated in multiple myeloma and increases heparanase expression by tumor cells. Oncotarget 2015, 6, 19647–19660. [Google Scholar] [CrossRef]
- Luan, Q.; Sun, J.; Li, C.; Zhang, G.; Lv, Y.; Wang, G.; Li, C.; Ma, C.; Gao, T. Mutual enhancement between heparanase and vascular endothelial growth factor: A novel mechanism for melanoma progression. Cancer Lett. 2011, 308, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Stoler-Barak, L.; Petrovich, E.; Aychek, T.; Gurevich, I.; Tal, O.; Hatzav, M.; Ilan, N.; Feigelson, S.W.; Shakhar, G.; Vlodavsky, I.; et al. Heparanase of murine effector lymphocytes and neutrophils is not required for their diapedesis into sites of inflammation. FASEB J. 2015, 29, 2010–2021. [Google Scholar] [CrossRef]
- Melo, C.M.; Nader, H.B.; Justo, G.Z.; Pinhal, M.A.S. Heparanase modulation by Wingless/INT (Wnt). Mol. Biol. Rep. 2021, 48. [Google Scholar] [CrossRef]
- An, X.F.; Zhou, L.; Jiang, P.J.; Yan, M.; Huang, Y.J.; Zhang, S.N.; Niu, Y.F.; Ten, S.C.; Yu, J.Y. Advanced glycation end-products induce heparanase expression in endothelial cells by the receptor for advanced glycation end products and through activation of the FOXO4 transcription factor. Mol. Cell. Biochem. 2011, 354, 47–55. [Google Scholar] [CrossRef]
- Naparstek, Y.; Cohen, I.R.; Fuks, Z.; Vlodavsky, I. Activated T lymphocytes produce a matrix-degrading heparan sulphate endoglycosidase. Nature 1984, 310, 241–244. [Google Scholar] [CrossRef]
- Fridman, R.; Lider, O.; Naparstek, Y.; Fuks, Z.; Vlodavsky, I.; Cohen, I.R. Soluble antigen induces T lymphocytes to secrete an endoglycosidase that degrades the heparan sulfate moiety of subendothelial extracellular matrix. J. Cell. Physiol. 1987, 130, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Eldor, A.; Haimovitz-Friedman, A.; Matzner, Y.; Lider, O.; Naparstek, Y.; Cohen, I.R.; Fuks, Z.; Ishai-Michaeli, R.; Lider, O.; et al. Expression of heparanase by platelets and circulating cells of the immune system: Possible involvement in diapedesis and extravasation. Invasion Metastasis 1992, 12, 112–127. [Google Scholar] [PubMed]
- Garsen, M.; Rops, A.L.; Li, J.; van Beneden, K.; van den Branden, C.; Berden, J.H.M.; Rabelink, T.J.; van der Vlag, J. Endothelial Nitric Oxide Synthase Prevents Heparanase Induction and the Development of Proteinuria. PLoS ONE 2016, 11, e0160894. [Google Scholar] [CrossRef] [PubMed]
- Elkin, M.; Cohen, I.; Zcharia, E.; Orgel, A.; Guatta-Rangini, Z.; Peretz, T.; Vlodavsky, I.; Kleinman, H.K. Regulation of Heparanase Gene Expression by Estrogen in Breast Cancer. Cancer Res. 2003, 63, 8821–8826. [Google Scholar] [PubMed]
- Singsuksawat, E.; Thuwajit, C.; Charngkaew, K.; Thuwajit, P. Increased ETV4 expression correlates with estrogen-enhanced proliferation and invasiveness of cholangiocarcinoma cells. Cancer Cell Int. 2018, 18, 25. [Google Scholar] [CrossRef] [PubMed]
- Cohen, I.; Maly, B.; Simon, I.; Meirovitz, A.; Pikarsky, E.; Zcharia, E.; Peretz, T.; Vlodavsky, I.; Elkin, M. Tamoxifen induces heparanase expression in estrogen receptor—Positive breast cancer. Clin. Cancer Res. 2007, 13, 4069–4077. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hermano, E.; Goldberg, R.; Rubinstein, A.M.; Sonnenblick, A.; Maly, B.; Nahmias, D.; Li, J.P.; Bakker, M.A.H.; van der Vlag, J.; Vlodavsky, I.; et al. Heparanase accelerates obesity-associated breast cancer progression. Cancer Res. 2019, 79, 5342–5354. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, Y.; Kim, M.S.; Puthanveetil, P.; Ghosh, S.; Luciani, D.S.; Johnson, J.D.; Abrahani, A.; Rodrigues, B. Glucose-induced endothelial heparanase secretion requires cortical and stress actin reorganization. Cardiovasc. Res. 2010, 87, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Luo, Y.; Zhu, X.; Li, T.; Hu, J.; Tang, S. Retinal heparanase expression in streptozotocin-induced diabetic rats. Can. J. Ophthalmol. 2010, 45, 46–51. [Google Scholar] [CrossRef]
- Rao, G.; Ding, H.G.; Huang, W.; Le, D.; Maxhimer, J.B.; Oosterhof, A.; van Kuppevelt, T.; Lum, H.; Lewis, E.J.; Reddy, V.; et al. Reactive oxygen species mediate high glucose-induced heparanase-1 production and heparan sulphate proteoglycan degradation in human and rat endothelial cells: A potential role in the pathogenesis of atherosclerosis. Diabetologia 2011, 54, 1527–1538. [Google Scholar] [CrossRef][Green Version]
- Laskov, R.; Michaeli, R.-I.; Sharir, H.; Yefenof, E.; Vlodavsky, I. Production of heparanase by normal and neoplastic murine B-lymphocytes. Int. J. Cancer 1991, 47, 92–98. [Google Scholar] [CrossRef]
- Zaza, G.; Masola, V.; Granata, S.; Pontrelli, P.; Sallustio, F.; Gesualdo, L.; Gambaro, G.; Grandaliano, G.; Lupo, A. Dialysis-related transcriptomic profiling: The pivotal role of heparanase. Exp. Biol. Med. 2014, 239, 52–64. [Google Scholar] [CrossRef]
- Kramer, A.; van den Hoven, M.; Rops, A.; Wijnhoven, T.; van den Heuvel, L.; Lensen, J.; van Kuppevelt, T.; van Goor, H.; van der Vlag, J.; Navis, G.; et al. Induction of Glomerular Heparanase Expression in Rats with Adriamycin Nephropathy Is Regulated by Reactive Oxygen Species and the Renin-Angiotensin System. J. Am. Soc. Nephrol. 2006, 17, 2513–2520. [Google Scholar] [CrossRef]
- Van den Hoven, M.J.; Waanders, F.; Rops, A.L.; Kramer, A.B.; van Goor, H.; Berden, J.H.; Navis, G.; van der Vlag, J. Regulation of glomerular heparanase expression by aldosterone, angiotensin II and reactive oxygen species. Nephrol. Dial. Transplant. 2009, 24, 2637–2645. [Google Scholar] [CrossRef]
- Garsen, M.; Sonneveld, R.; Rops, A.L.; Huntink, S.; van Kuppevelt, T.H.; Rabelink, T.J.; Hoenderop, J.G.; Berden, J.H.; Nijenhuis, T.; van der Vlag, J. Vitamin D attenuates proteinuria by inhibition of heparanase expression in the podocyte. J. Pathol. 2015, 237, 472–481. [Google Scholar] [CrossRef]
- Gallimidi, A.B.; Fischman, S.; Revach, B.; Bulvik, R.; Maliutina, A.; Rubinstein, A.M.; Nussbaum, G.; Elkin, M. Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget 2015, 6, 22613–22623. [Google Scholar] [CrossRef]
- Liu, L.-P.; Sheng, X.-P.; Shuai, T.-K.; Zhao, Y.-X.; Li, B.; Li, Y.-M. Helicobacter pylori promotes invasion and metastasis of gastric cancer by enhancing heparanase expression. World J. Gastroenterol. 2018, 24, 4565–4577. [Google Scholar] [CrossRef]
- Berk, R.S.; Dong, Z.; Alousi, S.; Kosir, M.A.; Wang, Y.; Vlodavsky, I. Murine ocular heparanase expression before and during infection with Pseudomonas aeruginosa. Investig. Ophthalmol. Vis. Sci. 2004, 45, 1182–1187. [Google Scholar] [CrossRef] [PubMed]
- Lapidot, M.; Barash, U.; Zohar, Y.; Geffen, Y.; Naroditsky, I.; Ilan, N.; Best, L.A.; Vlodavsky, I. Involvement of Heparanase in Empyema: Implication for Novel Therapeutic Approaches. J. Clin. Cell. Immunol. 2015, 6, 290. [Google Scholar] [CrossRef]
- Buijsers, B.; Yanginlar, C.; de Nooijer, A.; Grondman, I.; Maciej-Hulme, M.L.; Jonkman, I.; Janssen, N.A.F.; Rother, N.; de Graaf, M.; Pickkers, P.; et al. Increased Plasma Heparanase Activity in COVID-19 Patients. Front. Immunol. 2020, 11, 2572. [Google Scholar] [CrossRef]
- Stahl, K.; Gronski, P.A.; Kiyan, Y.; Seeliger, B.; Bertram, A.; Pape, T.; Welte, T.; Hoeper, M.M.; Haller, H.; David, S. Injury to the Endothelial Glycocalyx in Critically Ill Patients with COVID-19. Am. J. Respir. Crit. Care Med. 2020, 202, 1178–1181. [Google Scholar] [CrossRef] [PubMed]
- Puerta-Guardo, H.; Glasner, D.R.; Harris, E. Dengue Virus NS1 Disrupts the Endothelial Glycocalyx, Leading to Hyperpermeability. PLoS Pathog. 2016, 12, e1005738. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.R.; Chao, C.H.; Liu, C.C.; Ho, T.S.; Tsai, H.P.; Perng, G.C.; Lin, Y.S.; Wang, J.R.; Yeh, T.M. Macrophage migration inhibitory factor is critical for dengue NS1-induced endothelial glycocalyx degradation and hyperpermeability. PLoS Pathog. 2018, 14, e1007033. [Google Scholar] [CrossRef]
- Agelidis, A.M.; Hadigal, S.R.; Jaishankar, D.; Shukla, D. Viral Activation of Heparanase Drives Pathogenesis of Herpes Simplex Virus-1. Cell Rep. 2017, 20, 439–450. [Google Scholar] [CrossRef]
- Tao, C.; Wang, W.; Zhou, P.; Zhou, X.; Zhang, Q.; Liu, B. Molecular characterization, expression profiles of the porcine SDC2 and HSPG2 genes and their association with hematologic parameters. Mol. Biol. Rep. 2013, 40, 2549–2556. [Google Scholar] [CrossRef]
- Guo, C.; Zhu, Z.; Guo, Y.; Wang, X.; Yu, P.; Xiao, S.; Chen, Y.; Cao, Y.; Liu, X. Heparanase Upregulation Contributes to Porcine Reproductive and Respiratory Syndrome Virus Release. J. Virol. 2017, 91, e00625-17. [Google Scholar] [CrossRef]
- Bhattacharya, U.; Gutter-Kapon, L.; Kan, T.; Boyango, I.; Barash, U.; Yang, S.M.; Liu, J.J.; Gross-Cohen, M.; Sanderson, R.D.; Shaked, Y.; et al. Heparanase and chemotherapy synergize to drive macrophage activation and enhance tumor growth. Cancer Res. 2020, 80, 57–68. [Google Scholar] [CrossRef]
- Bartlett, M.R.; Underwood, P.A.; Parish, C.R. Comparative analysis of the ability of leucocytes, endothelial cells and platelets to degrade the subendothelial basement membrane: Evidence for cytokine dependence and detection of a novel sulfatase. Immunol. Cell Biol. 1995, 73, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Iriyama, S.; Matsunaga, Y.; Takahashi, K.; Matsuzaki, K.; Kumagai, N.; Amano, S. Activation of heparanase by ultraviolet B irradiation leads to functional loss of basement membrane at the dermal-epidermal junction in human skin. Arch. Dermatol. Res. 2011, 303, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.I.; Seong, J.; Park, Y.N.; Kim, W.W.; Oh, H.J.; Han, K.H. Identification of proteins indicating radiation-induced Hepatic Toxicity in cirrhotic rats. J. Radiat. Res. 2010, 51, 643–650. [Google Scholar] [CrossRef][Green Version]
- Khamaysi, I.; Singh, P.; Nasser, S.; Awad, H.; Chowers, Y.; Sabo, E.; Hammond, E.; Gralnek, I.; Minkov, I.; Noseda, A.; et al. The Role of Heparanase in the Pathogenesis of Acute Pancreatitis: A Potential Therapeutic Target. Sci. Rep. 2017, 7, 715. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, T.; Nakagawara, A. Role of p53 in cell death and human cancers. Cancers 2011, 3, 994–1013. [Google Scholar] [CrossRef] [PubMed]
- Nana-Sinkam, S.P.; Croce, C.M. MicroRNA regulation of tumorigenesis, cancer progression and interpatient heterogeneity: Towards clinical use. Genome Biol. 2014, 15, 445. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Zhou, S. The Functions of Heparanase in Human Diseases. Mini Rev. Med. Chem. 2017, 17, 541–548. [Google Scholar] [CrossRef]
- Lerner, I.; Hermano, E.; Zcharia, E.; Rodkin, D.; Bulvik, R.; Doviner, V.; Rubinstein, A.M.; Ishai-Michaeli, R.; Atzmon, R.; Sherman, Y.; et al. Heparanase powers a chronic inflammatory circuit that promotes colitis-associated tumorigenesis in mice. J. Clin. Investig. 2011, 121, 1709–1721. [Google Scholar] [CrossRef]
- Grimbacher, B.; Aicher, W.K.; Peter, H.H.; Eibel, H. TNF-α induces the transcription factor Egr-1, pro-inflammatory cytokines and cell proliferation in human skin fibroblasts and synovial lining cells. Rheumatol. Int. 1998, 17, 185–192. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, J.; Meng, X.; Wang, D. TNF-α induces early growth response gene-1 expression via ERK1/2 activation in endothelial cells. Acta Diabetol. 2013, 50, 27–31. [Google Scholar] [CrossRef]
- Ma, H.; Bernstein, L.; Ross, R.K.; Ursin, G. Hormone-related risk factors for breast cancer in women under age 50 years by estrogen and progesterone receptor status: Results from a case-control and a case-case comparison. Breast Cancer Res. 2006, 8, R39. [Google Scholar] [CrossRef] [PubMed]
- Moloney, J.N.; Cotter, T.G. ROS signalling in the biology of cancer. Semin. Cell Dev. Biol. 2018, 80, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Thakkar, N.; Yadavalli, T.; Jaishankar, D.; Shukla, D. Emerging Roles of Heparanase in Viral Pathogenesis. Pathogens 2017, 6, 43. [Google Scholar] [CrossRef] [PubMed]
- Koganti, R.; Suryawanshi, R.; Shukla, D. Heparanase, cell signaling, and viral infections. Cell. Mol. Life Sci. 2020, 77, 5059–5077. [Google Scholar] [CrossRef]
- Jackson, T.; Ellard, F.M.; Ghazaleh, R.A.; Brookes, S.M.; Blakemore, W.E.; Corteyn, A.H.; Stuart, D.I.; Newman, J.W.; King, A.M. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J. Virol. 1996, 70, 5282–5287. [Google Scholar] [CrossRef]
- Hallak, L.K.; Collins, P.L.; Knudson, W.; Peeples, M.E. Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology 2000, 271, 264–275. [Google Scholar] [CrossRef]
- Giroglou, T.; Florin, L.; Schäfer, F.; Streeck, R.E.; Sapp, M. Human Papillomavirus Infection Requires Cell Surface Heparan Sulfate. J. Virol. 2001, 75, 1565–1570. [Google Scholar] [CrossRef]
- Cooper, A.; Tal, G.; Lider, O.; Shaul, Y. Cytokine Induction by the Hepatitis B Virus Capsid in Macrophages Is Facilitated by Membrane Heparan Sulfate and Involves TLR2. J. Immunol. 2005, 175, 3165–3176. [Google Scholar] [CrossRef] [PubMed]
- Khanna, M.; Ranasinghe, C.; Jackson, R.; Parish, C.R. Heparan sulfate as a receptor for poxvirus infections and as a target for antiviral agents. J. Gen. Virol. 2017, 98, 2556–2568. [Google Scholar] [CrossRef]
- Bar-Sela, G.; Kaplan-Cohen, V.; Ilan, N.; Vlodavsky, I.; Ben-Izhak, O. Heparanase expression in nasopharyngeal carcinoma inversely correlates with patient survival. Histopathology 2006, 49, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Vornicova, O.; Boyango, I.; Feld, S.; Naroditsky, I.; Kazarin, O.; Zohar, Y.; Tiram, Y.; Ilan, N.; Ben-Izhak, O.; Vlodavsky, I.; et al. The prognostic significance of heparanase expression in metastatic melanoma. Oncotarget 2016, 7, 74678–74685. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Yu, J.; Gao, G.; Wang, X.; Liu, Y.; Zhu, S.; Gong, Z. Salivary Heparanase Level Is a Potential Biomarker to Diagnose and Prognose the Malignant Salivary Gland Tumor. PLoS ONE 2015, 10, e0143009. [Google Scholar] [CrossRef] [PubMed]
- Garcia, B.; Fernández-Vega, I.; García-Suárez, O.; Castañón, S.; Quirós, L.M. The Role of Heparan Sulfate Proteoglycans in Bacterial Infections. J. Med. Microbiol. Diagn. 2014, 03, e157. [Google Scholar] [CrossRef]
- García, B.; Merayo-Lloves, J.; Martin, C.; Alcalde, I.; Quirós, L.M.; Vazquez, F. Surface proteoglycans as mediators in bacterial pathogens infections. Front. Microbiol. 2016, 7, 220. [Google Scholar] [CrossRef]
- Rangarajan, S.; Richter, J.R.; Richter, R.P.; Bandari, S.K.; Tripathi, K.; Vlodavsky, I.; Sanderson, R.D. Heparanase-enhanced Shedding of Syndecan-1 and Its Role in Driving Disease Pathogenesis and Progression. J. Histochem. Cytochem. 2020, 68, 823–840. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ngo, J.A.; Wetzel, M.D.; Marchetti, D. Heparanase mediates a novel mechanism in lapatinib-resistant brain metastatic breast cancer. Neoplasia 2015, 17, 101–113. [Google Scholar] [CrossRef]
- Ramani, V.C.; Zhan, F.; He, J.; Barbieri, P.; Noseda, A.; Tricot, G.; Sanderson, R.D. Targeting heparanase overcomes chemoresistance and diminishes relapse in myeloma. Oncotarget 2016, 7, 1598–1607. [Google Scholar] [CrossRef]
- Wang, X.; Zuo, D.; Chen, Y.; Li, W.; Liu, R.; He, Y.; Ren, L.; Zhou, L.; Deng, T.; Ying, G.; et al. Shed Syndecan-1 is involved in chemotherapy resistance via the EGFR pathway in colorectal cancer. Br. J. Cancer 2014, 111, 1965–1976. [Google Scholar] [CrossRef]
- Goodall, K.J.; Poon, I.K.H.; Phipps, S.; Hulett, M.D. Soluble Heparan Sulfate Fragments Generated by Heparanase Trigger the Release of Pro-Inflammatory Cytokines through TLR-4. PLoS ONE 2014, 9, e109596. [Google Scholar] [CrossRef]
- Gutter-Kapon, L.; Alishekevitz, D.; Shaked, Y.; Li, J.-P.; Aronheim, A.; Ilan, N.; Vlodavsky, I. Heparanase is required for activation and function of macrophages. Proc. Natl. Acad. Sci. USA 2016, 113, E7808–E7817. [Google Scholar] [CrossRef]
- Song, T.; Spillmann, D. Transcriptomic analysis reveals cell apoptotic signature modified by heparanase in melanoma cells. J. Cell. Mol. Med. 2019, 23, 4559–4568. [Google Scholar] [CrossRef]
- Abboud-Jarrous, G.; Rangini-Guetta, Z.; Aingorn, H.; Atzmon, R.; Elgavish, S.; Peretz, T.; Vlodavsky, I. Site-directed mutagenesis, proteolytic cleavage, and activation of human proheparanase. J. Biol. Chem. 2005, 280, 13568–13575. [Google Scholar] [CrossRef] [PubMed]
- Abboud-Jarrous, G.; Atzmon, R.; Peretz, T.; Palermo, C.; Gadea, B.B.; Joyce, J.A.; Vlodavsky, I. Cathepsin L is responsible for processing and activation of proheparanase through multiple cleavages of a linker segment. J. Biol. Chem. 2008, 283, 18167–18176. [Google Scholar] [CrossRef]
- Temkin, V.; Aingorn, H.; Puxeddu, I.; Goldshmidt, O.; Zcharia, E.; Gleich, G.J.; Vlodavsky, I.; Levi-Schaffer, F. Eosinophil major basic protein: First identified natural heparanase-inhibiting protein. J. Allergy Clin. Immunol. 2004, 113, 703–709. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, E.A. Heparanase: A target for drug discovery in cancer and inflammation. Br. J. Pharmacol. 2007, 151, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, D.; Liu, S.; Spohn, W.C.; Carson, D.D. Heparanase and a synthetic peptide of heparan sulfate-interacting protein recognize common sites on cell surface and extracellular matrix heparan sulfate. J. Biol. Chem. 1997, 272, 15891–15897. [Google Scholar] [CrossRef] [PubMed]
- Levy-Adam, F.; Feld, S.; Cohen-Kaplan, V.; Shteingauz, A.; Gross, M.; Arvatz, G.; Naroditsky, I.; Ilan, N.; Doweck, I.; Vlodavsky, I. Heparanase 2 Interacts with Heparan Sulfate with High Affinity and Inhibits Heparanase Activity. J. Biol. Chem. 2010, 285, 28010–28019. [Google Scholar] [CrossRef]
- Mohan, C.D.; Hari, S.; Preetham, H.D.; Rangappa, S.; Barash, U.; Ilan, N.; Nayak, S.C.; Gupta, V.K.; Basappa; Vlodavsky, I.; et al. Targeting Heparanase in Cancer: Inhibition by Synthetic, Chemically modified and Natural Compounds. iScience 2019, 15, 360–390. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Ilan, N.; Naggi, A. Benito Casu Heparanase: Structure, Biological Functions, and Inhibition by Heparin-Derived Mimetics of Heparan Sulfate. Curr. Pharm. Des. 2007, 13, 2057–2073. [Google Scholar] [CrossRef]
- Cassinelli, G.; Torri, G.; Naggi, A. Non-Anticoagulant Heparins as Heparanase Inhibitors. Adv. Exp. Med. Biol. 2020, 1221, 493–522. [Google Scholar] [CrossRef] [PubMed]
- Van Den Hoven, M.J.; Rops, A.L.; Vlodavsky, I.; Levidiotis, V.; Berden, J.H.; van der Vlag, J. Heparanase in glomerular diseases. Kidney Int. 2007, 72, 543–548. [Google Scholar] [CrossRef]
- Coombe, D.R.; Gandhi, N.S. Heparanase: A Challenging Cancer Drug Target. Front. Oncol. 2019, 9, 1316. [Google Scholar] [CrossRef] [PubMed]
- Toyoshima, M.; Nakajima, M. Human heparanase. Purification, characterization, cloning, and expression. J. Biol. Chem. 1999, 274, 24153–24160. [Google Scholar] [CrossRef]
- Chen, L.; Sanderson, R.D. Heparanase Regulates Levels of Syndecan-1 in the Nucleus. PLoS ONE 2009, 4, e4947. [Google Scholar] [CrossRef]
- Purushothaman, A.; Hurst, D.R.; Pisano, C.; Mizumoto, S.; Sugahara, K.; Sanderson, R.D. Heparanase-mediated Loss of Nuclear Syndecan-1 Enhances Histone Acetyltransferase (HAT) Activity to Promote Expression of Genes That Drive an Aggressive Tumor Phenotype. J. Biol. Chem. 2011, 286, 30377–30383. [Google Scholar] [CrossRef]
- Yang, Y.; Gorzelanny, C.; Bauer, A.T.; Halter, N.; Komljenovic, D.; Bäuerle, T.; Borsig, L.; Roblek, M.; Schneider, S.W. Nuclear heparanase-1 activity suppresses melanoma progression via its DNA-binding affinity. Oncogene 2015, 34, 5832–5842. [Google Scholar] [CrossRef]
- Schubert, S.Y.; Ilan, N.; Shushy, M.; Ben-Izhak, O.; Vlodavsky, I.; Goldshmidt, O. Human heparanase nuclear localization and enzymatic activity. Lab. Investig. 2004, 84, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, T.C.F.; Gomes, A.M.; Paschoal, M.E.M.; Stelling, M.P.; Rumjanek, V.M.B.D.; do Junior, A.R.; Valiante, P.M.; Madi, K.; de Souza, H.S.P.; Pavão, M.S.G.; et al. Heparanase expression and localization in different types of human lung cancer. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 2599–2608. [Google Scholar] [CrossRef]
- He, Y.Q.; Sutcliffe, E.L.; Bunting, K.L.; Li, J.; Goodall, K.J.; Poon, I.K.A.; Hulett, M.D.; Freeman, C.; Zafar, A.; McInnes, R.L.; et al. The endoglycosidase heparanase enters the nucleus of T lymphocytes and modulates H3 methylation at actively transcribed genes via the interplay with key chromatin modifying enzymes. Transcription 2012, 3, 130–145. [Google Scholar] [CrossRef] [PubMed]
- Doweck, I.; Kaplan-Cohen, V.; Naroditsky, I.; Sabo, E.; Ilan, N.; Vlodavsky, I. Heparanase Localization and Expression by Head and Neck Cancer: Correlation with Tumor Progression and Patient Survival. Neoplasia 2006, 8, 1055–1061. [Google Scholar] [CrossRef]
- Cohen, E.; Doweck, I.; Naroditsky, I.; Ben-Izhak, O.; Kremer, R.; Best, L.A.; Vlodavsky, I.; Ilan, N. Heparanase is overexpressed in lung cancer and correlates inversely with patient survival. Cancer 2008, 113, 1004–1011. [Google Scholar] [CrossRef] [PubMed]
- Zetser, A.; Bashenko, Y.; Edovitsky, E.; Levy-Adam, F.; Vlodavsky, I.; Ilan, N. Heparanase induces vascular endothelial growth factor expression: Correlation with p38 phosphorylation levels and Src activation. Cancer Res. 2006, 66, 1455–1463. [Google Scholar] [CrossRef]
- Ramani, V.C.; Yang, Y.; Ren, Y.; Nan, L.; Sanderson, R.D. Heparanase plays a dual role in driving hepatocyte growth factor (HGF) signaling by enhancing HGF expression and activity. J. Biol. Chem. 2011, 286, 6490–6499. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Meng, X.; Hu, J.; Zhang, Y.; Dang, Y.; Wei, L.; Shi, M. Heparanase promotes radiation resistance of cervical cancer by upregulating hypoxia inducible factor 1. Am. J. Cancer Res. 2017, 7, 234–244. [Google Scholar]
- Masola, V.; Zaza, G.; Secchi, M.F.; Gambaro, G.; Lupo, A.; Onisto, M. Heparanase is a key player in renal fibrosis by regulating TGF-β expression and activity. Biochim. Biophys. Acta Mol. Cell Res. 2014, 1843, 2122–2128. [Google Scholar] [CrossRef]
- Abassi, Z.; Hamoud, S.; Hassan, A.; Khamaysi, I.; Nativ, O.; Heyman, S.N.; Muhammad, R.S.; Ilan, N.; Singh, P.; Hammond, E.; et al. Involvement of heparanase in the pathogenesis of acute kidney injury: Nephroprotective effect of PG545. Oncotarget 2017, 8, 34191–34204. [Google Scholar] [CrossRef]
- Xia, J.; Sheng, W.; Pei, L.; Li, N.; Zhang, Z.; Wang, J.; Zu, J.; Wang, N.; Wang, D. Effects of unfractionated heparin and rivaroxaban on the expression of heparanase and fibroblast growth factor 2 in human osteoblasts. Mol. Med. Rep. 2017, 16. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.; Fourgeaud, C.; Derieux, S.; Mirshahi, S.; Contant, G.; Pimpie, C.; lo Dico, R.; Soria, J.; Pocard, M.; Mirshahi, M. The close relationship between heparanase and epithelial mesenchymal transition in gastric signet-ring cell adenocarcinoma. Oncotarget 2018, 9, 33778–33787. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Masola, V.; Zaza, G.; Bellin, G.; Dall’Olmo, L.; Granata, S.; Vischini, G.; Secchi, M.F.; Lupo, A.; Gambaro, G.; Onisto, M.; et al. Heparanase regulates the M1 polarization of renal macrophages and their crosstalk with renal epithelial tubular cells after ischemia/reperfusion injury. FASEB J. 2018, 32, 742–756. [Google Scholar] [CrossRef]
- Tsunekawa, N.; Higashi, N.; Kogane, Y.; Waki, M.; Shida, H.; Nishimura, Y.; Adachi, H.; Nakajima, M.; Irimura, T. Heparanase augments inflammatory chemokine production from colorectal carcinoma cell lines. Biochem. Biophys. Res. Commun. 2016, 469, 878–883. [Google Scholar] [CrossRef] [PubMed]
- Purushothaman, A.; Chen, L.; Yang, Y.; Sanderson, R.D. Heparanase stimulation of protease expression implicates it as a master regulator of the aggressive tumor phenotype in myeloma. J. Biol. Chem. 2008, 283, 32628–32636. [Google Scholar] [CrossRef]
- Lv, B.; Zhang, B.; Hu, X.Y.; Zeng, Q.D. Heparanase regulates in vitro VEGF-C expression and its clinical significance to pancreatic ductal cell adenocarcinoma. Oncol. Lett. 2016, 11, 1327–1334. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, F.; Wang, Y.; Zhang, D.; Puthanveetil, P.; Johnson, J.D.; Rodrigues, B. Fatty Acid-Induced Nuclear Translocation of Heparanase Uncouples Glucose Metabolism in Endothelial Cells. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 406–414. [Google Scholar] [CrossRef]
- Okawa, T.; Naomoto, Y.; Nobuhisa, T.; Takaoka, M.; Motoki, T.; Shirakawa, Y.; Yamatsuji, T.; Inoue, H.; Ouchida, M.; Gunduz, M.; et al. Heparanase Is Involved in Angiogenesis in Esophageal Cancer through Induction of Cyclooxygenase-2. Clin. Cancer Res. 2005, 11, 7995. [Google Scholar] [CrossRef][Green Version]
- Yan, X.; Jin, S.; Li, S.; Gong, F.; Zhang, D.; Zhang, X.; Li, J. Heparanase Modulation of Early Growth Response Gene Expression. Zool. Sci. 2011, 28, 189–194. [Google Scholar] [CrossRef]
- Masola, V.; Bellin, G.; Vischini, G.; Dall’Olmo, L.; Granata, S.; Gambaro, G.; Lupo, A.; Onisto, M.; Zaza, G. Inhibition of heparanase protects against chronic kidney dysfunction following ischemia/reperfusion injury. Oncotarget 2018, 9, 36185–36201. [Google Scholar] [CrossRef][Green Version]
- Hu, X.; Zhang, L.; Jin, J.; Zhu, W.; Xu, Y.; Wu, Y.; Wang, Y.; Chen, H.; Webster, K.A.; Chen, H.; et al. Heparanase released from mesenchymal stem cells activates integrin beta1/HIF-2alpha/Flk-1 signaling and promotes endothelial cell migration and angiogenesis. Stem Cells 2015, 33, 1850–1862. [Google Scholar] [CrossRef]
- Chen, X.; Cheng, B.; Dai, D.; Wu, Y.; Feng, Z.; Tong, C.; Wang, X.; Zhao, J. Heparanase induces necroptosis of microvascular endothelial cells to promote the metastasis of hepatocellular carcinoma. Cell Death Discov. 2021, 7. [Google Scholar] [CrossRef]
- Huang, X.; Reye, G.; Momot, K.I.; Blick, T.; Lloyd, T.; Tilley, W.D.; Hickey, T.E.; Snell, C.E.; Okolicsanyi, R.K.; Haupt, L.M.; et al. Heparanase Promotes Syndecan-1 Expression to Mediate Fibrillar Collagen and Mammographic Density in Human Breast Tissue Cultured ex vivo. Front. Cell Dev. Biol. 2020, 8, 599. [Google Scholar] [CrossRef]
- Masola, V.; Gambaro, G.; Tibaldi, E.; Brunati, A.M.; Gastaldello, A.; D’Angelo, A.; Onisto, M.; Lupo, A. Heparanase and Syndecan-1 Interplay Orchestrates Fibroblast Growth Factor-2-induced Epithelial-Mesenchymal Transition in Renal Tubular Cells. J. Biol. Chem. 2012, 287, 1478–1488. [Google Scholar] [CrossRef]
- Singh, P.; Blatt, A.; Feld, S.; Zohar, Y.; Saadi, E.; Barki-Harrington, L.; Hammond, E.; Ilan, N.; Vlodavsky, I.; Chowers, Y.; et al. The Heparanase Inhibitor PG545 Attenuates Colon Cancer Initiation and Growth, Associating with Increased p21 Expression. Neoplasia 2017, 19, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Nadir, Y.; Brenner, B.; Zetser, A.; Ilan, N.; Shafat, I.; Zcharia, E.; Goldshmidt, O.; Vlodavsky, I. Heparanase induces tissue factor expression in vascular endothelial and cancer cells. J. Thromb. Haemost. 2006, 4, 2443–2451. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.; Gombert, A.; Chen, J.; Liebens, J.; Verleger, J.; Kalder, J.; Marx, G.; Jacobs, M.; Thiemermann, C.; Schuerholz, T. The ß-D-endoglucuronidase heparanase is a danger molecule that drives systemic inflammation and correlates with clinical course after open and endovascular thoracoabdominal aortic aneurysm repair: Lessons learnt from mice and men. Front. Immunol. 2017, 8, 681. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.R.; Sun, D.N.; Yang, H.; Yan, J.; Zhang, X.; Zheng, X.L.; Fu, X.H.; Geng, M.Y.; Huang, X.; Ding, J. CTC clusters induced by heparanase enhance breast cancer metastasis. Acta Pharmacol. Sin. 2018, 39, 1326–1337. [Google Scholar] [CrossRef]
- Crispel, Y.; Ghanem, S.; Attias, J.; Kogan, I.; Brenner, B.; Nadir, Y. Involvement of the heparanase procoagulant domain in bleeding and wound healing. J. Thromb. Haemost. 2017, 15, 1463–1472. [Google Scholar] [CrossRef]
- Blich, M.; Golan, A.; Arvatz, G.; Sebbag, A.; Shafat, I.; Sabo, E.; Cohen-Kaplan, V.; Petcherski, S.; Avniel-Polak, S.; Eitan, A.; et al. Macrophage activation by heparanase is mediated by TLR-2 and TLR-4 and associates with plaque progression. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 56–65. [Google Scholar] [CrossRef]
- Bitan, M.; Weiss, L.; Reibstein, I.; Zeira, M.; Fellig, Y.; Slavin, S.; Zcharia, E.; Nagler, A.; Vlodavsky, I. Heparanase upregulates Th2 cytokines, ameliorating experimental autoimmune encephalitis. Mol. Immunol. 2010, 47, 1890–1898. [Google Scholar] [CrossRef]
- Agelidis, A.; Turturice, B.A.; Suryawanshi, R.K.; Yadavalli, T.; Jaishankar, D.; Ames, J.; Hopkins, J.; Koujah, L.; Patil, C.D.; Hadigal, S.R.; et al. Disruption of innate defense responses by endoglycosidase HPSE promotes cell survival. JCI Insight 2021, 6, e144255. [Google Scholar] [CrossRef] [PubMed]
- Abeyrathna, P.; Su, Y. The critical role of Akt in cardiovascular function. Vascul. Pharmacol. 2015, 74, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Gingis-Velitski, S.; Zetser, A.; Flugelman, M.Y.; Vlodavsky, I.; Ilan, N. Heparanase Induces Endothelial Cell Migration via Protein Kinase B/Akt Activation. J. Biol. Chem. 2004, 279, 23536–23541. [Google Scholar] [CrossRef] [PubMed]
- Ben-Zaken, O.; Gingis-Velitski, S.; Vlodavsky, I.; Ilan, N. Heparanase induces Akt phosphorylation via a lipid raft receptor. Biochem. Biophys. Res. Commun. 2007, 361, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Kaplan, V.; Jrbashyan, J.; Yanir, Y.; Naroditsky, I.; Ben-Izhak, O.; Ilan, N.; Doweck, I.; Vlodavsky, I. Heparanase Induces Signal Transducer and Activator of Transcription (STAT) Protein Phosphorylation. J. Biol. Chem. 2012, 287, 6668–6678. [Google Scholar] [CrossRef] [PubMed]
- Ramos, J.W. The regulation of extracellular signal-regulated kinase (ERK) in mammalian cells. Int. J. Biochem. Cell Biol. 2008, 40, 2707–2719. [Google Scholar] [CrossRef]
- Xiong, A.; Kundu, S.; Forsberg, M.; Xiong, Y.; Bergström, T.; Paavilainen, T.; Kjellén, L.; Li, J.-P.P.; Forsberg-Nilsson, K. Heparanase confers a growth advantage to differentiating murine embryonic stem cells, and enhances oligodendrocyte formation. Matrix Biol. 2017, 62, 92–104. [Google Scholar] [CrossRef]
- Oda, K.; Matsuoka, Y.; Funahashi, A.; Kitano, H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol. Syst. Biol. 2005, 1, 2005-0010. [Google Scholar] [CrossRef]
- Cohen-Kaplan, V.; Doweck, I.; Naroditsky, I.; Vlodavsky, I.; Ilan, N. Heparanase augments epidermal growth factor receptor phosphorylation: Correlation with head and neck tumor progression. Cancer Res. 2008, 68, 10077–10085. [Google Scholar] [CrossRef]
- Vlodavsky, I.; Singh, P.; Boyango, I.; Gutter-Kapon, L.; Elkin, M.; Sanderson, R.D.; Ilan, N. Heparanase: From basic research to therapeutic applications in cancer and inflammation. Drug Resist. Updat. 2016, 29, 54–75. [Google Scholar] [CrossRef]
| Protein | Technique | Reference |
|---|---|---|
| Amyloid peptide β (1–42) | Surface plasmon resonance | [13] |
| Amyloid precursor protein | Fluorescence lifetime imaging microscopy | [14] |
| Annexin V | Affinity chromatography | [15] |
| Basic fibroblast growth factor (bFGF) (FGF-2) | Iodinated-bFGF and specific activity following heparanase addition; Affinity chromatography; Iodinated-bFGF and specific activity; Cross-linking of iodinated-bFGF following heparitinase treatment | [16,17,18,19] |
| Collagen I | Affinity chromatography; Surface plasmon resonance | [13,20] |
| Collagen V | Solid phase binding assay; Surface plasmon resonance | [21,22] |
| Collagenase IV | Antibody-linked detection assay; Surface plasmon resonance | [13,23] |
| Collagen VI | Surface plasmon resonance | [13] |
| chemokine (C-X-C motif) ligand (CXCL1) (KC) | Surface plasmon resonance | [24] |
| CXCL2 (MIP-2) | Surface plasmon resonance | [24] |
| CXCL6 (GCP-2) | Surface plasmon resonance | [24] |
| CXCL10 (IP-10) | Alkaline phosphatase-conjucated IP-10; Surface plasmon resonance | [24,25] |
| CXCL11 (I-TAC) | Surface plasmon resonance | [24] |
| CXCL13 | Surface plasmon resonance | [26] |
| Endostatin | Alkaline phosphatase-endostatin binding assay; Filter-binding assay; Surface plasmon resonance | [13,27,28] |
| FGFR4 | Affinity chromatography | [29] |
| Fibronectin | Affinity chromatography; Antibody-linked detection assay | [7,8,23] |
| HGF | Affinity chromatography | [30] |
| Histidine-rich glycoprotein | Flow cytometry after heparanase treatment | [31] |
| High mobility group protein B1 | Biotinylation and streptadivin-HRP detection | [32] |
| Integrin α5β1 | Surface plasmon resonance | [13] |
| Interferon-β (IFN-β) | Filter binding assay | [33] |
| Interleukin-8 (IL-8) | Affinity co-electrophoresis | [34] |
| Laminin-1 | Antibody-linked detection assay; Surface plasmon resonance | [13,23] |
| L-selectin | Heparinase treatment of 35SO4-labeled L-selectin ligands and SDS-PAGE; Affinity chromatography | [35,36] |
| Monocyte chemoattractant protein-1 (MCP-1) | Competitive binding to 3H-heparin by nitrocellulose membrane filtration and liquid scintillation | [37] |
| Macrophage migration inhibitory factor (MIF) | Flow cytometry of fluorescently labeled MIF on A549 cells after heparinase treatment | [38] |
| Macrophage inflammatory protein-1α (MIP-1α) | Affinity chromatography after heparinase treatment | [39] |
| NKp46 | ELISA | [40] |
| Platelet-derived growth factor (PDGF) | Surface plasmon resonance; Affinity chromatography | [41,42] |
| Platelet Factor 4 | Affinity co-electrophoresis | [34] |
| P-selectin | Affinity chromatography | [36] |
| Receptor for advanced glycation end products (RAGE) | Biotinylation and streptadivin-HRP detection | [32,43] |
| Regulated on activation normal T cell expressed and secreted (RANTES) (CCL5) (oligomerized, filamentous) | Surface plasmon resonance | [24,44] |
| Receptor protein-tyrosine phosphatase-σ (RPTP-σ) | Blot overlay assay probing agrin and collagen with cPTP-σ-conditioned medium following heparinase digestion | [45] |
| Stromal cell-derived factor-1 (SDF-1) | Flow cytometry of endothelial cells after heparinase treatment for bound SDF-1 | [46] |
| Transglutaminase-2 | Surface plasmon resonance | [13] |
| Thrombospondin-1 | Surface plasmon resonance | [13] |
| Vascular endothelial growth factor (VEGF) | Metabolic labeling of pHEBO cells overexpressing VEGF189 followed by heparinase treatment, immunoprecipitation, and SDS PAGE | [47] |
| Agent | Species | Findings | Reference |
|---|---|---|---|
| Transcription factors | |||
| c-Myc | Human | hTERT enabled binding of c-Myc to the heparanase promoter and increased heparanase mRNA expression | [65] |
| Early growth response 1 (EGR1) | Human and mouse | Direct binding to the heparanase promoter resulted in activation of the heparanase promoter in PC-3, COLO397, and MCF-7 cells, and repression of the heparanase promoter in MM170 cells; EGR1 was recruited to the heparanase promoter upon glucose treatment of HEK293T cells | [61,62,66,67,68] |
| Erythroblast Transformation Specific 1 (ETS1) and ETS2 | Human | Direct binding to the heparanase promoter increased heparanase mRNA expression | [69] |
| GA-binding protein (GABP) | Human | Direct binding to the heparanase promoter increased heparanase promoter activity | [70,71] |
| NF-κB | Human and mouse | NF-κB-deficient lung carcinoma cells produced less heparanase; Inhibiting canonical NF-κB signaling blocked tumor necrosis factor-α (TNF-α)-induced upregulation of heparanase in endothelial cells; Chemotherapy treatment of multiple myeloma cells, hypoxia induction in pancreatic cancer cells, or infection with herpes simplex virus-1 (HSV-1) activated NF-κB to upregulate heparanase | [72,73,74,75,76] |
| p53 | Human and mouse | Direct binding to the heparanase promoter reduced heparanase mRNA expression | [77] |
| Snail | Mouse | Overexpression of Snail in B16F1 cells increased heparanase mRNA expression | [78] |
| specificity protein 1 (SP1) and SP3 | Human | Direct binding to the heparanase promoter increased heparanase promoter activity | [70] |
| MicroRNA | |||
| miR-1258 | Human | miRNA-1258 suppressed heparanase expression in breast cancer cells | [79] |
| miR-1252-5p | Overexpression of miR-1252-5p in multiple myeloma cells reduced heparanase mRNA and protein expression and activity | [80] | |
| Cytokines | |||
| IFN-γ | Human | Treatment of endothelial cells with IFN-γ increased heparanase mRNA expression and activity | [81] |
| IL-1β | Human and mouse | Treatment of endothelial cells with IL-1β increased heparanase mRNA expression | [73,82] |
| IL-2 | Mouse | Treatment of purified NK cells with IL-2 induced expression of both pro-heparanase and the catalytically active heparanase protein, more so when also cultured with IL-15 | [50] |
| IL-10 | Human | IL-10 treatment of SUM149 breast cancer cells modestly increased heparanase mRNA expression | [83] |
| IL-15 | Mouse | Treatment of purified NK cells with IL-15 induced expression of both pro-heparanase and the catalytically active heparanase protein, more so when also cultured with IL-12 and IL-18 | [50] |
| IL-17 | Human | Treatment of cervical cancer cells with IL-17 increased heparanase mRNA expression, and IL-17 knockdown reduced heparanase expression | [84] |
| MCP-1 | Mouse | MCP-1 inhibition reduced glomerular heparanase expression | [85] |
| TNF-α | Human, mouse and bovine | Treatment of endothelial cells, U937 macrophages and colon cancer cells with TNF-α increased heparanase mRNA and protein expression | [73,82,86,87,88] |
| Growth factors | |||
| bFGF | Human | Treatment of lung cancer cells with bFGF induced heparanase mRNA expression | [89] |
| HGF | Human | Treatment of lung cancer cells with HGF induced heparanase mRNA expression; HGF activated the PI3K/Akt/NF-κB signaling pathway and upregulated heparanase mRNA and protein expression | [89,90] |
| FGF23 | Human | Treatment of multiple myeloma cells with FGF23 increased heparanase mRNA expression, likely via Egr1 upregulation which was also upregulated | [91] |
| PDGF | Human | Treatment of lung cancer cells with PDGF induced heparanase mRNA expression | [89] |
| VEGF | Human | Treatment of endothelial cells with VEGF reduced heparanase expression; Treatment of melanoma cells with recombinant VEGF increased heparanase mRNA expression, and VEGF knockdown decreased heparanase expression | [82,92] |
| Pathways | |||
| CD3/CD28 activation | Human and mouse | Heparanase mRNA and protein increased with anti-CD3 and anti-CD28 antibody stimulation in mouse splenic (CD4/CD8) and human PBMC-derived T cells | [62,93] |
| MEK/ERK pathway | Human | Activation of the MEK/ERK pathway increased heparanase expression | [66] |
| Wnt signaling | Hamster and Zebrafish | CHO-K1 cells treated with lithium chloride (LiCl; a Wnt signaling activator) modestly increased heparanase protein expression. Zebrafish embryos treated with LiCl increased heparanase mRNA expression | [94] |
| Other biological molecules | |||
| Heparin | Hamster | CHO-K1 cells treated with heparin increased heparanase mRNA, protein, and enzymatic activity. Authors propose this may be via Wnt signaling | [94] |
| Lumican | Mouse | Treatment of B16F1 cells with recombinant lumican increased heparanase mRNA and protein expression; Treatment of Snail-overexpressing B16F1 cells (which resulted in increased heparanase expression) with recombinant lumican decreased heparanase mRNA expression but did not change protein expression | [78] |
| Hormones, metabolites and other signaling molecules | |||
| Advanced Glycation End Products (AGEs) | Human | Exposure of HMVECs to AGEs increased heparanase mRNA and protein expression | [95] |
| Basic protein | Rat | T lymphocytes degraded heparan sulfate (HS) after stimulation with Con A or basic protein | [96,97,98] |
| BRAF | Human | Wild tpye BRAF suppressed ETS1 family of transcription factors, which suppressed heparanase promoter activity and mRNA expression. Mutant BRAF lost repression ability and heparanase mRNA expression was upregulated | [71] |
| Endothelial nitric oxide synthase (eNOS) | Mouse | In a rat model of adriamycin nephropathy, the deletion or inhibition of eNOS induced heparanase mRNA and protein expression | [99] |
| Estrogen | Human and mouse | Estrogen treatment of ER-positive MCF-7 cells increased heparanase mRNA and protein expression via estrogen receptor signaling and estrogen response elements in the heparanase promoter. Upregulation of heparanase occured more so in low levels than high levels of estrogen; Estrogen supplementation in MCF-7 implanted tumors in mice increased heparanase protein expression; Treatment of cholangiocarcinoma cells with an estrogenic inducer upregulated heparanase mRNA expression; Estrogen treatment of ER-positive EO771 breast cancer cells increased heparanase mRNA expression | [100,101,102,103] |
| High glucose | Human and bovine | Glucose-treated cells modestly increased heparanase protein expression. Heparanase mRNA expression, secretion, and activity increased upon glucose treatment | [104,105,106] |
| Hypoxia (1% O2) | Human | Hypoxia-induced activation of NF-κB upregulated heparanase mRNA and protein expression | [74] |
| Low density lipoprotein (LDL) | Human | Treatment of endothelial cells with LDL increased heparanase mRNA expression | [82] |
| Lipopolysaccharide (LPS) | Human and mouse | Stimulation of B cells with LPS increased heparanase activity. Activating toll-like receptor 4 (TLR4) with LPS on PBMCs and cord blood cells increased heparanase mRNA expression; LPS stimulation of PBMCs increased heparanase mRNA expression; LPS stimulation of endothelial cells increased levels of enzymatically active heparanase | [63,98,107,108,109] |
| Oleic acid | Bovine | Treatment of endothelial cells increased heparanase mRNA and protein expression, and was likely via Sp1 | [82] |
| Reactive oxygen species (ROS) | Human and rat | In a rat model of adriamycin nephropathy, the depletion of hydroxyl radicals with DMTU reduced heparanase expression. Inducing mouse podocytes to generate free radicals and ROS increased heparanase mRNA and protein expression. Treatment of endothelial cells with ROS scavengers perturbed glucose-mediated heparanase expression | [106,109,110] |
| Vitamin D | Rat and mouse | Vitamin D treatment reduced heparanase mRNA expression via initiating direct binding of the vitamin D receptor to the heparanase promoter | [111] |
| Pathogens | |||
| Fusobacterium nucleatum | Human | Co-culture of SSC-25 oral cancer cells with F. nucleatum increased heparanase expression | [112] |
| Helicobacter pylori | H. pylori infection of gastric cancer cells induced an upregulation of heparanase protein, which was dependent on MAPK signaling | [113] | |
| Pseudomonas aeruginosa | Mouse | P. aeruginosa intracorneal infection in mice induced an upregulation of heparanase mRNA and enzymatically active protein in the cornea. This was from both infiltrating immune cells as well as from the corneal epithelium | [114] |
| Streptococcus pneumoniae | Mouse | Intranasal S. pneumoniae infection in mice increased heparanase protein expression | [115] |
| Bovine herpes virus | Human | Heparanase mRNA was upregulated upon epithelial cell infection in vitro | [75] |
| SARS-CoV-2 | Human | COVID-19 patients displayed elevated heparanase activity and soluble HS levels in the plasma; Increase shed syndecan-1 was observed | [116,117] |
| Cytomegalovirus | Human | Heparanase mRNA was upregulated upon fibroblast cell infection in vitro | [75] |
| Dengue virus | Human | Dengue virus protein NS1 upregulated heparanase protein in endothelial cells, and upregulation was found to be macrophage inhibitory factor-dependent | [118,119] |
| HSV-1 | Human | Heparanase mRNA and protein were upregulated upon HSV-1 infection through NF-κB activation | [75,120] |
| HSV-2 | Human | Heparanase mRNA was upregulated upon epithelial cell infection in vitro | [75] |
| Porcine reproductive and respiratory syndrome virus | Pig | Piglets infected with PRSSV in vivo increased heparanase mRNA expression in alveolar macrophages Cells infected in vitro with PRSSV increased heparanase mRNA and protein expression | [121,122] |
| Pseudorabies virus | Human | Heparanase mRNA was upregulated upon epithelial cell infection in vitro | [75] |
| Therapeutics | |||
| Bortezomib | Human | Treatment of myeloma cells increased heparanase mRNA and protein expression | [76] |
| Carfilzomib | Human | Treatment of myeloma cells increased heparanase mRNA expression | [76] |
| Cisplatin | Human | Treatment of mesothelioma cells, gastric cancer cells, and J774 macrophages increased heparanase mRNA expression | [123] |
| Clarithromycin | Human | Clarithromycin blocked the upregulation of heparanase mRNA induced by bFGF | [89] |
| Doxorubicin | Human | Treatment of myeloma cells, gastric cancer cells, and J774 macrophages increased heparanase protein expression | [76,123] |
| Erythromycin | Human | Erythromycin blocked the upregulation of heparanase mRNA induced by PDGF and HGF | [89] |
| Paclitaxel | Human | Treatment of gastric cancer cells with paclitaxel increased heparanase mRNA expression | [123] |
| phorbol-12-myristate-13-acetate (PMA) | Human and mouse | Heparanase mRNA expression increased upon stimulation with PMA ionomycin in EL4 T lymphocytes. HS degradation increased after PMA stimulation in neutrophils, human umbilical vein endothelial cells (HUVECs), and platelets; Heparanase mRNA, protein, and activity increased in human NK cells after activation with B-LCL cells, IL-2 and PMA, and ionomycin | [50,61,124] |
| Poly(I:C) | Mouse | Poly(I:C) stimulation in vivo increased heparanase activity in splenic NK cells | [50] |
| Radiation | Human and rat | Human epidermal keratinocytes exposed to UVB radiation exhibited increased heparanase enzymatic activity and detectable levels of the 50 kDa active subunit; Rats with experimental liver cirrhosis showed an increase in heparanase precursor protein in liver and serum after treatment with partial liver radiation | [125,126] |
| Tamoxifen | Human | Treatment of MCF-7 cells with high concentration of tamoxifen inhibited estrogen-induced heparanase expression; Tamoxifen treatment of MCF-7 cells and T47D cells increased heparanase mRNA expression | [100,102] |
| Miscellaneous | |||
| Cerulein | Mouse | Injection of cerulein into mice increased heparanase mRNA expression and enzymatic activity in pancreatic tissue extracts | [127] |
| Gene/Protein | Observation/Mechanism | Related Disease/Function | Reference |
|---|---|---|---|
| Genes | |||
| Aromatase | The expression of aromatase was decreased in heparanase- knockout obese mice. Heparanase was required for the activation of fatty acid-stimulated macrophages to induce aromatase in adipose stromal cells | Obesity-associated breast cancer progression | [103] |
| Bcl-XL (Bcl2l1) | Increased expression of Bcl-XL in heparanase overexpressing transgenic mice with dextran sulfate sodium (DSS)-induced colitis was regulated by NF-κB | Ulcerative colitis | [131] |
| Caspase-1 | Silence of heparanase and heparanase inhibitor (SST0001) blocked caspase 1 expression in human kidney cells | Acute kidney injury/M1 macrophage polarization | [182] |
| Cathepsin L | Induction of acute kidney injury in heparanase-transgenic mice enhanced the expression of cathepsin L mRNA. Pre-treatment with heparanase inhibitor PG545 reduced the expression of cathepsin L | Epithelial-mesenchymal transition (EMT)/Acute kidney injury | [179] |
| CD44 | siRNA knockdown of heparanase in SUM149 breast cancer cells reduced mRNA expression of CD44 | Breast cancer | [83] |
| c-Fos (AP-1) | The expression of c-Fos was decreased in heparanase-knockout macrophages and adding exogenous heparanase enhanced c-Fos expression. Heparanase regulated the gene expression of c-Fos through Erk, p38, and JNK signaling pathway | Tumor/Induction of cytokine expression | [153] |
| Collagen-I | Treatment of KATO-III gastric cancer cells with heparanase inhibitor suramin exhibited reduced expression of collagen-I | EMT/Gastric ring cell adenocarcinoma | [181] |
| Cox-2 | Cox-2 mRNA expression was increased in heparanase overexpressing transgenic mice with DSS-induced colitis and was regulated by NF-κB | Ulcerative colitis | [131] |
| Heparanase upregulated the mRNA expression of Cox-2 in cancer cells | Tumor/Promoting angiogenesis | [187] | |
| CXCR-4 | mRNA expression of CXCR-4 was decreased in gastric cancer cell KATO-III treated with heparanase inhibitor suramin | EMT/Gastric ring cell adenocarcinoma | [181] |
| EGR1 | Overexpression of heparanase increased Egr1 mRNA expression | Modulation of EGR gene expression | [188] |
| EGR2 | Overexpression of heparanase increased Egr2 mRNA expression | Modulation of EGR gene expression | [188] |
| E-Cadherin | The expression of Epithelial marker E-cadherin was increased in KATO-III gastric cancer cells treated with heparanase inhibitor suramin | EMT/Gastric ring cell adenocarcinoma | [181] |
| Endothelin-1 (ET-1) | Induction of acute kidney injury in heparanase-transgenic mice enhanced the expression of ET-1mRNA. Pre-treatment with heparanase inhibitior PG545 reduced the expression of ET-1 | EMT/Acute kidney injury | [179] |
| FGF/bFGF | Heparanase activated HIF1 pathway which led to reduced mRNA expression level of bFGF in heparanase knockdown cells and elevated mRNA expression level of bFGF in heparanase overexpressing-cells | Cervical cancer | [177] |
| FGF-2 | Treatment of human osteoblasts with heparin, a heparanase inhibitor, inhibited mRNA FGF2 expression | Growth of osteoblasts | [180] |
| Fibronectin (FN) | Heparanase-transgenic mice displayed remarkable upregulation of FN during acute kidney injury. Pre-treatment with heparanase inhibitor PG545 abolished the increased expression of FN in heparanase-transgenic mice | EMT/Acute kidney injury | [179] |
| Heparanase-silenced cells showed reduced FN expression; Renal tissue extracts from mice with acute kidney injury treated with Roneparstat showed reduced FN expression | EMT/Acute kidney injury | [182,189] | |
| Hepatocyte growth factor (HGF) | Addition of either recombinant or chemotherapy-generated soluble heparanase increased HGF mRNA expression. Immunodepletion or addition of heparanase inhibitor diminished the increased expression of HGF gene. Upregulation of HGF expression by heparanase was independent of heparanase enzyme activity | Tumor progression | [76,176] |
| HIF-1 | mRNA expression level of HIF1 was reduced in heparanase knockdown cells and increased in heparanase-overexpressing cells | Cervical cancer | [177] |
| HIF-2α | Knockdown of heparanase in HUVEC cells reduced HIF-2α expression | Tumor angiogenesis | [190] |
| IL-1β | HS fragments generated by heparanase activated TLR4, MyD88, and NF-κB to upregulate IL-1β mRNA | Inflammation | [120,152] |
| The expression of IL-1β in macrophages isolated from heparanase-knockout mice was significantly reduced compared to macrophages isolated from wild type mice. Heparanase regulated IL-1β expression through Erk, p38, and JNK signaling pathway | Tumor/Regulation of cytokine expression in macrophage | [153] | |
| Increased expression of IL-1β in heparanase overexpressing transgenic mice with colitis-associated carcinoma | Colitis-associated tumor/Induction of NK-κB activation/Macrophage activation | [131] | |
| Heparanase upregulated the expression of IL-1β in PMA-activated U937 macrophages. Treatment cells with heparanase inhibitor SST0001 reduced IL-1β expression | Acute kidney injury/M1 macrophage polarization | [182] | |
| IL-5 | House dust mite (HDM)-induced allergic inflammation in heparanase deficient mice reduced mRNA expression of IL-5 in lung cells | Allergic inflammation/Recruitment of eosinophils and mucus-secreting airway epithelial cells | [51] |
| IL-6 | HS fragments generated by heparanase activated TLR4, MyD88, and NF-κB to upregulate IL-6 | Inflammation | [152] |
| The expression of IL-6 in macrophages isolated from heparanase deficient mice was significantly reduced compared to macrophages isolated from wild type mice. Heparanase regulated IL-6 expression through Erk, p38, and JNK signaling pathways | Tumor/Regulation of cytokine expression in macrophage | [153] | |
| IL-6 mRNA expression was increased in heparanase transgenic mice with DSS-inducedcolitis. LPS-treated mouse peritoneal macrophages increased mRNA expression of IL-6 in the presence of recombinant enzymatically active heparanase | Ulcerative colitis/Induction of NK-κB activation/Macrophage recruitment and activation | [131] | |
| Induction of acute kidney injury in heparanase-transgenic mice enhanced the expression of mRNA IL-6. Pre-treatment with heparanase inhibitior PG545 reduced the expression of IL-6 | EMT/Acute kidney injury | [179] | |
| Heparanase upregulated the expression of IL-6 in PMA-activated U937 macrophage cells. Treatment of cells with heparanase inhibitor SST0001 reduced IL-6 expression | Acute kidney injury/M1 macrophage polarization | [182] | |
| Heparanase induced the expression of IL-6 by fatty acid-stimulated macrophages in a dose-dependent manner | Obesity-associated breast cancer | [103] | |
| IL-6 expression was increased in heparanase-knockout macrophages treated with exogenous heparanase and chemotherapy | Tumor Growth/Induction of pro-inflammatory cytokine expression by chemotherapy-treated macrophage | [123] | |
| IL-8 | HS fragments generated by heparanase activated TLR4, MyD88, and NF-κB to upregulate IL-8 | Inflammation | [152,183] |
| IL-10 | IL-10 mRNA expression was reduced in chemotherapy-treated macrophages isolated from heparanase knockout mice | Tumor Growth/Induction of pro-inflammatory cytokine expression by chemotherapy-treated macrophage | [123] |
| HS fragments generated by heparanase activated TLR4, MyD88, and NF-κB to upregulate IL-10 | Inflammation | [152] | |
| The expression of IL-10 in macrophages isolated from heparanase deficient mice was significantly reduced compared to macrophages isolated from wild type mice. Heparanase regulated IL-10 expression through Erk, p38, and JNK signaling pathway | Tumor/Regulation of cytokine expression in macrophage | [153] | |
| Inhibition of heparanase with SST0001 reduced IL-10 mRNA expression in macrophages | Acute kidney injury/M1 macrophage polarization | [182] | |
| IL-13 | (HDM-induced allergic inflammation in heparanase deficient mice reduced mRNA expression of IL-13 in lung cells | Allergic inflammation/Recruitment of eosinophils and mucus-secreting airway epithelial cells | [51] |
| IL-12p53 | LPS-treated mouse peritoneal macrophages increased mRNA expression of IL-12p53 in the presence of recombinant enzymatically active heparanase | Ulcerative colitis/Macrophage activation | [131] |
| IL-17A | Silencing of heparanase resulted in a significant decrease in the mRNA expression of IL-17A in human cervical cancer cell lines HeLa and SiHa | Promoting tumor angiogenesis, cell proliferation, and invasion in cervical cancer | [84] |
| Inducible nitric oxide synthase (iNOS) | Heparanase upregulated the expression of iNOS in PMA-activated U937 cells. Treatment of cells with heparanase inhibitor SST0001 suppressed iNOS expression | Acute kidney injury/M1 macrophage polarization | [182] |
| Lysozyme 1 | Heparanase-knockout mice showed less lysozyme 1 expression | Tumor/Macrophage cytotoxic activity is decreased in the absence of heparanase | [153] |
| MCP-1/CCL-2 | Non-enzymatic heparanase in colorectal cancer cell lines could upregulate the expression of MCP-1 | Promoting extravasation of colon carcinoma cells | [183] |
| MIP-2 (CXCL2) | Macrophages isolated from heparanase-deficient mice and mice treated with heparanase-neutralizing antibodies exhibited reduced MIP-2 expression | Tumor/Regulation of cytokine expression in macrophage | [153] |
| MIP-2 mRNA expression was reduced in chemotherapy-treated macrophages isolated from heparanase knockout mice | Tumor Growth/Induction of pro-inflammatory cytokine expression by chemotherapy-treated macrophage | [123] | |
| Mixed Lineage Kinase Domain Like Pseudokinase (MLKL) | Transwell co-culture of heparanase-silenced hepatocellular carcinoma (HCC) cells with HUVECs protected HUVECs from MLKL mRNA and protein upregulation and necroptosis In transwell co-cultures of heparanase-overexpressing HCC cells and HUVECs, HUVECs displayed higher MLKL protein expression after co-culture compared to controls | Necroptosis | [191] |
| matrix metalloprotease-2 (MMP-2) | The mRNA expression of MMP-2 was decreased in the kidney of heparanase deficient mice | Allergen-induced inflammation/DC migration | [51] |
| Human melanoma cells deficient in heparanase exhibited increased MMP-2 expression | Melanoma progression | [169] | |
| Inhibiting heparanase with either PG545 or PI-88 in patient-derived explants of normal mammary tissue increased MMP-2 mRNA expression | Tissue density and breast cancer | [192] | |
| siRNA knockdown of heparanase in SUM149 breast cancer cells reduced MMP-2 mRNA expression | Breast cancer | [83] | |
| MMP-9 | Addition of recombinant or chemotherapy-generated soluble heparanase elevated the expression of MMP-9 in myeloma cells. Chemotherapeutic induction of MMP-9 required heparanase through Erk phosphorylation | Tumor progression | [76,79,184] |
| The gene expression level of MMP-9 in heparanase-silenced human kidney 2 (HK2) cells was lower than wild type cells | Renal fibrosis | [193] | |
| Heparanase upregulated the expression of MMP-9 by its HS-degrading activity and stimulating HAT activity | Myeloma tumor/Upregulation of HAT activity | [168] | |
| Human melanoma cells deficient in heparanase exhibited increased MMP-9 expression | Melanoma progression | [169] | |
| MMP-14 | The mRNA expression of MMP-14 was decreased in the liver of heparanase deficient mice | Allergen-induced inflammation/DC migration | [51] |
| Inhibiting heparanase with either PG545 or PI-88 in patient-derived explants of normal mammary tissue increased MMP-14 mRNA expression | Tissue density and breast cancer | [192] | |
| MMP-25 | The mRNA expression of MMP25 was increased in the spleen but decreased in mouse bone marrow-derived DCs and Langerhans cells from heparanase deficient mice | Allergen-induced inflammation/DC migration | [51] |
| NANOG | mRNA expression of NANOG was decreased in KATO-III gastric cancer cells treated with heparanase inhibitor suramin | EMT/Gastric ring cell adenocarcinoma | [181] |
| OCT3/4 | mRNA expression of OCT3/4 was decreased in KATO-III gastric cancer cells treated with heparanase inhibitor suramin | EMT/Gastric ring cell adenocarcinoma | [181] |
| P21 | Heparanase downregulated p21 in colon carcinoma cells through its enzymatic activity and involved TLRs and NF-κB signaling | Colon carcinoma/Modification of cell cycle | [194] |
| P38 | In transwell co-cultures of heparanase-silenced HCC cells and HUVECs, HUVECs displayed lower p38 mRNA and phosphorylated protein expression after co-culture compared to controls | Necroptosis | [191] |
| PDK2 | Nuclear heparanase regulated the mRNA expression of PDK2 through HAT activation. Depletion of heparanase reduced the expression of PDK2 mRNA | Glucose metabolism | [186] |
| Pentraxin 3 (PTX-3) | Human melanoma cells deficient in heparanase exhibited increased PTX-3 expression | Melanoma progression | [169] |
| Receptor interacting protein kinase 1 (RIPK1) and RIPK3 | Transwell co-cultures of heparanase-silenced HCC cells with HUVECs protected HUVECs from RIPK1 and RIPK3 mRNA and protein upregulation and necroptosis. In transwell co-cultures of heparanase-overexpressing HCC cells and HUVECs, HUVECs displayed higher RIPK1 and RIPK3 protein expression after co-culture compared to controls | Necroptosis | [191] |
| SDF-1 (CXCL-12) | SDF-1 expression was reduced in heparanase-deficient macrophages. Heparanase regulated SDF-1 expression through Erk, p38, and JNK signaling pathway | Tumor/Promoting phagocytic capacity of macrophages | [153] |
| SERPINE1 | Heparanase regulated HAT activity, leading to upregulation of SERPINE1 | Inflammation | [186] |
| Slug | KATO-III gastric cancer cells treated with heparanase inhibitor suramin reduced Slug mRNA expression | EMT/Gastric ring cell adenocarcinoma | [181] |
| α-SMA | Treatment of KATO-III gastric cancer cells with heparanase inhibitor suramin exhibited reduced expression of α-SMA | EMT/Gastric ring cell adenocarcinoma | [181] |
| Heparanase-overexpressing micedisplayed remarkable upregulation of α-SMA during acute kidney injury. Pre-treatment with heparanase inhibitor PG545 abolished the increased expression of α-SMA in hpse-tg mice | EMT/Acute kidney injury | [179] | |
| Heparanase-silenced cells showed reduced α-SMA expression | EMT/Acute kidney injury | [182,189] | |
| Snail | Heparanase-silenced cells showed reduced Snail expression | EMT/Acute kidney injury | [182] |
| Syndecan-1 | Inhibiting heparanase with either PG545 or PI-88 in patient-derived explants of normal mammary tissue reduced syndecan-1 mRNA expression | Tissue density and breast cancer | [192] |
| In transwell co-cultures of heparanase-silenced HCC cells and HUVECs, HUVECs displayed lower syndecan-1 mRNA and protein expression after co-culture compared to controls. In transwell co-cultures of heparanase-overexpressing HCC cells and HUVECs, HUVECs displayed higher syndecan-1 mRNA and protein expression after co-culture compared to controls | Necroptosis | [191] | |
| Tissue factor (TF) | mRNA expression levels of TF were elevated in heparanase transfected breast carcinoma cells and transgenic mice over-expressing heparanase. Exogenous addition of heparanase also induced TF expression in human promyelocytic leukemia cells. Heparanase induced TF expression via inducing p38 signaling non-enzymatically | Blood coagulation | [195] |
| Human melanoma cells deficient in heparanase exhibited increased TF expression | Melanoma progression | [169] | |
| Transforming growth factor (TGF)-β/TGFβ1 | Gene expression levels of TGF-β was decreased in the heparanase-silenced tubular cells | EMT/Renal fibrosis | [178,189] |
| Induction of acute kidney injury in heparanase-transgenic mice enhanced the expression of TGF-β mRNA. Pre-treatment with heparanase inhibitior PG545 abolished the elevation in TGF-β | EMT/Acute kidney injury | [179] | |
| Heparanase inhibitor suramin down-regulated TGFβ-1 expression in KATO-III gastric cancer cells | EMT/Gastric ring cell adenocarcinoma | [181] | |
| TLR-2 | The expression of TLR-2 in macrophages isolated from heparanase deficient mice and in macrophages isolated from mice treated with heparanase-neutralizing antibodies was significantly reduced. Heparanase regulated TLR2 expression through Erk, p38, and JNK signaling pathway | Tumor/Macrophage activation and function in tumorigenesis | [153] |
| TLR-4 | The expression of TLR-4 on macrophages was upregulated in the presence of heparanase but was reduced when cells were treated with heparanase inhibitor SST0001 | Acute kidney injury/Regulation of macrophage polarization | [182] |
| TNF-α | TNF-α expression was reduced in macrophages isolated from heparanase-knockout mice and in macrophages isolated from mice treated with heparanase-neutralizing antibodies. Heparanase regulated TNF-α expression through Erk, p38, and JNK signaling pathway | Tumor/Macrophage activation and function in tumorigenesis | [153] |
| Heparanase overexpressing transgenic mice expressed more TNF-α during DSS-induced colitis through NF-κB signaling. LPS-treated mouse peritoneal macrophages increased mRNA expression of TNF-α in the presence of recombinant enzymatically active heparanase | Ulcerative colitis/Induction of NK-κB activation/Macrophage recruitment and activation | [131] | |
| Induction of acute kidney injury in heparanase-transgenic mice enhanced the expression of TNF-α mRNA. Pre-treatment with heparanase inhibitior PG545 reduced the expression of TNF-α | EMT/Acute kidney injury | [179] | |
| Human melanoma cells deficient in heparanase exhibited increased TNF-α expression | Melanoma progression | [169] | |
| Heparanase upregulated the expression of TNF-α in PMA-activated U937 macrophage cells. Treatment of cells with heparanase inhibitor SST0001 reduced TNF-α expression | Acute kidney injury/M1 macrophage polarization | [182] | |
| HS fragments generated by heparanase activated TLR4, MyD88, and NF-κB to upregulate TNF-α | Inflammation | [152] | |
| TNF-α mRNA expression was reduced in chemotherapy-treated macrophages isolated from heparanase knockout mice In transwell co-cultures of heparanase-silenced HCC cells and HUVECs, HUVECs displayed lower TNF-α mRNA and protein expression compared to controls | Tumor Growth/Induction of pro-inflammatory cytokine expression by chemotherapy-treated macrophage | [123] | |
| In transwell co-cultures of heparanase-overexpressing HCC cells and HUVECs, HUVECs displayed higher TNF-α mRNA and protein expression after co-culture compared to controls | Necroptosis | [191] | |
| TNF-α receptor (TNFR) | In transwell co-cultures of heparanase-silenced HCC cells and HUVECs, HUVECs displayed lower TNFR mRNA and protein expression compared to controls | Necroptosis | [191] |
| TNFR-associated death domain protein (TRADD) | In transwell co-cultures of heparanase-silenced HCC cells and HUVECs, HUVECs displayed lower TRADD mRNA and protein expression after co-culture compared to controls | Necroptosis | [191] |
| Vascular cell adhesion molecule 1 (VCAM-1) | Heparanase regulated HAT activity, leading to upregulation of VCAM-1 | Inflammation | [186] |
| VEGF | Heparanase overexpression or exogenous addition led to the enhanced expression of VEGF. Heparanase regulated the expression of VEGF by mediating the activation of SRC family members | Promoting angiogenesis in tumor | [175] |
| Heparanase upregulated the expression of VEGF through its HS-degrading activity and stimulating the HAT activity | Tumor phenotype/Upregulation of HAT activity | [168] | |
| Heparanase regulated the expression of VEGF via activating HIF1 pathway | Cervical cancer | [177] | |
| Addition of recombinant or chemotherapy-generated soluble heparanase elevated the expression of VEGF in myeloma | Tumor progression | [76] | |
| Heparanase overexpression in melanoma cell lines increased the expression of VEGF mRNA. Downregulation of heparanase via anti-heparanase siRNA transfection resulted in a significant reduction of VEGF mRNA expression in melanoma cell lines | Melanoma progression | [92] | |
| VEGF-A | Reduced VEGF-A expression was observed in macrophages isolated from heparanase-knockout mice and in macrophages isolated from mice treated with heparanase-neutralizing antibodies | Tumor/Macrophage activation and function in tumorigenesis | [153] |
| Heparanase regulated HAT activity, leading to upregulation of VEGF-A | Atherosclerosis/Glucose Metabolism | [186] | |
| VEGF-C | Overexpression of heparanase increased VEGF-C mRNA expression | Pancreatic cancer/Facilitating cell invasion | [185] |
| Vimentin | KATO-III gastric cancer cells exhibited reduced Vimentin expression after treating with heparanase inhibitor suramin | EMT/Gastric ring cell adenocarcinoma | [181] |
| Heparanase-overexpressing mice displayed remarkable upregulation of vimentin during acute kidney injury. Pre-treatment with heparanase inhibition abolished the increased expression of vimentin in heparanase-overexpressing mice. | EMT/Acute kidney injury | [179] | |
| Heparanase-silenced cells reduced vimentin expression | EMT/Acute kidney injury | [189] | |
| WDR5 | Upon paclitaxel treatment, WDR5 expression was induced in wild type but not heparanase-knockout macrophages, but could be rescued with exogenous heparanase Heparanase was required for the expression of WDR5 in macrophages | Tumor Growth/Induction of pro-inflammatory cytokine expression by chemotherapy-treated macrophage | [123] |
| Proteins | |||
| bFGF | bFGF protein expression was decreased in heparanase knockdown cell and increased in heparanase overexpressing cells via activating HIF1 pathway | Cervical cancer | [177] |
| BLC | BLC expression was reduced in macrophages isolated from heparanase-knockout mice | Tumor/Macrophage activation and function in tumorigenesis | [153] |
| Caspase-1 | Heparanase-silenced and heparanase inhibitor SST0001-treated cells reduced caspase-1 expression | Acute kidney injury/M1 macrophage polarization | |
| Cox-2 | Heparanase upregulated the mRNA expression of Cox-2 in cancer cells | Tumor/Promoting angiogenesis | [187] |
| CXCL1 (KC) | Administration of heparanase increased CXCL1 level in mouse serum | Thoracoabdominal aortic aneurysm/Systemic Inflammation | [196] |
| CXCL1 expression was reduced in macrophages isolated from heparanase-knockout mice | Tumor/Macrophage activation and function in tumorigenesis | [153] | |
| Heparanase-stimulated colon cancer cells released CXCL1 | Colon cancer | [183] | |
| FGF21 | Heparanase-overexpressing mice had higher FGF21 expression in the blood plasma compared to wild type mice | Diabetes/Glucose homeostasis | [197] |
| Fibrinogen | High dose heparanase-derived peptides induced a decrease in the level of fibrinogen | Coagulopathy and wound healing/Activation of the coagulation system | [198] |
| Fibronectin | Protein expression of fibronectin was increased in heparanase-overexpressing mice with acute kidney injury but decreased when pre-treating the mice with heparanase inhibitor PG545 | EMT/Acute kidney injury | [179] |
| FXa | Heparanase-derived peptides enhanced the level of FXa probable through interacting with TF | Coagulopathy and wound healing/Activation of the coagulation system | [198] |
| Hepatocyte growth factor (HGF) | Addition of soluble heparanase or increased heparanase expression upregulated HGF expression in myeloma cell lines. Knockdown of heparanase reduced HGF expression | Tumor progression | [76,176] |
| HIF1 | HIF1 protein expression was decreased in heparanase knockdown cells and increased in heparanase overexpressing cells via HIF1 pathway | Cervical cancer | [177] |
| ICAM-1 | ICAM-1 expression was significantly increased in heparanase overexpressing human breast cancer cell lines. Likewise, the expression of ICAM-1 was decreased in heparanase-knockout cell lines | Cancer metastasis/Promotion of cell cluster formation by modulating adhesion molecules | [197] |
| IL-1 | Addition or overexpression of heparanase upregulated the expression of IL-1 | Atherosclerosis/Macrophage activation | [199] |
| IL-1β | Administration of heparanase increased IL-1β level in mouse serum | Thoracoabdominal aortic aneurysm/Systemic Inflammation | [196] |
| Heparanase upregulated the expression of IL-1β in macrophages. Treatment of cells with heparanase inhibitor SST0001 reduced IL-1β expression | Acute kidney injury/M1 macrophage polarization | [182] | |
| Heparanase via its enzymatic activity upregulated IL-1β through TLR4 signaling | Inflammation | [152] | |
| IL-4 | IL-4 expression was reduced in lung cells isolated from heparanase deficient mice with HDM-induced allergic inflammation | Allergic inflammation/Recruitment of eosinophils and mucus-secreting airway epithelial cells | [51] |
| Administration of heparanase upregulated the expression of IL-4 in mouse immune cells | Autoimmune encephalitis/inhibition of inflammation | [200] | |
| IL-5 | IL-5 expression was reduced in lung cells isolated from heparanase deficient mice with HDM-induced allergic inflammation | Allergic inflammation/Recruitment of eosinophils and mucus-secreting airway epithelial cells | [51] |
| IL-6 | Administration of heparanase increased IL-6 level in mouse serum | Thoracoabdominal aortic aneurysm/Systemic Inflammation | [196] |
| Heparanase via its enzymatic activity upregulated IL-6 through TLR4 signaling | Inflammation | [152] | |
| Administration of heparanase upregulated the expression of IL-6 in mouse immune cells | Autoimmune encephalitis/Inhibition of inflammation | [200] | |
| Addition of heparanase enhanced the expression of IL-6 in fatty acid-stimulated macrophages | Obesity-associated breast cancer | [103] | |
| IL-8 | Heparanase enhanced IL-8 expression | Colon cancer | [183] |
| Heparanase upregulated IL-8 expression via its enzymatic activity | Inflammation | [152] | |
| IL-10 | Administration of heparanase increased IL-10 level in mouse serum | Thoracoabdominal aortic aneurysm/Systemic Inflammation | [196] |
| Heparanase upregulated IL-10 expression via its enzymatic activity | Inflammation | [152] | |
| Administration of heparanase upregulated the expression of IL-10 in mouse immune cells | Autoimmune encephalitis/Inhibition of inflammation | [200] | |
| IL-12 | Administration of heparanase downregulated the expression of IL-12 in mouse immune cells | Autoimmune encephalitis/Inhibition of inflammation | [200] |
| IL-17A | Silencing of heparanase resulted in a significant decrease in protein expression of IL-17A in human cervical cancer cell lines HeLa and SiHa | Promoting tumor angiogenesis, cell proliferation, and invasion in cervical cancer | [84] |
| iNOS | Heparanase upregulated the expression of iNOS in macrophages. Treatment of cells with the heparanase inhibitor SST0001 reduced iNOS expression | Acute kidney injury/M1 macrophage polarization | [182] |
| MCP-1 | Addition or overexpression of heparanase upregulated the expression of MCP-1 | Atherosclerosis/Macrophage activation Thoracoabdominal aortic aneurysm/Systemic | [199] |
| Administration of heparanase increased MCP-1 level in mouse serum | Inflammation | [196] | |
| Heparanase-stimulated colon cancer cells released MCP-1 | Colon cancer | [183] | |
| Heparanase upregulated MCP-1 via TLR4 signaling | Inflammation | [152] | |
| Obese heparanase knockout mice showed less MCP-1 expression compared to obese wild type mice | Obesity-associated breast cancer progression | [103] | |
| MIP-2 (CXCL2) | MIP-2 expression was reduced in macrophages isolated from heparanase-knockout mice | Tumor/Macrophage activation and function in tumorigenesis | [153] |
| MMP-9 | Addition or overexpression of heparanase upregulated the expression of MMP-9 | Atherosclerosis/Macrophage activation | [199] |
| NF-κB (p65) | Knockdown of heparanase led to increased expression of nuclear NF-κB in melanoma cell lines | Melanoma progression | [169] |
| P21 | Heparanase downregulated p21 in colon carcinoma cells through its enzymatic activity and involved TLRs and NF-κB signaling | Colon carcinoma/Modification of cell cycle | [194] |
| α-SMA | Protein expression of α-SMA was increased in heparanase-overexpressing mice with acute kidney injury but decreased when pre-treating the mice with heparanase inhibitor PG545 | EMT/Acute kidney injury | [179] |
| TLR2 | Heparanase knockout cells expressed less TLR2 protein | Tumor/Macrophage activation and function in tumorigenesis | [153] |
| TNF-α | TNF-α expression was reduced in macrophages isolated from heparanase-knockout mice | Tumor/Macrophage activation and function in tumorigenesis | [153] |
| Increased expression of TNF-α in heparanase overexpressing transgenic mice with DSS-induced colitis | Ulcerative colitis/Induction of NK-κB activation | [131] | |
| Addition or overexpression of heparanase increased the expression of TNF-α | Atherosclerosis/Macrophage activation | [199] | |
| Heparanase upregulated TNF-α via TLR4 signaling. Heparanase deficiency reduced the expression of TNF-α in macrophages | Inflammation Obesity-associated breast cancer progression | [103,152] | |
| VEGF | VEGF protein expression was decreased in heparanase knockdown cells and increased in heparanase overexpressing cells via activating the HIF1 pathway | Cervical cancer | [177] |
| Heparanase overexpression led to the enhanced expression of VEGF. Heparanase regulated the expression of VEGF by mediating the activation of SRC | Tumor vascularity | [175] | |
| VEGF expression was increased in heparanase overexpressing melanoma cell lines and decreased in heparanase downregulated cells | Melanoma progression | [92] | |
| Vimentin | Protein expression of vimentin was increased in heparanase-overexpressing mice with acute kidney injury but decreased when pre-treating the mice with heparanase inhibitor PG545 | EMT/Acute kidney injury | [179] |
| Protein | Observation/Mechanism | Related Disease/Function | Reference |
|---|---|---|---|
| AKT | Inhibition of heparanase reduced AKT phosphorylation | Breast Cancer Brain Metastasis | [79] |
| High expression of heparanase in myeloma cell lines led to increased AKT phosphorylation. This was blocked by treating cells with heparanase inhibitor SST0001 | Tumor progression | [76] | |
| Epidermal growth factor receptor (EGFR) | Inhibition of heparanase reduced EGFR phosphorylation | Breast Cancer Brain Metastasis | [79] |
| Heparanase enhanced the phosphorylation level of EGFR in carcinoma cells | Tumor progression | [205] | |
| Heparanase released HS via shedding syndecan-1 which induced EGFR phosphorylation | Colorectal cancer | [151] | |
| ERK | The level of phosphorylated ERK was increased in heparanase overexpressing neural stem and progenitor cells during differentiation | Promoting Embryonic stem cell differentiation into Oligodendrocytes | [207] |
| Addition of exogenous heparanase induced ERK phosphorylation in macrophages | Inducing cytokine expression in macrophage | [153] | |
| High expression of heparanase in myeloma cell lines led to increased ERK phosphorylation. The increased phosphorylation of ERK was blocked in cells treated with heparanase inhibitor SST0001 | Tumor progression | [76] | |
| Focal-adhesion kinase (FAK) | The phosphorylation of FAK was elevated in heparanase-overexpressing breast cancer cell lines. Likewise, the phosphorylation of FAK was decreased in heparanase-knockout cell lines. Heparanase promoted cell cluster formation by regulating FAK-Src-paxillin pathway | Promotion of cell cluster formation/Tumor metastasis | [197] |
| IκBα/IκB | Heparanase enhanced phosphorylation of IκBα in heparanase overexpressing mice suffering colitis-associated tumors | Ulcerative colitis/Induction of NK-κB activation | [131] |
| IκB phosphorylation was decreased in pancreas tissues of heparanase-overexpressing mice treated with heparanase inhibitor PG545 | Acute pancreatitis | [127] | |
| JNK | Addition of exogenous heparanase induced JNK phosphorylation in macrophages | Inducing cytokine expression in macrophage | [153] |
| JNK phosphorylation was decreased in macrophages isolated from heparanase knockout mice | Tumor Growth/Induction of pro-inflammatory cytokine expression by chemotherapy-treated macrophage | [123] | |
| MEK | Heparanase induced MEK phosphorylation via releasing HS of syndecan-1 | Colorectal cancer | [151] |
| p38 | Addition of exogenous heparanase enhanced p38 phosphorylation in macrophages | Inducing cytokine expression in macrophage | [153] |
| Heparanase-overexpressing cells induced p38 phosphorylation | Promoting tumor angiogenesis | [175] | |
| p65 NF-κB | Increased nuclear p65 phosphorylation was detected in heparanase overexpressing mice treated with DSS to induce colitis-associated tumors | Ulcerative colitis/Induction of NK-κB activation | [131] |
| Paxillin | The phosphorylation of paxillin was elevated in heparanase-overexpressing breast cancer cell lines. In contrast, the phosphorylation of paxillin was decreased in heparanase-knockout cell lines. Heparanase promoted cell cluster formation by regulating FAK-Src-paxillin pathway | Promotion of cell cluster formation/Tumor metastasis | [197] |
| SRC | The phosphorylation of SRC was increased in heparanase-overexpressing breast cancer cell lines. In contrast, the level of SRC phosphorylation was decreased in heparanase-knockout cell lines. Heparanase promoted cell cluster formation by regulating FAK-Src-paxillin pathway | Promotion of cell cluster formation/Tumor metastasis | [197] |
| Inactive heparanase stimulated SRC phosphorylation | Tumor angiogenesis | [175] | |
| Heparanase enhanced the phosphorylation level of SRC in carcinoma cells | Tumor progression | [205] | |
| Signal Transducer and Activator of Transcription (STAT) | Heparanase increased nuclear STAT phosphorylation | Tumor progression | [205] |
| STAT3 | Higher number of cells positive for nuclear-localized pSTAT3 were observed in heparanase-overexpressing transgenic mice | Modulator of tumor-promoting chronic inflammation | [131] |
| Heparanase enhanced STAT3 phosphorylation | Tumor progression | [205] | |
| Reduced STAT3 phosphorylation was observed in obese heparanase knockout mice | Obesity-associated breast cancer progression | [103] | |
| STAT5b | Heparanase enhanced STAT5b phosphorylation | Tumor progression | [205] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mayfosh, A.J.; Nguyen, T.K.; Hulett, M.D. The Heparanase Regulatory Network in Health and Disease. Int. J. Mol. Sci. 2021, 22, 11096. https://doi.org/10.3390/ijms222011096
Mayfosh AJ, Nguyen TK, Hulett MD. The Heparanase Regulatory Network in Health and Disease. International Journal of Molecular Sciences. 2021; 22(20):11096. https://doi.org/10.3390/ijms222011096
Chicago/Turabian StyleMayfosh, Alyce J., Tien K. Nguyen, and Mark D. Hulett. 2021. "The Heparanase Regulatory Network in Health and Disease" International Journal of Molecular Sciences 22, no. 20: 11096. https://doi.org/10.3390/ijms222011096
APA StyleMayfosh, A. J., Nguyen, T. K., & Hulett, M. D. (2021). The Heparanase Regulatory Network in Health and Disease. International Journal of Molecular Sciences, 22(20), 11096. https://doi.org/10.3390/ijms222011096






