Abstract
The conversion of adenosine to inosine in RNA editing (A-to-I RNA editing) is recognized as a critical post-transcriptional modification of RNA by adenosine deaminases acting on RNAs (ADARs). A-to-I RNA editing occurs predominantly in mammalian and human central nervous systems and can alter the function of translated proteins, including neurotransmitter receptors and ion channels; therefore, the role of dysregulated RNA editing in the pathogenesis of neurological diseases has been speculated. Specifically, the failure of A-to-I RNA editing at the glutamine/arginine (Q/R) site of the GluA2 subunit causes excessive permeability of α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptors to Ca2+, inducing fatal status epilepticus and the neurodegeneration of motor neurons in mice. Therefore, an RNA editing deficiency at the Q/R site in GluA2 due to the downregulation of ADAR2 in the motor neurons of sporadic amyotrophic lateral sclerosis (ALS) patients suggests that Ca2+-permeable AMPA receptors and the dysregulation of RNA editing are suitable therapeutic targets for ALS. Gene therapy has recently emerged as a new therapeutic opportunity for many heretofore incurable diseases, and RNA editing dysregulation can be a target for gene therapy; therefore, we reviewed neurological diseases associated with dysregulated RNA editing and a new therapeutic approach targeting dysregulated RNA editing, especially one that is effective in ALS.
1. Introduction
Amyotrophic lateral sclerosis (ALS), also known as Lou Gehrig’s disease in the USA, is an adult-onset fatal motor neuron disease characterized by the degeneration of the upper (cortical) and lower (spinal and brain stem) motor neurons, resulting in progressive muscular weakness and ultimately death due to respiratory failure []. Approximately 90% of ALS cases are sporadic, with ALS not being present among their blood relatives, and approximately only 10% of the cases are familial. Recent surveillance has identified more than 30 ALS-linked genes, including superoxide dismutase 1 (SOD-1), transactive response DNA/RNA binding protein of 43 kDa (TDP-43), fused in sarcoma (FUS), and chromosome 9 open reading frame 72 (C9ORF72) []. The incidence of ALS is 2 to 3 people per a 100,000-person population and is increasing in developed countries due to an increase in life expectancy. Although various plausible mechanisms such as excitotoxicity due to the dysregulation of glutamatergic signaling, axon transport impairment, neuroinflammation, protein aggregation, or oxidative stress have been proposed as the etiology of ALS, no one has been successful in elucidating the mechanisms underlying the molecular and morphological changes in the degenerating motor neurons of ALS patients [,]. Although two drugs, riluzole and edaravone, have been approved for the treatment of ALS, neither can significantly extend the lives of patients [,].
Neurons in the central nervous system (CNS) use glutamate, an excitatory neurotransmitter. Depolarization of pre-synaptic neurons (upper motor neurons) leads to the vesicular release of glutamate into the synaptic cleft; after secretion from the axon terminal of the upper motor neurons, glutamate activates the post-synaptic neurons (lower motor neurons) through specific membrane glutamate receptors []. Glutamate receptors (GluRs) are classified into two major receptor families: ionotropic GluRs and metabotropic GluRs (mGluRs) []. Ionotropic GluRs are ligand-gated ion channels that are composed of four subunits that mediate an immediate influx of extracellular Na+ and/or Ca2+, regulating membrane depolarization and diverse signal transduction events []. Alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptors regulate the membrane potential and mediate the vast majority of fast excitatory neurotransmission, whereas N-methyl-D-aspartate (NMDA) receptors have a voltage-dependent ion channel that mediates excitatory neurotransmission by passing large amounts of Ca2+, Na+, and K+ during depolarization [,,]. In contrast, mGluRs are G protein-coupled receptors and are subclassified into three groups based on sequence homology. Groups I, II, and III include mGluR1 and mGluR5; mGluR2 and mGluR3; and mGluR4, mGluR6, mGluR7, and mGluR8, respectively. In pre-synaptic neurons, Group I mGluRs promote glutamate release, whereas Group II and III mGluRs inhibit glutamate release. In post-synaptic neurons, mGluRs act relatively slowly compared to ionotropic GluRs, and Group I mGluRs increase intracellular Ca2+ influx via G protein signaling cascades [,].
Excitatory signals are terminated by the uptake of glutamate released into the surrounding neurons and/or astrocytes through the glutamate transporters and excitatory amino acid transporters (EAATs) [,]. Five members, EAAT1 to EAAT5, are known in the EAAT family: EAAT1 or GLAST is the major glutamate transporter and is primarily expressed in the Bergmann glial cells of the cerebellar cortex; EAAT2, or GLT-1, is responsible for more than 90% of glutamate uptake and is expressed primarily in the astrocytes of the cerebral cortex and hippocampus; EAAT3, or excitatory amino acid carrier 1 (EAAC1), is primarily expressed in the pre-synaptic neurons of the cerebral cortex and basal ganglia, while EAAT4 and EAAT5 are low-capacity glutamate transporters expressed in the post-synaptic dendrites of the Purkinje cells of the cerebellar cortex and the amacrine cells of the retina, respectively []. These transporters rapidly re-uptake the released glutamate, thereby maintaining its concentration in the synaptic cleft at sufficiently low levels.
Exaggerated activation of the glutamatergic system leads to the hyperexcitation of neurons and ultimately excitotoxicity. Elevated glutamate levels in the post-mortem tissue and cerebrospinal fluid of ALS patients [,,,] and the loss of high-affinity glutamate uptake [] makes excitotoxicity an attractive therapeutic target for ALS. Although an earlier trial using branched-chain amino acids, a modifier of glutamate metabolism, turned out to be ineffective [], riluzole, an inhibitor of glutamate release, was approved for use as an ALS drug. Riluzole improved one-year survival rates and progressive muscle weakness [,,], especially in the late stages of ALS []. However, the effects of riluzole are limited. The one-year survival was only 9%, and the median survival was prolonged by only 2–3 months []. In addition, studies using transcranial magnetic stimulation (TMS) or threshold tracking nerve conduction studies (TTNCSs) have shown cortical and spinal motor neuron hyperexcitability in sporadic and familial ALS patients, and cortical hyperexcitability due to increased glutamatergic drive has been hypothesized [,,,]. Ezogabine, an activator of Kv7 potassium channels, reduced neuronal excitability and improved the in vitro survival of differentiated motor neurons from induced pluripotent stem cells (iPSCs) derived from ALS patients and decreased cortical and spinal motor neuron excitability, as evaluated by TMS and TTNCSs in ALS patients [,]. However, it is unclear whether a reduction in cortical hyperexcitability prolongs survival in patients with ALS. Other potential ALS drugs targeting neuronal hyperactivity or excitotoxicity, including memantine, a non-competitive antagonist of NMDA receptors, lamotrigine, which inhibits glutamate release and inactivates voltage-gated calcium channels, and talampanel, a non-competitive antagonist of AMPA receptors, were found to be unsuccessful in clinical trials involving patients with ALS [,]. Therefore, different therapeutic approaches are needed to effectively prevent neuronal death in ALS.
Mechanistically, exaggerated Ca2+ influx through glutamate receptors plays a pivotal role in excitotoxicity []. Ca2+ plays an important role in physiological neuronal functions, including those related to synaptic plasticity, presynaptic transmitter release, and postsynaptic responses as a second messenger; however, exaggerated Ca2+ influx results in brain damage (Figure 1) [,]. Exaggerated activation of NMDA receptors is involved in rapid cell death in diseases such as epilepsy and encephalitis [,]. Though most AMPA receptors do not mediate large Ca2+ influx, a small proportion are Ca2+-permeable, and these AMPA receptors mediate the slow death of motor neurons, which is reminiscent of the death of motor neurons in ALS [,].
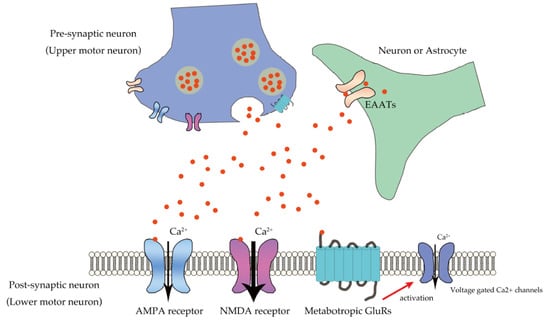
Figure 1.
Ca2+ influx in lower motor neurons through glutamatergic neurotransmission. Glutamate secreted from pre-synaptic neuron (upper motor neuron) into the synaptic cleft binds to the glutamate receptors, including the AMPA receptor, NMDA receptor, and mGluRs expressed in the postsynaptic neurons (lower motor neurons). AMPA receptors mediate excitatory neurotransmission by regulating membrane potential, whereas NMDA receptors predominantly mediate large amounts of Ca2+ influx after depolarization, and mGluRs mediate Ca2+ influx via voltage-gated Ca2+ channels. Glutamate transporters, EAATs expressed in neurons or astrocytes, regulate glutamate concentration in the synaptic cleft.
AMPA receptors are comprised of homo- or hetero-tetramers of GluA1, GluA2, GluA3, and GluA4, of which only GluA2 is subjected to adenosine-to-inosine (A-to-I) RNA editing at the glutamine/arginine (Q/R) site [,]. GluA2 is the determinant of the Ca2+ permeability of AMPA receptors, and all of the GluA2 expressed in the neurons is edited at the Q/R site by adenosine deaminase acting on RNA 2 (ADAR2) []. AMPA receptors are Ca2+-impermeable when they contain edited GluA2 in their subunit assembly, whereas AMPA receptors devoid of GluA2 or containing unedited GluA2 are Ca2+-permeable []. Additionally, editing at the Q/R site in GluA2 reduces the AMPA receptor tetramerization and trafficking of GluA2 to the synaptic membrane, suggesting that Q/R site-unedited GluA2 is more prone to being integrated into functional AMPA receptors than edited GluA2 is, enhancing Ca2+ influx through the AMPA receptors []. Moreover, auxiliary subunits of AMPA receptors, such as transmembrane AMPA receptor regulatory protein (TARP) and cornichon family AMPA receptor auxiliary protein 2 (CNIH2), have a regulatory role in Ca2+ influx through AMPA receptors by influencing gate dilation [,].
In this report, we summarized current knowledge regarding the failure of A-to-I RNA editing in neurological diseases. Moreover, as some recent excellent reviews have described the association of RNA editing with neurodegenerative diseases or current and promising gene therapies for neurodegeneration in general [,,,], we focused on ALS-related excitotoxicity due to the dysfunction of glutamatergic signaling resulting from dysregulated RNA editing at the GluA2 Q/R site, which are promising future targets in ALS therapy.
2. RNA Editing and Neurological Diseases
A-to-I RNA editing, or site-specific and post-transcriptional modification of RNA by ADARs, occurs in various classes of RNA, including both protein-coding and non-coding mRNAs, microRNAs (miRNAs), and circular RNAs (circRNAs) []. Among the millions of A-to-I sites, the vast majority reside in the non-coding regions of RNA, including in the 5′- and 3′-untranslated regions (UTRs) and the Alu repetitive elements or short interspersed nuclear element (SINE) sequences [], whereas less than 1 % reside in the coding regions in mammals and humans [,,]. A-to-I sites are the most abundant in the RNAs expressed in the CNS, and several mRNAs that code neuronal ion channels and receptors have A-to-I sites in the coding regions. The editing efficiency varies widely from less than 1% to 100% among different RNAs; at the same, the A-to-I site in the same RNA differs among the various tissues, developmental stages, environmental conditions, and cell types []. A-to-I RNA editing can influence the efficacy of RNA splicing, translation, localization, and the stability of RNA and biogenesis of non-coding RNAs, including miRNA and circRNA [,,]. Moreover, RNA editing within protein-coding sequences can form mRNA that carries a codon that is different from the original DNA, thereby altering protein structure and function [,]. There are three ADAR proteins in humans: ADAR1, ADAR2, and ADAR3, which are are encoded by ADAR, ADARB1, and ADARB2, respectively. ADAR1 and ADAR2 have editing activity in vitro and in vivo, whereas ADAR3 has no natural substrate RNA and has a possible role as a negative regulator of RNA editing by sequestering the editing substrate of ADAR1 and ADAR2 [,,]. ADAR1 is widely expressed across tissues, and ADAR1p110, a 110 kDa isoform protein, localizes primarily in the nucleus, while ADAR1p150, a 150 kDa isoform protein, localizes in the cytoplasm []. ADAR2 is also widely expressed, but its expression level in the CNS is higher than that of ADAR1; ADAR3 is expressed exclusively in the brain, in the glial cells in particular [,,].
ADARs have distinct roles in the development and expression of normal phenotypes in mammals and humans. ADAR1 is crucial for developing non-nervous tissues, including the liver and spleen, and mice lacking ADAR1 are lethal to embryos due to widespread apoptosis []. Mutations in ADAR are principally associated with dyschromatosis symmetrica hereditaria and Aicardi–Goutieres syndrome [,]. In contrast, mice lacking ADAR2 develop normally but die by three weeks of age from recurrent epileptic seizures []. Infants carrying bi-allelic variants of ADARB1, the gene encoding ADAR2, develop either epileptic encephalopathy or microcephaly associated with intellectual disability and seizures [,,]. In addition to the roles in developing tissues, the conditional targeting of ADARB1 in mouse motor neurons led to their slow death [], indicating that A-to-I RNA editing plays an important role in proper neuronal functions in mature CNS []. Therefore, the dysregulation of A-to-I RNA editing has been suggested to be associated with several adult- or adolescent-onset neurological diseases, including ALS, epilepsy, and schizophrenia [,,,,] (Table 1).

Table 1.
CNS diseases linked to RNA editing.
Among the neurological diseases associated with defects in RNA editing, the pathogenic roles of A-to-I RNA editing dysfunction in the GluA2 Q/R site due to the downregulation of ADAR2 have been extensively demonstrated in sporadic ALS, as described in the next section [,]. A comprehensive study on RNA editing in the tissues of post-mortem Alzheimer’s disease (AD) patients reported a reduction in RNA editing activity at various A-to-I sites in the hippocampus, temporal lobe, and frontal lobe [], and neuronal death in AD has been suggested to be associated with disturbance of RNA editing at the GluA2 Q/R site [,]. A comprehensive analysis of RNA editing alterations in glioblastoma or Grade IV glial cell tumors reported significant changes in editing efficiencies at a large number of RNA editing sites. Reduced A-to-I RNA editing at the Q/R site of GluA2 mRNA has a role in the proliferation and migration of glioblastoma cells via the activation of the Akt pathway [,,]. The reduction in GluA2 Q/R site RNA editing in the glioblastoma cells results from the overexpression of ADAR1 and ADAR3, which may inhibit the homodimerization of ADAR2 rather than from a reduction in ADAR2 expression level []. Malignant glioma is also associated with a reduction in RNA editing at various A-to-I sites, including the isoleucine/methionine (I/M) site in gamma-aminobutyric acid type A receptor subunit alpha 3 (GABRA3) [], five editing sites in cell division cycle 14 B (CDC14B) [], the +9 site of miR-336, and the +6 site of miR-589-3p [,].
Another neurological disease associated with deficient RNA editing is brain ischemia, in which an aberrant Ca2+ influx through Q/R site-unedited GluA2 containing Ca2+-permeable AMPA receptors has been suspected to play a crucial role in the neuronal death of CA1 pyramidal neurons in mice [,]. RNA editing at the arginine/glycine (R/G) site in GluA2 mRNA is reduced in the core and penumbra of acute spinal cord injury []. Moreover, the RNA editing at site D in 5-hydroxytryptamine receptor 2C (5-HT2C), a subtype of serotonin receptors, and the isoleucine/valine (I/V) site in Kv1.1 have been reported in injured rat spinal cords []. Although the factors relevant to abnormal RNA editing in spinal cord injury are unclear, these changes in RNA may influence postsynaptic excitatory responses to glutamate, culminating in the modulation of cell death progression [].
Since severe epileptic seizures induce brain damage, the role of aberrant RNA editing in patients with epilepsy has been investigated. Epilepsy is characterized by abnormal neuronal hyperexcitability and affects over 70 million people worldwide []. Although mice expressing Q/R site-unedited GluA2 exhibited recurrent epileptic seizures and early postnatal death [], RNA editing at this site was not reduced in the brains of postmortem patients [,]. In contrast, increased editing efficiency has been recognized at the R/G site in GluA2, the Q/R site in glutamate ionotropic receptor kainite type subunit 1 (GluK1), and the Q/R site in GluK2 in surgically excised brain tissues of patients with epilepsy [,,]. These changes enhance the response to glutamate and modulate neuronal excitability in the mouse brain [,,].
Changes in monoamines such as serotonin and dopamine have been implicated in common psychiatric disorders, including depression and schizophrenia []. Although the influence of changes in editing efficiencies at each of the five editing sites in 5-HT2C were inconsistent among previous reports [,,,,,,], the proportion of site A-edited 5-HT2C was higher, especially in the postmortem brains of suicide victims than in non-suicidal patients with both depression and schizophrenia [,]. Interestingly, editing efficiencies at sites B, C, and E in phosphodiesterase 8A (PDE8A), a key modulator of signal transduction downstream of 5-HT2C, were decreased in the brain and blood of patients with depression [,]. A comprehensive study on the alteration of RNA editing has demonstrated a higher overall RNA editing level and significant changes in editing efficiencies at many editing sites in the brains of patients with schizophrenia [].
Comprehensive studies on alterations in RNA editing have demonstrated changes in editing efficiencies at many editing sites in the brains of patients with autism spectrum disorder, a common neurodevelopmental disorder characterized by social communication deficits and repetitive sensorimotor behaviors []; however, no specific editing site relevant to the pathogenesis could be identified [,]. In addition, studies in mice and rats have shown negative effects of alcohol and/or cocaine abuse/abstinence on the RNA editing efficiency of ADAR2 [,].
3. Excitotoxicity in ALS Due to Excessive Ca2+ Influx through Unedited GluA2-Conaining AMPA Receptors
As described in the introduction section, the role of excitotoxicity, mediated by Ca2+-permeable AMPA receptors, has been proposed in ALS pathogenesis [,,] (Figure 2). AMPA receptors are Ca2+-permeable when they either lack GluA2 or contain Q/R site-unedited GluA2 in their subunit assembly.
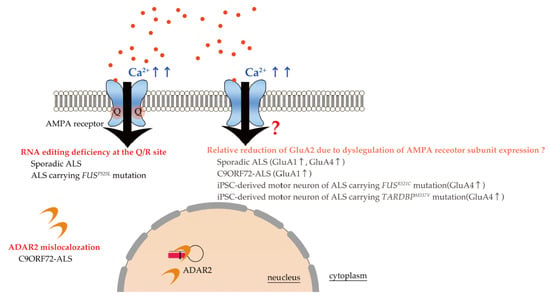
Figure 2.
Excessive Ca2+ influx through Ca2+-permeable AMPA receptors in ALS. Ca2+ permeability of AMPA receptors (arrows) is determined by the presence or absence of Q/R site-edited GluA2 in the subunit assembly. RNA editing deficiency at the Q/R site in GluA2 due to downregulation of ADAR2 has been demonstrated in the motor neurons of patients with sporadic ALS and those carrying the FUSP525L mutation. Furthermore, ADAR2 mislocalization with loss of RNA editing activity has been reported in C9ORF72-ALS patients. Alterations in the expression of various AMPA subunits has been reported in sporadic ALS, C9ORF72 patients, and familial ALS patients carrying the FUSR521C and TARDBPM337V mutations; however, loss of GluA2 does not induce neuronal death in GluA2 knockout mice. Therefore, a relative reduction of GluA2 per se may not be pathological in causing motor neuron death in ALS. Then, the question mark near the arrow was added.
Studies on mutant mice have demonstrated that the expression of unedited GluA2 induces fatal epilepsy in mice [] and that the expression level of unedited GluA2 correlates with the level of Ca2+ influx and the severity of neurological dysfunctions [,,,]. Conversely, a study on GluA2 knockout mice indicated that expression of GluA2-lacking Ca2+-permeable AMPA receptors per se does not cause the death of motor neurons []. Notably, in conditional ADAR2 knockout (ADAR2flox/flox/VAChT.Cre or AR2) mice, the motor neurons undergo slow progressive death when the process of RNA editing at the Q/R site of GluA2 is ablated selectively in these cells []; this suggests that the expression of Q/R site-unedited GluA2 rather than a decreased expression of GluA2 plays a central role in neuronal death, particularly motor neuron death, mediated by Ca2+-permeable AMPA receptors. In the spinal motor neurons of patients with sporadic ALS, the Q/R site-unedited GluA2 is expressed due to downregulation of ADAR2 [,], suggesting the pathogenic role of unedited GluA2 expression in ALS. However, there have been several contradictory reports. The lack of expression of unedited GluA2 in the whole spinal cord tissue of sporadic ALS patients [] may be due to the masking of the small proportion of unedited GluA2 by the abundantly expressed edited GluA2; moreover, the proportions of unedited GluA2 differ markedly (from 0% to 100%) among individual motor neurons in ALS patients when analyzed at the single-cell level [,]. The lack of unedited GluA2 in iPSC-derived motor neurons from sporadic ALS or familial ALS patients carrying FUSH517D mutations [,] may be due to either or both of the following reasons: the considerably shorter life time of iPSC-derived differentiated motor neurons in culture than that of the degenerating motor neurons in ALS patients or the large difference in the cellular environment between the in vitro culture condition and the in vivo microenvironmental conditions surrounding the degenerating motor neurons in patients.
Moreover, the pathogenic role of Q/R site-unedited GluA2 in ALS is demonstrated by its role in TDP-43 pathology, the pathological hallmark of ALS. TDP-43 pathology is observed exclusively in motor neurons lacking ADAR2 in patients with sporadic ALS []. Studies on conditional ADAR2 knockout AR2 mice have demonstrated that TDP-43 mislocalizes from the nucleus to the cytoplasm in a manner that is dependent on increased Ca2+ influx due to the expression of unedited GluA2 in the motor neurons lacking ADAR2 []. Mechanistically, an increase in intracellular Ca2+ levels activates calpain, which cleaves TDP-43 into aggregation-prone fragments; the resultant aggregates progressively enlarge with continued calpain activation caused by elevated intracellular Ca2+ resulting from the expression of Ca2+-permeable AMPA receptors [,]. These results suggest that progressive ADAR2 downregulation determines the initiation and progression of sporadic ALS (ADAR2-GluA2 hypothesis, []) (Figure 3).

Figure 3.
ADAR2-GluA2 hypothesis: specific molecular abnormalities beginning with a reduction in ADAR2 activity due to downregulation of ADAR2 in sporadic ALS or ADAR2 mislocalization in C9ORF72 ALS. The numbers and arrows indicate the order of molecular cascade based on the hypothesis. Uncertain parts were indicated by question marks.
In addition to sporadic ALS, some ALS-linked genes are involved in excitotoxicity due to the dysregulation of glutamatergic signaling or to A-to-I RNA editing.
FUS is involved in RNA processing, and it has been demonstrated that excitotoxicity affects FUS translocation from the nucleus to the cytoplasm and increases FUS-mediated dendritic GluA2 mRNA expression []. Additionally, glutamate release from brain synaptosomes and glutamate uptake were increased in transgenic mice with mutant FUS lacking a nuclear localization signal motif []. Regarding RNA editing, ADAR2 downregulation with a concomitant expression of Q/R site-unedited GluA2 mRNA was reported in the motor neurons of ALS patients carrying the FUSP525L mutation [] but not in motor neurons differentiated from iPSCs derived from ALS patients carrying FUSH517D mutation [].
Furthermore, reduced ADAR2 activity due to the loss of nuclear ADAR2 rather than a decrease in ADAR2 expression was reported the cause of a widespread reduction in RNA editing with ADAR2 cytoplasmic mislocalization in the motor neurons and differentiated motor neurons, which were generated from iPSCs derived from ALS patients carrying C9ORF72 with enhanced hexanucleotide repeat expansion []. Moreover, it has been demonstrated that dipeptide repeat proteins derived from the hexanucleotide repeat expansion of C9ORF72 bind to ADAR1 and ADAR2, thereby inhibiting their RNA editing activity in vitro []. However, one study reported that all GluA2 was edited at the Q/R site in the motor neurons differentiated from iPSCs derived from ALS patients carrying C9ORF72 with enhanced hexanucleotide repeat expansion [].
Although TDP-43 mislocalizes from the nucleus to the cytoplasm in the primary neuronal culture exposed to excitotoxicity [] and may have a modulatory role in ADAR1-mediated A-to-I RNA editing in the cell lines [], no studies have demonstrated aberrant RNA editing at the Q/R site in GluA2 in familial ALS carrying TARDBP mutations. In addition, possible roles of excitotoxicity in ALS-linked SOD-1 mutations have been suggested in SOD-1 transgenic mice [,], but all of the GluA2 mRNA expressed in the motor neurons of mutated human SOD-1 transgenic rats were edited at the Q/R site [].
The close association of reduced ADAR2 activity with both motor neuron death and TDP-43 pathology in sporadic and some forms of familial ALS cases lends further support to the ADAR2-GluA2 hypothesis in ALS pathogenesis.
TDP-43 pathology was also observed in the motor neurons of TARDBP-linked ALS patients. However, a reduction in ADAR2 activity does not appear to be involved in the mechanism underlying the death of motor neurons or TDP-43 pathology. In the motor neurons, mutant TDP-43 is more vulnerable to calpain-dependent cleavage than wild-type TDP-43 is and is readily cleaved by activated calpain; GluA2-lacking Ca2+-permeable AMPA receptors are expressed more abundantly in the motor neurons than in other neuronal classes, such as cortical neurons []. Thus, Ca2+-permeable AMPA receptors are involved in the pathogenic mechanism underlying TDP-43 pathology in TARDBP-linked ALS in a way different from that in other ALS types.
An increase in the proportion of GluA2-lacking Ca2+-permeable AMPA receptors is associated with hyperactivity in ALS motor neurons (Figure 2); this hyperactivity was associated with an increase in the expression levels of GluA1 mRNA in the spinal motor neurons of C9ORF72 ALS patients [] and in FUS knockdown mice [,], an increase in the expression levels of GluA1 mRNA and GluA3 mRNA in the iPSC-derived motor neurons of C9ORF72 ALS patients [,], a decrease in GluA2 mRNA in FUS knockdown mice or differentiated motor neurons derived from the embryonic stem cells of FUSP525L knock-in mice [,], and an increase in GluA4 mRNA expression in the iPSC-derived motor neurons from familial ALS patients carrying the FUSR521C and TARDBPM337V mutations []. However, as GluA2 knockout mice do not exhibit any neuronal loss [], it is unclear whether a relative reduction of GluA2, among the other AMPA receptor subunits, is mechanistically associated with motor neuron death in ALS.
Therefore, these results indicate that excessive Ca2+ influx through Ca2+-permeable AMPA receptors containing unedited GluA2 is critically involved in the pathogenesis of both sporadic and familial ALS.
4. Promising Therapy Targeting RNA Editing Dysregulation in Neurological Diseases
As a reduction in RNA editing activity is involved in the pathogenesis of neurological diseases (Table 1), the normalization of RNA editing activity in the motor neurons is a therapeutic strategy for not only sporadic ALS and some familial ALS but also various neurological diseases for which no cure can be achieved by current therapies (Figure 4).
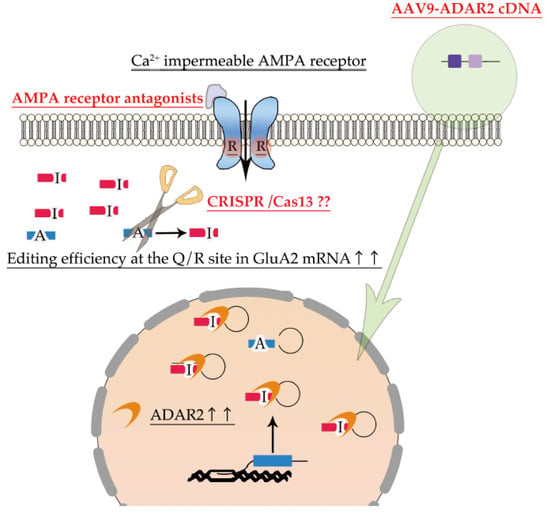
Figure 4.
Promising therapy targeting dysregulation of ADAR2 RNA editing. AMPA receptor antagonists are potential therapeutic drugs for normalizing excessive Ca2+ influx through Ca2+-permeable AMPA receptors. Delivery of ADAR2 cDNA to motor neurons using the adeno-associated virus type 9 (AAV-9) vector is a therapeutic approach to improve the expression of ADAR2 in the degenerating neurons (green arrow). Furthermore, RNA base editing of unedited GluA2 using CRISPR-Cas13 could also be a potential therapeutic approach to increase the proportion of edited GluA2. As RNA base editing therapy have not been established, the question marks were added.
Restoration of ADAR2 activity is a promising fundamental therapy for sporadic ALS and some forms of familial ALS. Elevated ADAR1 expression promotes cancer growth and metastasis [], whereas in ADARB1 transgenic mice, elevated ADAR2 expression is associated with simple obesity due to chronic hyperphagia, but apparently normal motor, lung, and heart functions []. Indeed, the delivery of ADAR2 cDNA to the motor neurons of conditional ADAR2 knockout AR2 mice via adeno-associated virus serotype 9 (AAV-9) robustly prevented progressive motor dysfunction and motor neuron death and also improved TDP-43 mislocalization without adverse behavioral or pathologic effects []. An increasing number of clinical trials using gene therapy for human diseases, including neurological diseases, have been conducted, and some of them have been approved for clinical use [,]. Although gene therapy is associated with several problems, such as daunting costs and regulatory policies, AAV9-ADAR2 therapy is a promising fundamental treatment for sporadic ALS in the future.
RNA base editing using clustered regularly interspaced short palindromic repair (CRISPR)-Cas13 could also be a potential therapeutic approach for deficient RNA editing-associated neurological diseases, including ALS. Programmable A-to-I RNA editing using CRISPR-Cas13-fused hyperactive mutant ADAR2 has proven to be successful in the site-directed A-to-I editing of point mutations in human disease genes such as methyl-CpF binding protein 2 (MECP2) (311G>A) in Rett syndrome and survival motor neuron (SMN1) (305G>A) in spinal muscular atrophy in vitro. The development of more efficient and safer RNA base editors [,,] as well as more efficient delivery systems targeting diseased cells—such as AAV vectors [], will realize the use of programmable A-to-I RNA editing using CRISPR-Cas13-fused hyperactive ADAR2 mutant as a future therapeutic strategy for deficient RNA editing-associated neurological diseases, including ALS.
Since the death of ADAR2-deficient motor neurons in AR2 mice is specifically mediated by Ca2+-permeable AMPA receptors, AMPA receptor antagonists are promising candidates to treat ALS. Non-competitive AMPA receptors have been demonstrated to confer protection against motor neuron death caused by excitotoxicity and 1,2,3,4-tetrahydro-6-nitro-2,3-dioxo-benzo[f]quinoxaline-7-sulfonamide disodium salt (NBQX)—the selective AMPA/kainate receptor antagonist, as well as prolong the survival of transgenic SOD1G93A mutant mice [,]. Moreover, in other studies, perampanel, an FDA-approved non-competitive AMPA receptor antagonist to manage epilepsy, has prevented ALS-like progressive motor dysfunction in conditional ADAR2 knockout mice and increased the cortical excitability threshold [,]. However, with the exception of an improvement in the manual muscle testing score, perampanel did not effectively inhibit disease progression in a recent phase 2 clinical trial on sporadic ALS patients [,]. The clinical use of AMPA receptor antagonists is limited by their frequent adverse effects due to the suppression of the physiological function of the CNS neurons; therefore, the development of more sophisticated drugs, such as receptor subunit-specific antagonists, are required in the future.
A-to-I RNA editing-based therapeutic strategies would effectively rescue the death of lower motor neurons in the majority of ALS patients, including patients with both sporadic and some with familial ALS that are associated with the dysregulation of A-to-I RNA editing. However, as the roles of dysregulation of RNA editing in the upper motor neuron death remain elusive, further studies are needed for the elucidation of the pathogenic mechanism underlying the death of the upper motor neurons in ALS and, more broadly, the death of the cortical neurons in frontotemporal lobar degeneration, which is closely associated with ALS. When such a new therapeutic strategy is realized in ALS, disease-specific biomarkers will also be increasingly required. Since biomarkers linked to disease-specific molecular changes can specify ALS, the demonstration of deficient ADAR2 activity non-invasively will help immensely in the evaluation of treatment efficacy. As the RNA editing efficiency at the ADAR2 sites in extracellular RNAs reflects intracellular ADAR2 activity in vitro [], demonstrating a reduction in the editing levels of these RNAs in the cerebrospinal fluid would become a biomarker for ALS.
5. Conclusions
We reviewed the mechanistic roles of defective RNA editing in neurological diseases and the potential therapeutic strategies targeting RNA editing in these diseases, especially for ALS. As excitotoxicity mediated by excessive Ca2+ influx through Ca2+-permeable AMPA receptors seems to be a plausible pathomechanism in ALS, the development of therapy based on this underlying pathomechanism would change the currently incurable ALS to a treatable disease. Moreover, we will be able to evaluate therapeutic response noninvasively by measuring alterations in editing efficiency in the RNAs in body fluids.
Author Contributions
H.T. and S.K. supervised the project. All authors wrote the main text, produced the figures, discussed the results, and reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by JSPS KAKENHI Grant Number 19K23957 and the Uehara Memorial Foundation.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The authors thank Harumi Tobita at the University of Tsukuba for providing technical assistance.
Conflicts of Interest
The authors declare no competing financial interests.
References
- Rowland, L.P.; Shneider, N.A. Amyotrophic lateral sclerosis. N. Engl. J. Med. 2001, 344, 1688–1700. [Google Scholar] [CrossRef] [PubMed]
- Chia, R.; Chiò, A.; Traynor, B.J. Novel genes associated with amyotrophic lateral sclerosis: Diagnostic and clinical implications. Lancet Neurol. 2018, 17, 94–102. [Google Scholar] [CrossRef]
- Van Damme, P.; Dewil, M.; Robberecht, W.; Van Den Bosch, L. Excitotoxicity and amyotrophic lateral sclerosis. Neurodegener. Dis. 2005, 2, 147–159. [Google Scholar] [CrossRef] [PubMed]
- King, A.E.; Woodhouse, A.; Kirkcaldie, M.T.; Vickers, J.C. Excitotoxicity in ALS: Overstimulation, or overreaction? Exp. Neurol. 2016, 275, 162–171. [Google Scholar] [CrossRef]
- Brown, R.H.; Al-Chalabi, A. Amyotrophic Lateral Sclerosis. N. Engl. J. Med. 2017, 377, 162–172. [Google Scholar] [CrossRef]
- Rodriguez-Campuzano, A.G.; Ortega, A. Glutamate transporters: Critical components of glutamatergic transmission. Neuropharmacology 2021, 192, 108602. [Google Scholar] [CrossRef]
- Reiner, A.; Levitz, J. Glutamatergic Signaling in the Central Nervous System: Ionotropic and Metabotropic Receptors in Concert. Neuron 2018, 98, 1080–1098. [Google Scholar] [CrossRef]
- Traynelis, S.F.; Wollmuth, L.P.; McBain, C.J.; Menniti, F.S.; Vance, K.M.; Ogden, K.K.; Hansen, K.B.; Yuan, H.; Myers, S.J.; Dingledine, R. Glutamate receptor ion channels: Structure, regulation, and function. Pharmacol. Rev. 2010, 62, 405–496. [Google Scholar] [CrossRef]
- Kerchner, G.A.; Nicoll, R.A. Silent synapses and the emergence of a postsynaptic mechanism for LTP. Nat. Rev. Neurosci. 2008, 9, 813–825. [Google Scholar] [CrossRef]
- Mayer, M.L.; Westbrook, G.L.; Guthrie, P.B. Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature 1984, 309, 261–263. [Google Scholar] [CrossRef]
- Niswender, C.M.; Conn, P.J. Metabotropic glutamate receptors: Physiology, pharmacology, and disease. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 295–322. [Google Scholar] [CrossRef] [PubMed]
- Plaitakis, A.; Constantakakis, E.; Smith, J. The neuroexcitotoxic amino acids glutamate and aspartate are altered in the spinal cord and brain in amyotrophic lateral sclerosis. Ann. Neurol. 1988, 24, 446–449. [Google Scholar] [CrossRef] [PubMed]
- Rothstein, J.D.; Tsai, G.; Kuncl, R.W.; Clawson, L.; Cornblath, D.R.; Drachman, D.B.; Pestronk, A.; Stauch, B.L.; Coyle, J.T. Abnormal excitatory amino acid metabolism in amyotrophic lateral sclerosis. Ann. Neurol. 1990, 28, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Shaw, P.J.; Forrest, V.; Ince, P.G.; Richardson, J.P.; Wastell, H.J. CSF and plasma amino acid levels in motor neuron disease: Elevation of CSF glutamate in a subset of patients. Neurodegeneration 1995, 4, 209–216. [Google Scholar] [CrossRef]
- Spreux-Varoquaux, O.; Bensimon, G.; Lacomblez, L.; Salachas, F.; Pradat, P.F.; Le Forestier, N.; Marouan, A.; Dib, M.; Meininger, V. Glutamate levels in cerebrospinal fluid in amyotrophic lateral sclerosis: A reappraisal using a new HPLC method with coulometric detection in a large cohort of patients. J. Neurol. Sci. 2002, 193, 73–78. [Google Scholar] [CrossRef]
- Rothstein, J.D.; Martin, L.J.; Kuncl, R.W. Decreased glutamate transport by the brain and spinal cord in amyotrophic lateral sclerosis. N. Engl. J. Med. 1992, 326, 1464–1468. [Google Scholar] [CrossRef]
- Plaitakis, A.; Smith, J.; Mandeli, J.; Yahr, M.D. Pilot trial of branched-chain aminoacids in amyotrophic lateral sclerosis. Lancet 1988, 1, 1015–1018. [Google Scholar] [CrossRef]
- Martin, D.; Thompson, M.A.; Nadler, J.V. The neuroprotective agent riluzole inhibits release of glutamate and aspartate from slices of hippocampal area CA1. Eur. J. Pharmacol. 1993, 250, 473–476. [Google Scholar] [CrossRef]
- Bensimon, G.; Lacomblez, L.; Meininger, V. A controlled trial of riluzole in amyotrophic lateral sclerosis. ALS/Riluzole Study Group. N. Engl. J. Med. 1994, 330, 585–591. [Google Scholar] [CrossRef]
- Lacomblez, L.; Bensimon, G.; Leigh, P.N.; Guillet, P.; Meininger, V. Dose-ranging study of riluzole in amyotrophic lateral sclerosis. Amyotrophic Lateral Sclerosis/Riluzole Study Group II. Lancet 1996, 347, 1425–1431. [Google Scholar] [CrossRef]
- Fang, T.; Al Khleifat, A.; Meurgey, J.-H.; Jones, A.; Leigh, P.N.; Bensimon, G.; Al-Chalabi, A. Stage at which riluzole treatment prolongs survival in patients with amyotrophic lateral sclerosis: A retrospective analysis of data from a dose-ranging study. Lancet Neurol. 2018, 17, 416–422. [Google Scholar] [CrossRef]
- Miller, R.G.; Mitchell, J.D.; Moore, D.H. Riluzole for amyotrophic lateral sclerosis (ALS)/motor neuron disease (MND). Cochrane Database Syst. Rev. 2012, 65, CD001447. [Google Scholar] [CrossRef]
- van Zundert, B.; Izaurieta, P.; Fritz, E.; Alvarez, F.J. Early pathogenesis in the adult-onset neurodegenerative disease amyotrophic lateral sclerosis. J. Cell. Biochem. 2012, 113, 3301–3312. [Google Scholar] [CrossRef]
- Vucic, S.; Nicholson, G.A.; Kiernan, M.C. Cortical hyperexcitability may precede the onset of familial amyotrophic lateral sclerosis. Brain 2008, 131, 1540–1550. [Google Scholar] [CrossRef]
- Menon, P.; Geevasinga, N.; Yiannikas, C.; Howells, J.; Kiernan, M.C.; Vucic, S. Sensitivity and specificity of threshold tracking transcranial magnetic stimulation for diagnosis of amyotrophic lateral sclerosis: A prospective study. Lancet Neurol. 2015, 14, 478–484. [Google Scholar] [CrossRef]
- Schanz, O.; Bageac, D.; Braun, L.; Traynor, B.J.; Lehky, T.J.; Floeter, M.K. Cortical hyperexcitability in patients with C9ORF72 mutations: Relationship to phenotype. Muscle Nerve 2016, 54, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Wainger, B.J.; Kiskinis, E.; Mellin, C.; Wiskow, O.; Han, S.S.; Sandoe, J.; Perez, N.P.; Williams, L.A.; Lee, S.; Boulting, G.; et al. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell. Rep. 2014, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wainger, B.J.; Macklin, E.A.; Vucic, S.; McIlduff, C.E.; Paganoni, S.; Maragakis, N.J.; Bedlack, R.; Goyal, N.A.; Rutkove, S.B.; Lange, D.J.; et al. Effect of Ezogabine on Cortical and Spinal Motor Neuron Excitability in Amyotrophic Lateral Sclerosis: A Randomized Clinical Trial. JAMA Neurol. 2021, 78, 186–196. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho, M.; Pinto, S.; Costa, J.; Evangelista, T.; Ohana, B.; Pinto, A. A randomized, placebo-controlled trial of memantine for functional disability in amyotrophic lateral sclerosis. Amyotroph. Lateral. Scler. 2010, 11, 456–460. [Google Scholar] [CrossRef]
- Ryberg, H.; Askmark, H.; Persson, L.I. A double-blind randomized clinical trial in amyotrophic lateral sclerosis using lamotrigine: Effects on CSF glutamate, aspartate, branched-chain amino acid levels and clinical parameters. Acta Neurol. Scand. 2003, 108, 1–8. [Google Scholar] [CrossRef]
- Rothstein, J.D.; Jin, L.; Dykes-Hoberg, M.; Kuncl, R.W. Chronic inhibition of glutamate uptake produces a model of slow neurotoxicity. Proc. Natl. Acad. Sci. USA 1993, 90, 6591–6595. [Google Scholar] [CrossRef]
- Van Den Bosch, L.; Van Damme, P.; Bogaert, E.; Robberecht, W. The role of excitotoxicity in the pathogenesis of amyotrophic lateral sclerosis. Biochim. Biophys. Acta 2006, 1762, 1068–1082. [Google Scholar] [CrossRef]
- Kwak, S.; Weiss, J.H. Calcium-permeable AMPA channels in neurodegenerative disease and ischemia. Curr. Opin. Neurobiol. 2006, 16, 281–287. [Google Scholar] [CrossRef]
- Rosenthal, J.J.; Seeburg, P.H. A-to-I RNA editing: Effects on proteins key to neural excitability. Neuron 2012, 74, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Sommer, B.; Köhler, M.; Sprengel, R.; Seeburg, P.H. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 1991, 67, 11–19. [Google Scholar] [CrossRef]
- Hideyama, T.; Yamashita, T.; Aizawa, H.; Tsuji, S.; Kakita, A.; Takahashi, H.; Kwak, S. Profound downregulation of the RNA editing enzyme ADAR2 in ALS spinal motor neurons. Neurobiol. Dis. 2012, 45, 1121–1128. [Google Scholar] [CrossRef]
- Henley, J.M.; Wilkinson, K.A. Synaptic AMPA receptor composition in development, plasticity and disease. Nat. Rev. Neurosci. 2016, 17, 337–350. [Google Scholar] [CrossRef]
- Greger, I.H.; Khatri, L.; Ziff, E.B. RNA editing at arg607 controls AMPA receptor exit from the endoplasmic reticulum. Neuron 2002, 34, 759–772. [Google Scholar] [CrossRef]
- Herguedas, B.; Watson, J.F.; Ho, H.; Cais, O.; Garcia-Nafria, J.; Greger, I.H. Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP gamma8. Science 2019, 364, 9011. [Google Scholar] [CrossRef]
- Zhang, D.; Watson, J.F.; Matthews, P.M.; Cais, O.; Greger, I.H. Gating and modulation of a hetero-octameric AMPA glutamate receptor. Nature 2021, 594, 454–458. [Google Scholar] [CrossRef]
- Singh, M. Dysregulated A to I RNA editing and non-coding RNAs in neurodegeneration. Front. Genet. 2012, 3, 326. [Google Scholar] [CrossRef]
- Costa Cruz, P.H.; Kawahara, Y. RNA Editing in Neurological and Neurodegenerative Disorders. Methods Mol. Biol. 2021, 2181, 309–330. [Google Scholar] [CrossRef]
- Zhu, X.; Zhang, Y.; Yang, X.; Hao, C.; Duan, H. Gene Therapy for Neurodegenerative Disease: Clinical Potential and Directions. Front. Mol. Neurosci. 2021, 14, 618171. [Google Scholar] [CrossRef]
- Sun, J.; Roy, S. Gene-based therapies for neurodegenerative diseases. Nat. Neurosci. 2021, 24, 297–311. [Google Scholar] [CrossRef]
- Nishikura, K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell. Biol. 2016, 17, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Elbarbary, R.A.; Lucas, B.A.; Maquat, L.E. Retrotransposons as regulators of gene expression. Science 2016, 351, aac7247. [Google Scholar] [CrossRef] [PubMed]
- Picardi, E.; Manzari, C.; Mastropasqua, F.; Aiello, I.; D’Erchia, A.M.; Pesole, G. Profiling RNA editing in human tissues: Towards the inosinome Atlas. Sci. Rep. 2015, 5, 14941. [Google Scholar] [CrossRef] [PubMed]
- Picardi, E.; Horner, D.S.; Pesole, G. Single-cell transcriptomics reveals specific RNA editing signatures in the human brain. RNA 2017, 23, 860–865. [Google Scholar] [CrossRef]
- Tan, M.H.; Li, Q.; Shanmugam, R.; Piskol, R.; Kohler, J.; Young, A.N.; Liu, K.I.; Zhang, R.; Ramaswami, G.; Ariyoshi, K.; et al. Dynamic landscape and regulation of RNA editing in mammals. Nature 2017, 550, 249–254. [Google Scholar] [CrossRef]
- Heraud-Farlow, J.E.; Walkley, C.R. What do editors do? Understanding the physiological functions of A-to-I RNA editing by adenosine deaminase acting on RNAs. Open Biol. 2020, 10, 200085. [Google Scholar] [CrossRef]
- Yang, W.; Chendrimada, T.P.; Wang, Q.; Higuchi, M.; Seeburg, P.H.; Shiekhattar, R.; Nishikura, K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006, 13, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell. Rep. 2015, 10, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Rueter, S.M.; Dawson, T.R.; Emeson, R.B. Regulation of alternative splicing by RNA editing. Nature 1999, 399, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Solomon, O.; Di Segni, A.; Cesarkas, K.; Porath, H.T.; Marcu-Malina, V.; Mizrahi, O.; Stern-Ginossar, N.; Kol, N.; Farage-Barhom, S.; Glick-Saar, E.; et al. RNA editing by ADAR1 leads to context-dependent transcriptome-wide changes in RNA secondary structure. Nat. Commun. 2017, 8, 1440. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, E.; Levanon, E.Y. A-to-I RNA editing-immune protector and transcriptome diversifier. Nat. Rev. Genet. 2018, 19, 473–490. [Google Scholar] [CrossRef]
- Lorenzini, I.; Moore, S.; Sattler, R. RNA Editing Deficiency in Neurodegeneration. Adv. Neurobiol. 2018, 20, 63–83. [Google Scholar] [CrossRef] [PubMed]
- Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010, 79, 321–349. [Google Scholar] [CrossRef]
- Kawahara, Y.; Ito, K.; Sun, H.; Kanazawa, I.; Kwak, S. Low editing efficiency of GluR2 mRNA is associated with a low relative abundance of ADAR2 mRNA in white matter of normal human brain. Eur. J. Neurosci. 2003, 18, 23–33. [Google Scholar] [CrossRef]
- Hartner, J.C.; Schmittwolf, C.; Kispert, A.; Muller, A.M.; Higuchi, M.; Seeburg, P.H. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004, 279, 4894–4902. [Google Scholar] [CrossRef]
- Miyamura, Y.; Suzuki, T.; Kono, M.; Inagaki, K.; Ito, S.; Suzuki, N.; Tomita, Y. Mutations of the RNA-specific adenosine deaminase gene (DSRAD) are involved in dyschromatosis symmetrica hereditaria. Am. J. Hum. Genet. 2003, 73, 693–699. [Google Scholar] [CrossRef]
- Rice, G.I.; Kasher, P.R.; Forte, G.M.; Mannion, N.M.; Greenwood, S.M.; Szynkiewicz, M.; Dickerson, J.E.; Bhaskar, S.S.; Zampini, M.; Briggs, T.A.; et al. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat. Genet. 2012, 44, 1243–1248. [Google Scholar] [CrossRef]
- Higuchi, M.; Maas, S.; Single, F.N.; Hartner, J.; Rozov, A.; Burnashev, N.; Feldmeyer, D.; Sprengel, R.; Seeburg, P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 2000, 406, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.Y.; Sedmik, J.; Fitzgerald, M.P.; Halevy, R.S.; Keegan, L.P.; Helbig, I.; Basel-Salmon, L.; Cohen, L.; Straussberg, R.; Chung, W.K.; et al. Bi-allelic ADARB1 Variants Associated with Microcephaly, Intellectual Disability, and Seizures. Am. J. Hum. Genet. 2020, 106, 467–483. [Google Scholar] [CrossRef] [PubMed]
- Maroofian, R.; Sedmik, J.; Mazaheri, N.; Scala, M.; Zaki, M.S.; Keegan, L.P.; Azizimalamiri, R.; Issa, M.; Shariati, G.; Sedaghat, A.; et al. Biallelic variants in ADARB1, encoding a dsRNA-specific adenosine deaminase, cause a severe developmental and epileptic encephalopathy. J. Med. Genet. 2020, 11, 36. [Google Scholar] [CrossRef]
- Hideyama, T.; Yamashita, T.; Suzuki, T.; Tsuji, S.; Higuchi, M.; Seeburg, P.H.; Takahashi, R.; Misawa, H.; Kwak, S. Induced loss of ADAR2 engenders slow death of motor neurons from Q/R site-unedited GluR2. J. Neurosci. 2010, 30, 11917–11925. [Google Scholar] [CrossRef]
- Hwang, T.; Park, C.K.; Leung, A.K.; Gao, Y.; Hyde, T.M.; Kleinman, J.E.; Rajpurohit, A.; Tao, R.; Shin, J.H.; Weinberger, D.R. Dynamic regulation of RNA editing in human brain development and disease. Nat. Neurosci. 2016, 19, 1093–1099. [Google Scholar] [CrossRef]
- Tariq, A.; Jantsch, M.F. Transcript diversification in the nervous system: A to I RNA editing in CNS function and disease development. Front. Neurosci. 2012, 6, 99. [Google Scholar] [CrossRef]
- Gal-Mark, N.; Shallev, L.; Sweetat, S.; Barak, M.; Billy Li, J.; Levanon, E.Y.; Eisenberg, E.; Behar, O. Abnormalities in A-to-I RNA editing patterns in CNS injuries correlate with dynamic changes in cell type composition. Sci. Rep. 2017, 7, 43421. [Google Scholar] [CrossRef]
- Tran, S.S.; Jun, H.I.; Bahn, J.H.; Azghadi, A.; Ramaswami, G.; Van Nostrand, E.L.; Nguyen, T.B.; Hsiao, Y.E.; Lee, C.; Pratt, G.A.; et al. Widespread RNA editing dysregulation in brains from autistic individuals. Nat. Neurosci. 2019, 22, 25–36. [Google Scholar] [CrossRef]
- Breen, M.S.; Dobbyn, A.; Li, Q.; Roussos, P.; Hoffman, G.E.; Stahl, E.; Chess, A.; Sklar, P.; Li, J.B.; Devlin, B.; et al. Global landscape and genetic regulation of RNA editing in cortical samples from individuals with schizophrenia. Nat. Neurosci. 2019, 22, 1402–1412. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; Ito, K.; Sun, H.; Aizawa, H.; Kanazawa, I.; Kwak, S. Glutamate receptors: RNA editing and death of motor neurons. Nature 2004, 427, 801. [Google Scholar] [CrossRef] [PubMed]
- Khermesh, K.; D’Erchia, A.M.; Barak, M.; Annese, A.; Wachtel, C.; Levanon, E.Y.; Picardi, E.; Eisenberg, E. Reduced levels of protein recoding by A-to-I RNA editing in Alzheimer’s disease. RNA 2016, 22, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Akbarian, S.; Smith, M.A.; Jones, E.G. Editing for an AMPA receptor subunit RNA in prefrontal cortex and striatum in Alzheimer’s disease, Huntington’s disease and schizophrenia. Brain Res. 1995, 699, 297–304. [Google Scholar] [CrossRef]
- Gaisler-Salomon, I.; Kravitz, E.; Feiler, Y.; Safran, M.; Biegon, A.; Amariglio, N.; Rechavi, G. Hippocampus-specific deficiency in RNA editing of GluA2 in Alzheimer’s disease. Neurobiol. Aging 2014, 35, 1785–1791. [Google Scholar] [CrossRef]
- Maas, S.; Patt, S.; Schrey, M.; Rich, A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl. Acad. Sci. USA 2001, 98, 14687–14692. [Google Scholar] [CrossRef]
- Ishiuchi, S.; Yoshida, Y.; Sugawara, K.; Aihara, M.; Ohtani, T.; Watanabe, T.; Saito, N.; Tsuzuki, K.; Okado, H.; Miwa, A.; et al. Ca2+-permeable AMPA receptors regulate growth of human glioblastoma via Akt activation. J. Neurosci. 2007, 27, 7987–8001. [Google Scholar] [CrossRef]
- Cenci, C.; Barzotti, R.; Galeano, F.; Corbelli, S.; Rota, R.; Massimi, L.; Di Rocco, C.; O’Connell, M.A.; Gallo, A. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J. Biol. Chem. 2008, 283, 7251–7260. [Google Scholar] [CrossRef]
- Patil, V.; Pal, J.; Mahalingam, K.; Somasundaram, K. Global RNA editome landscape discovers reduced RNA editing in glioma: Loss of editing of gamma-amino butyric acid receptor alpha subunit 3 (GABRA3) favors glioma migration and invasion. PeerJ 2020, 8, e9755. [Google Scholar] [CrossRef]
- Galeano, F.; Rossetti, C.; Tomaselli, S.; Cifaldi, L.; Lezzerini, M.; Pezzullo, M.; Boldrini, R.; Massimi, L.; Di Rocco, C.M.; Locatelli, F.; et al. ADAR2-editing activity inhibits glioblastoma growth through the modulation of the CDC14B/Skp2/p21/p27 axis. Oncogene 2013, 32, 998–1009. [Google Scholar] [CrossRef]
- Choudhury, Y.; Tay, F.C.; Lam, D.H.; Sandanaraj, E.; Tang, C.; Ang, B.T.; Wang, S. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J. Clin. Investig. 2012, 122, 4059–4076. [Google Scholar] [CrossRef]
- Cesarini, V.; Silvestris, D.A.; Tassinari, V.; Tomaselli, S.; Alon, S.; Eisenberg, E.; Locatelli, F.; Gallo, A. ADAR2/miR-589-3p axis controls glioblastoma cell migration/invasion. Nucleic Acids. Res. 2018, 46, 2045–2059. [Google Scholar] [CrossRef]
- Liu, S.; Lau, L.; Wei, J.; Zhu, D.; Zou, S.; Sun, H.S.; Fu, Y.; Liu, F.; Lu, Y. Expression of Ca(2+)-permeable AMPA receptor channels primes cell death in transient forebrain ischemia. Neuron 2004, 43, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Peng, P.L.; Zhong, X.; Tu, W.; Soundarapandian, M.M.; Molner, P.; Zhu, D.; Lau, L.; Liu, S.; Liu, F.; Lu, Y. ADAR2-dependent RNA editing of AMPA receptor subunit GluR2 determines vulnerability of neurons in forebrain ischemia. Neuron 2006, 49, 719–733. [Google Scholar] [CrossRef]
- Barbon, A.; Fumagalli, F.; Caracciolo, L.; Madaschi, L.; Lesma, E.; Mora, C.; Carelli, S.; Slotkin, T.A.; Racagni, G.; Di Giulio, A.M.; et al. Acute spinal cord injury persistently reduces R/G RNA editing of AMPA receptors. J. Neurochem. 2010, 114, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Di Narzo, A.F.; Kozlenkov, A.; Ge, Y.; Zhang, B.; Sanelli, L.; May, Z.; Li, Y.; Fouad, K.; Cardozo, C.; Koonin, E.V.; et al. Decrease of mRNA Editing after Spinal Cord Injury is Caused by Down-regulation of ADAR2 that is Triggered by Inflammatory Response. Sci. Rep. 2015, 5, 12615. [Google Scholar] [CrossRef] [PubMed]
- Thijs, R.D.; Surges, R.; O’Brien, T.J.; Sander, J.W. Epilepsy in adults. Lancet 2019, 393, 689–701. [Google Scholar] [CrossRef]
- Kortenbruck, G.; Berger, E.; Speckmann, E.J.; Musshoff, U. RNA editing at the Q/R site for the glutamate receptor subunits GLUR2, GLUR5, and GLUR6 in hippocampus and temporal cortex from epileptic patients. Neurobiol. Dis. 2001, 8, 459–468. [Google Scholar] [CrossRef]
- Vollmar, W.; Gloger, J.; Berger, E.; Kortenbruck, G.; Kohling, R.; Speckmann, E.J.; Musshoff, U. RNA editing (R/G site) and flip-flop splicing of the AMPA receptor subunit GluR2 in nervous tissue of epilepsy patients. Neurobiol. Dis. 2004, 15, 371–379. [Google Scholar] [CrossRef]
- Krestel, H.; Raffel, S.; von Lehe, M.; Jagella, C.; Moskau-Hartmann, S.; Becker, A.; Elger, C.E.; Seeburg, P.H.; Nirkko, A. Differences between RNA and DNA due to RNA editing in temporal lobe epilepsy. Neurobiol. Dis. 2013, 56, 66–73. [Google Scholar] [CrossRef]
- Bernard, A.; Ferhat, L.; Dessi, F.; Charton, G.; Represa, A.; Ben-Ari, Y.; Khrestchatisky, M. Q/R editing of the rat GluR5 and GluR6 kainate receptors in vivo and in vitro: Evidence for independent developmental, pathological and cellular regulation. Eur. J. Neurosci. 1999, 11, 604–616. [Google Scholar] [CrossRef]
- Caracciolo, L.; Barbon, A.; Palumbo, S.; Mora, C.; Toscano, C.D.; Bosetti, F.; Barlati, S. Altered mRNA editing and expression of ionotropic glutamate receptors after kainic acid exposure in cyclooxygenase-2 deficient mice. PLoS ONE 2011, 6, e19398. [Google Scholar] [CrossRef] [PubMed]
- Malhi, G.S.; Mann, J.J. Depression. Lancet 2018, 392, 2299–2312. [Google Scholar] [CrossRef]
- Niswender, C.M.; Herrick-Davis, K.; Dilley, G.E.; Meltzer, H.Y.; Overholser, J.C.; Stockmeier, C.A.; Emeson, R.B.; Sanders-Bush, E. RNA editing of the human serotonin 5-HT2C receptor. alterations in suicide and implications for serotonergic pharmacotherapy. Neuropsychopharmacology 2001, 24, 478–491. [Google Scholar] [CrossRef]
- Gurevich, I.; Tamir, H.; Arango, V.; Dwork, A.J.; Mann, J.J.; Schmauss, C. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron 2002, 34, 349–356. [Google Scholar] [CrossRef]
- Iwamoto, K.; Kato, T. RNA editing of serotonin 2C receptor in human postmortem brains of major mental disorders. Neurosci. Lett. 2003, 346, 169–172. [Google Scholar] [CrossRef]
- Zhu, H.; Urban, D.J.; Blashka, J.; McPheeters, M.T.; Kroeze, W.K.; Mieczkowski, P.; Overholser, J.C.; Jurjus, G.J.; Dieter, L.; Mahajan, G.J.; et al. Quantitative analysis of focused a-to-I RNA editing sites by ultra-high-throughput sequencing in psychiatric disorders. PLoS ONE 2012, 7, e43227. [Google Scholar] [CrossRef]
- Weissmann, D.; van der Laan, S.; Underwood, M.D.; Salvetat, N.; Cavarec, L.; Vincent, L.; Molina, F.; Mann, J.J.; Arango, V.; Pujol, J.F. Region-specific alterations of A-to-I RNA editing of serotonin 2c receptor in the cortex of suicides with major depression. Transl. Psychiatry 2016, 6, e878. [Google Scholar] [CrossRef]
- Sodhi, M.S.; Burnet, P.W.; Makoff, A.J.; Kerwin, R.W.; Harrison, P.J. RNA editing of the 5-HT(2C) receptor is reduced in schizophrenia. Mol. Psychiatry 2001, 6, 373–379. [Google Scholar] [CrossRef][Green Version]
- Dracheva, S.; Patel, N.; Woo, D.A.; Marcus, S.M.; Siever, L.J.; Haroutunian, V. Increased serotonin 2C receptor mRNA editing: A possible risk factor for suicide. Mol. Psychiatry 2008, 13, 1001–1010. [Google Scholar] [CrossRef]
- Chimienti, F.; Cavarec, L.; Vincent, L.; Salvetat, N.; Arango, V.; Underwood, M.D.; Mann, J.J.; Pujol, J.F.; Weissmann, D. Brain region-specific alterations of RNA editing in PDE8A mRNA in suicide decedents. Transl. Psychiatry 2019, 9, 91. [Google Scholar] [CrossRef]
- Salvetat, N.; Chimienti, F.; Cayzac, C.; Dubuc, B.; Checa-Robles, F.; Dupre, P.; Mereuze, S.; Patel, V.; Genty, C.; Lang, J.P.; et al. Phosphodiesterase 8A to discriminate in blood samples depressed patients and suicide attempters from healthy controls based on A-to-I RNA editing modifications. Transl. Psychiatry 2021, 11, 255. [Google Scholar] [CrossRef]
- Lord, C.; Elsabbagh, M.; Baird, G.; Veenstra-Vanderweele, J. Autism spectrum disorder. Lancet 2018, 392, 508–520. [Google Scholar] [CrossRef]
- Eran, A.; Li, J.B.; Vatalaro, K.; McCarthy, J.; Rahimov, F.; Collins, C.; Markianos, K.; Margulies, D.M.; Brown, E.N.; Calvo, S.E.; et al. Comparative RNA editing in autistic and neurotypical cerebella. Mol. Psychiatry 2013, 18, 1041–1048. [Google Scholar] [CrossRef][Green Version]
- Schmidt, H.D.; McFarland, K.N.; Darnell, S.B.; Huizenga, M.N.; Sangrey, G.R.; Cha, J.H.; Pierce, R.C.; Sadri-Vakili, G. ADAR2-dependent GluA2 editing regulates cocaine seeking. Mol. Psychiatry 2015, 20, 1460–1466. [Google Scholar] [CrossRef]
- Tanaka, M.; Watanabe, Y. RNA Editing of Serotonin 2C Receptor and Alcohol Intake. Front. Neurosci 2019, 13, 1390. [Google Scholar] [CrossRef]
- Starr, A.; Sattler, R. Synaptic dysfunction and altered excitability in C9ORF72 ALS/FTD. Brain Res. 2018, 1693, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Hosaka, T.; Yamashita, T.; Tamaoka, A.; Kwak, S. Extracellular RNAs as Biomarkers of Sporadic Amyotrophic Lateral Sclerosis and Other Neurodegenerative Diseases. Int. J. Mol. Sci. 2019, 20, 3148. [Google Scholar] [CrossRef] [PubMed]
- Burnashev, N.; Monyer, H.; Seeburg, P.H.; Sakmann, B. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron 1992, 8, 189–198. [Google Scholar] [CrossRef]
- Feldmeyer, D.; Kask, K.; Brusa, R.; Kornau, H.C.; Kolhekar, R.; Rozov, A.; Burnashev, N.; Jensen, V.; Hvalby, O.; Sprengel, R.; et al. Neurological dysfunctions in mice expressing different levels of the Q/R site-unedited AMPAR subunit GluR-B. Nat. Neurosci. 1999, 2, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Seeburg, P.H. A-to-I editing: New and old sites, functions and speculations. Neuron 2002, 35, 17–20. [Google Scholar] [CrossRef]
- Jia, Z.; Agopyan, N.; Miu, P.; Xiong, Z.; Henderson, J.; Gerlai, R.; Taverna, F.A.; Velumian, A.; MacDonald, J.; Carlen, P.; et al. Enhanced LTP in mice deficient in the AMPA receptor GluR2. Neuron 1996, 17, 945–956. [Google Scholar] [CrossRef]
- D’Erchia, A.M.; Gallo, A.; Manzari, C.; Raho, S.; Horner, D.S.; Chiara, M.; Valletti, A.; Aiello, I.; Mastropasqua, F.; Ciaccia, L.; et al. Massive transcriptome sequencing of human spinal cord tissues provides new insights into motor neuron degeneration in ALS. Sci. Rep. 2017, 7, 10046. [Google Scholar] [CrossRef]
- Ichiyanagi, N.; Fujimori, K.; Yano, M.; Ishihara-Fujisaki, C.; Sone, T.; Akiyama, T.; Okada, Y.; Akamatsu, W.; Matsumoto, T.; Ishikawa, M.; et al. Establishment of In Vitro FUS-Associated Familial Amyotrophic Lateral Sclerosis Model Using Human Induced Pluripotent Stem Cells. Stem Cell Reports 2016, 6, 496–510. [Google Scholar] [CrossRef] [PubMed]
- Gregory, J.M.; Livesey, M.R.; McDade, K.; Selvaraj, B.T.; Barton, S.K.; Chandran, S.; Smith, C. Dysregulation of AMPA receptor subunit expression in sporadic ALS post-mortem brain. J. Pathol. 2020, 250, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Aizawa, H.; Sawada, J.; Hideyama, T.; Yamashita, T.; Katayama, T.; Hasebe, N.; Kimura, T.; Yahara, O.; Kwak, S. TDP-43 pathology in sporadic ALS occurs in motor neurons lacking the RNA editing enzyme ADAR2. Acta Neuropathol. 2010, 120, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Kwak, S. The molecular link between inefficient GluA2 Q/R site-RNA editing and TDP-43 pathology in motor neurons of sporadic amyotrophic lateral sclerosis patients. Brain Res. 2014, 1584, 28–38. [Google Scholar] [CrossRef]
- Yamashita, T.; Hideyama, T.; Hachiga, K.; Teramoto, S.; Takano, J.; Iwata, N.; Saido, T.C.; Kwak, S. A role for calpain-dependent cleavage of TDP-43 in amyotrophic lateral sclerosis pathology. Nat. Commun 2012, 3, 1307. [Google Scholar] [CrossRef]
- Hideyama, T.; Kwak, S. When Does ALS Start? ADAR2-GluA2 Hypothesis for the Etiology of Sporadic ALS. Front. Mol. Neurosci. 2011, 4, 33. [Google Scholar] [CrossRef]
- Tischbein, M.; Baron, D.M.; Lin, Y.C.; Gall, K.V.; Landers, J.E.; Fallini, C.; Bosco, D.A. The RNA-binding protein FUS/TLS undergoes calcium-mediated nuclear egress during excitotoxic stress and is required for GRIA2 mRNA processing. J. Biol. Chem. 2019, 294, 10194–10210. [Google Scholar] [CrossRef]
- Grigoriev, V.V.; Efimova, A.D.; Ustyugov, A.A.; Shevchenko, V.P.; Bachurin, S.O.; Myasoedov, N.F. Glutamate release and uptake processes are altered in a new mouse model of amyotrophic lateral sclerosis. Dokl. Biochem. Biophys. 2016, 468, 165–167. [Google Scholar] [CrossRef]
- Aizawa, H.; Hideyama, T.; Yamashita, T.; Kimura, T.; Suzuki, N.; Aoki, M.; Kwak, S. Deficient RNA-editing enzyme ADAR2 in an amyotrophic lateral sclerosis patient with a FUS(P525L) mutation. J. Clin. Neurosci. 2016, 32, 128–129. [Google Scholar] [CrossRef]
- Moore, S.; Alsop, E.; Lorenzini, I.; Starr, A.; Rabichow, B.E.; Mendez, E.; Levy, J.L.; Burciu, C.; Reiman, R.; Chew, J.; et al. ADAR2 mislocalization and widespread RNA editing aberrations in C9orf72-mediated ALS/FTD. Acta Neuropathol. 2019, 19, 1999. [Google Scholar] [CrossRef]
- Suzuki, H.; Matsuoka, M. Proline-arginine poly-dipeptide encoded by the C9orf72 repeat expansion inhibits adenosine deaminase acting on RNA. J. Neurochem. 2021, 158, 753–765. [Google Scholar] [CrossRef]
- Selvaraj, B.T.; Livesey, M.R.; Zhao, C.; Gregory, J.M.; James, O.T.; Cleary, E.M.; Chouhan, A.K.; Gane, A.B.; Perkins, E.M.; Dando, O.; et al. C9ORF72 repeat expansion causes vulnerability of motor neurons to Ca(2+)-permeable AMPA receptor-mediated excitotoxicity. Nat. Commun. 2018, 9, 347. [Google Scholar] [CrossRef]
- Quinones-Valdez, G.; Tran, S.S.; Jun, H.I.; Bahn, J.H.; Yang, E.W.; Zhan, L.; Brummer, A.; Wei, X.; Van Nostrand, E.L.; Pratt, G.A.; et al. Regulation of RNA editing by RNA-binding proteins in human cells. Commun. Biol. 2019, 2, 19. [Google Scholar] [CrossRef] [PubMed]
- Ohgomori, T.; Yamasaki, R.; Takeuchi, H.; Kadomatsu, K.; Kira, J.I.; Jinno, S. Differential involvement of vesicular and glial glutamate transporters around spinal alpha-motoneurons in the pathogenesis of SOD1(G93A) mouse model of amyotrophic lateral sclerosis. Neuroscience 2017, 356, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Bonifacino, T.; Provenzano, F.; Gallia, E.; Ravera, S.; Torazza, C.; Bossi, S.; Ferrando, S.; Puliti, A.; Van Den Bosch, L.; Bonanno, G.; et al. In-vivo genetic ablation of metabotropic glutamate receptor type 5 slows down disease progression in the SOD1(G93A) mouse model of amyotrophic lateral sclerosis. Neurobiol. Dis. 2019, 129, 79–92. [Google Scholar] [CrossRef]
- Kawahara, Y.; Sun, H.; Ito, K.; Hideyama, T.; Aoki, M.; Sobue, G.; Tsuji, S.; Kwak, S. Underediting of GluR2 mRNA, a neuronal death inducing molecular change in sporadic ALS, does not occur in motor neurons in ALS1 or SBMA. Neurosci. Res. 2006, 54, 11–14. [Google Scholar] [CrossRef] [PubMed]
- Udagawa, T.; Fujioka, Y.; Tanaka, M.; Honda, D.; Yokoi, S.; Riku, Y.; Ibi, D.; Nagai, T.; Yamada, K.; Watanabe, H.; et al. FUS regulates AMPA receptor function and FTLD/ALS-associated behaviour via GluA1 mRNA stabilization. Nat. Commun. 2015, 6, 7098. [Google Scholar] [CrossRef]
- Capauto, D.; Colantoni, A.; Lu, L.; Santini, T.; Peruzzi, G.; Biscarini, S.; Morlando, M.; Shneider, N.A.; Caffarelli, E.; Laneve, P.; et al. A Regulatory Circuitry Between Gria2, miR-409, and miR-495 Is Affected by ALS FUS Mutation in ESC-Derived Motor Neurons. Mol. Neurobiol. 2018, 55, 7635–7651. [Google Scholar] [CrossRef]
- Dafinca, R.; Barbagallo, P.; Farrimond, L.; Candalija, A.; Scaber, J.; Ababneh, N.A.; Sathyaprakash, C.; Vowles, J.; Cowley, S.A.; Talbot, K. Impairment of Mitochondrial Calcium Buffering Links Mutations in C9ORF72 and TARDBP in iPS-Derived Motor Neurons from Patients with ALS/FTD. Stem Cell Rep. 2020, 14, 892–908. [Google Scholar] [CrossRef]
- Bursch, F.; Kalmbach, N.; Naujock, M.; Staege, S.; Eggenschwiler, R.; Abo-Rady, M.; Japtok, J.; Guo, W.; Hensel, N.; Reinhardt, P.; et al. Altered calcium dynamics and glutamate receptor properties in iPSC-derived motor neurons from ALS patients with C9orf72, FUS, SOD1 or TDP43 mutations. Hum. Mol. Genet. 2019, 28, 2835–2850. [Google Scholar] [CrossRef]
- Xu, L.D.; Ohman, M. ADAR1 Editing and its Role in Cancer. Genes 2018, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Zimmerman, M.B.; Beltz, T.G.; Johnson, A.K. Affect-related behaviors in mice misexpressing the RNA editing enzyme ADAR2. Physiol. Behav. 2009, 97, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Chai, H.L.; Teramoto, S.; Tsuji, S.; Shimazaki, K.; Muramatsu, S.; Kwak, S. Rescue of amyotrophic lateral sclerosis phenotype in a mouse model by intravenous AAV9-ADAR2 delivery to motor neurons. EMBO Mol. Med. 2013, 5, 1710–1719. [Google Scholar] [CrossRef] [PubMed]
- High, K.A.; Roncarolo, M.G. Gene Therapy. N. Engl. J. Med. 2019, 381, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Kariyawasam, D.; Alexander, I.E.; Kurian, M.; Farrar, M.A. Great expectations: Virus-mediated gene therapy in neurological disorders. J. Neurol. Neurosurg. Psychiatry 2020, 91, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Qin, S.; Pu, Q.; Wang, Z.; Wu, Q.; Gao, P.; Schettler, J.; Guo, K.; Li, R.; Li, G.; et al. CRISPR-Cas13 Inhibitors Block RNA Editing in Bacteria and Mammalian Cells. Mol. Cell. 2020, 78, 850–861.e5. [Google Scholar] [CrossRef]
- Xu, C.; Zhou, Y.; Xiao, Q.; He, B.; Geng, G.; Wang, Z.; Cao, B.; Dong, X.; Bai, W.; Wang, Y.; et al. Programmable RNA editing with compact CRISPR-Cas13 systems from uncultivated microbes. Nat. Methods 2021, 18, 499–506. [Google Scholar] [CrossRef]
- Van Den Bosch, L.; Vandenberghe, W.; Klaassen, H.; Van Houtte, E.; Robberecht, W. Ca(2+)-permeable AMPA receptors and selective vulnerability of motor neurons. J. Neurol. Sci. 2000, 180, 29–34. [Google Scholar] [CrossRef]
- Van Damme, P.; Leyssen, M.; Callewaert, G.; Robberecht, W.; Van Den Bosch, L. The AMPA receptor antagonist NBQX prolongs survival in a transgenic mouse model of amyotrophic lateral sclerosis. Neurosci. Lett. 2003, 343, 81–84. [Google Scholar] [CrossRef]
- Akamatsu, M.; Yamashita, T.; Hirose, N.; Teramoto, S.; Kwak, S. The AMPA receptor antagonist perampanel robustly rescues amyotrophic lateral sclerosis (ALS) pathology in sporadic ALS model mice. Sci. Rep. 2016, 6, 28649. [Google Scholar] [CrossRef] [PubMed]
- Oskarsson, B.; Mauricio, E.A.; Shah, J.S.; Li, Z.; Rogawski, M.A. Cortical excitability threshold can be increased by the AMPA blocker Perampanel in amyotrophic lateral sclerosis. Muscle Nerve 2021, 64, 215–219. [Google Scholar] [CrossRef]
- Pascuzzi, R.M.; Shefner, J.; Chappell, A.S.; Bjerke, J.S.; Tamura, R.; Chaudhry, V.; Clawson, L.; Haas, L.; Rothstein, J.D. A phase II trial of talampanel in subjects with amyotrophic lateral sclerosis. Amyotroph. Lateral. Scler. 2010, 11, 266–271. [Google Scholar] [CrossRef]
- Aizawa, H.; Kato, H.; Oba, K.; Kawahara, T.; Okubo, Y.; Saito, T.; Naito, M.; Urushitani, M.; Tamaoka, A.; Nakamagoe, K.; et al. Randomized phase 2 study of perampanel for sporadic amyotrophic lateral sclerosis. J. Neurol. 2021, 7, 21. [Google Scholar] [CrossRef]
- Hosaka, T.; Yamashita, T.; Teramoto, S.; Hirose, N.; Tamaoka, A.; Kwak, S. ADAR2-dependent A-to-I RNA editing in the extracellular linear and circular RNAs. Neurosci. Res. 2018, 11, 5. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).