Characterization of Pathogenicity-Associated V2 Protein of Tobacco Curly Shoot Virus
Abstract
1. Introduction
2. Results
2.1. TbCSV V2 Inhibited Both Local and Systemic RNA Silencing Triggered by Positive-Sense GFP RNA
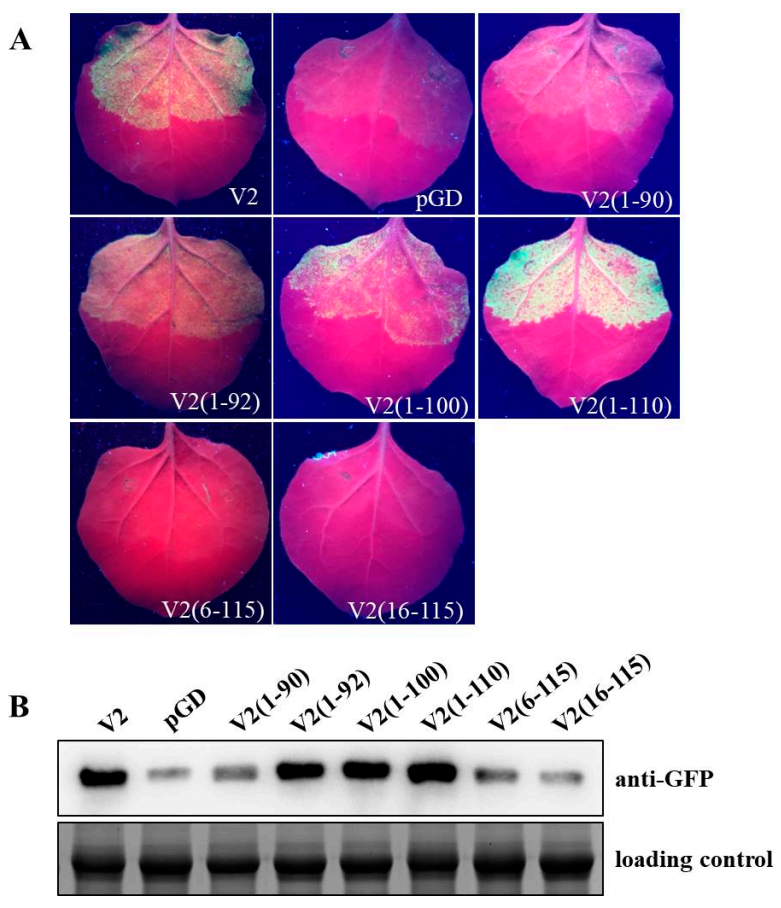
2.2. Residues 1–92 Are Necessary for VSR Activity of V2
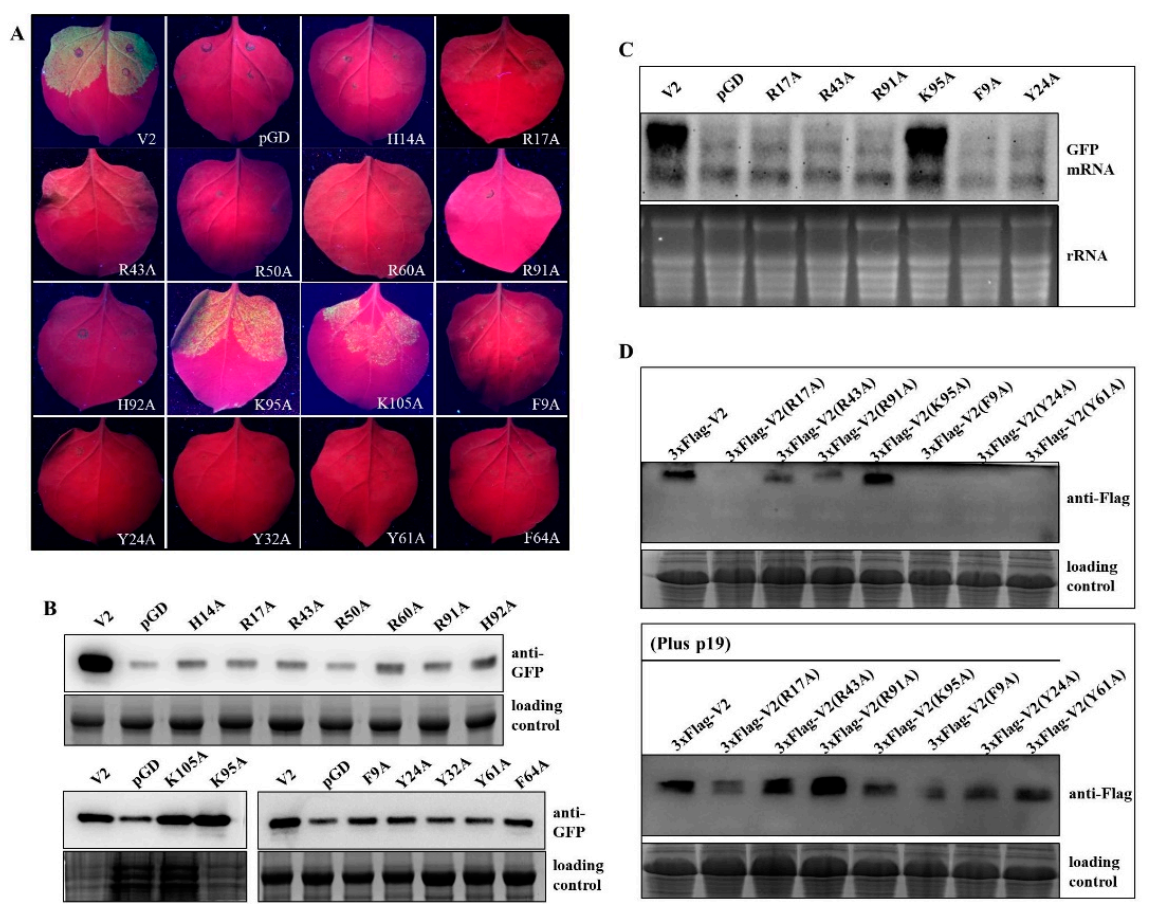
2.3. Basic and Ring-Structured Amino Acids Located in the Conserved Region Were Required for V2 Suppressor Activity
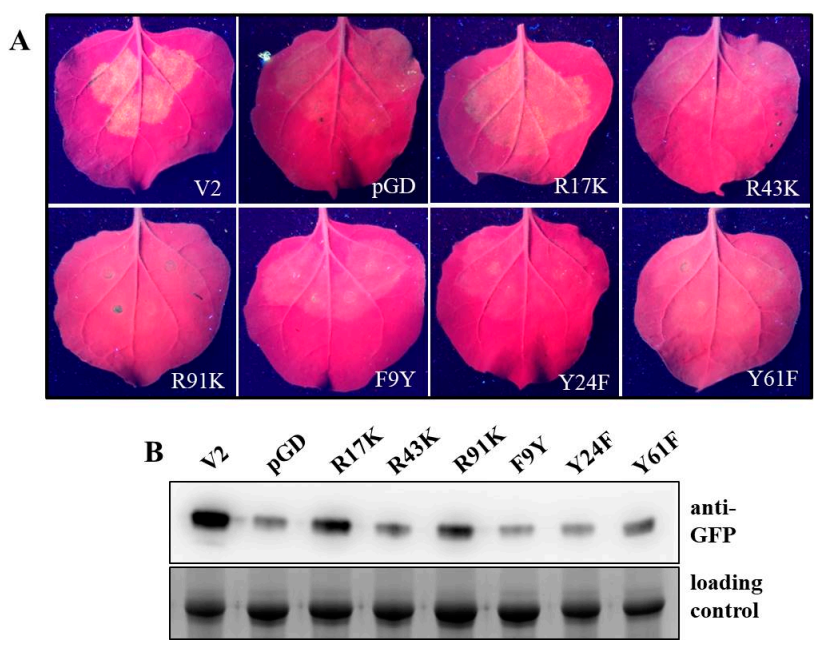
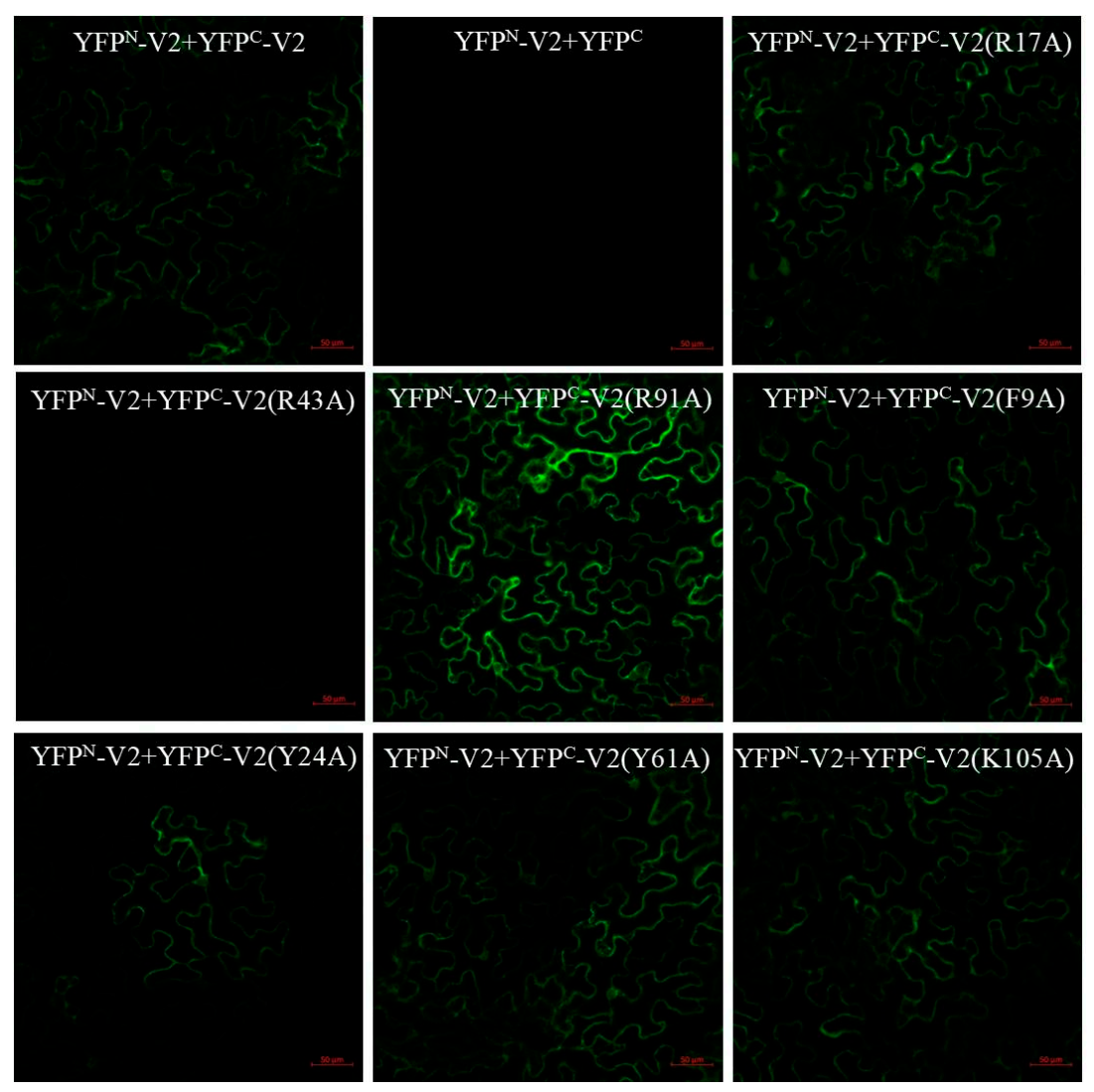
2.4. Specific Amino Acids with Positive Charge or Those Required for V2 Self-Interaction Are Critical for RNA Silencing Suppression
2.5. The Suppressor Activity of V2 Is Dispensable for Symptom Aggravation of Heterogenous PVX Infection
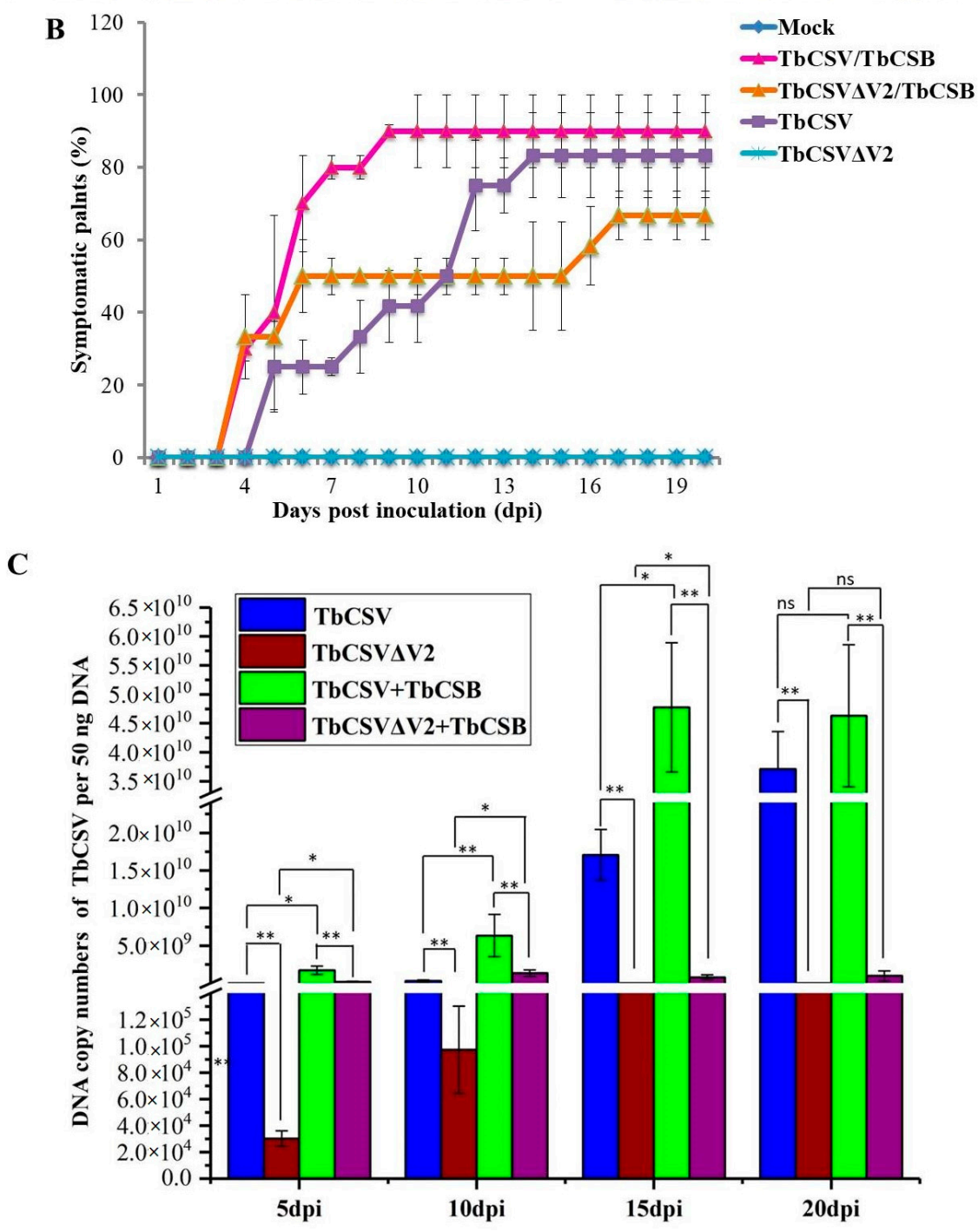
2.6. V2 Is Critical for TbCSV Infection and Symptom Development
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Plasmid Construction
4.3. Agroinfiltration and GFP Fluorescence Imaging
4.4. Western Blot Analyses
4.5. RNA Extraction and Northern Blot Analyses
4.6. BiFC Assay
4.7. qPCR and qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TbCSV | tobacco curly shoot virus |
| TbCSB | betasatellite of TbCSV |
| PV | potato virus X |
| TBSV | tomato bushy stunt virus |
| VSR | viral suppressor of RNA silencing |
| dpi | days postinoculation |
| dpa | days postagroinfiltration |
| GFP | green fluorescence protein |
References
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Carrington, J.C. Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol. 2015, 27, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.W.; Lu, R. Virus-derived siRNAs and piRNAs in immunity and pathogenesis. Curr. Opin. Virol. 2011, 1, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wu, Z.; Li, Y.; Wu, J. Biogenesis, function, and applications of virus-derived small RNAs in plants. Front Microbiol. 2015, 6, 1237. [Google Scholar] [CrossRef] [PubMed]
- Smith, N.A.; Eamens, A.L.; Wang, M.B. Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog. 2011, 7, e1002022. [Google Scholar] [CrossRef]
- Yang, Z.; Li, Y. Dissection of RNAi-based antiviral immunity in plants. Curr. Opin. Virol. 2018, 32, 88–99. [Google Scholar] [CrossRef]
- Ding, S.W.; Voinnet, O. Antiviral immunity directed by small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef]
- Burgyan, J.; Havelda, Z. Viral suppressors of RNA silencing. Trends Plant Sci. 2011, 16, 265–272. [Google Scholar] [CrossRef]
- Kasschau, K.D.; Carrington, J.C. A counterdefensive strategy of plant viruses: Suppression of posttranscriptional gene silencing. Cell 1998, 95, 461–470. [Google Scholar] [CrossRef]
- Pumplin, N.; Voinnet, O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013, 11, 745–760. [Google Scholar] [CrossRef]
- Csorba, T.; Kontra, L.; Burgyan, J. viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 479–480, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Lozsa, R.; Hutvagner, G.; Burgyan, J. Polerovirus protein P0 prevents the assembly of small RNA-containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J. 2010, 62, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Chiu, M.H.; Chen, I.H.; Baulcombe, D.C.; Tsai, C.H. The silencing suppressor P25 of Potato virus X interacts with Argonaute1 and mediates its degradation through the proteasome pathway. Mol. Plant Pathol. 2010, 11, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Merai, Z.; Kerenyi, Z.; Molnar, A.; Barta, E.; Valoczi, A.; Bisztray, G.; Havelda, Z.; Burgyan, J.; Silhavy, D. Aureusvirus P14 is an efficient RNA silencing suppressor that binds double-stranded RNAs without size specificity. J. Virol. 2005, 79, 7217–7226. [Google Scholar] [CrossRef] [PubMed]
- Silhavy, D.; Molnar, A.; Lucioli, A.; Szittya, G.; Hornyik, C.; Tavazza, M.; Burgyan, J. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002, 21, 3070–3080. [Google Scholar] [CrossRef] [PubMed]
- Giner, A.; Lakatos, L.; Garcia-Chapa, M.; Lopez-Moya, J.J.; Burgyan, J. Viral protein inhibits RISC activity by argonaute binding through conserved WG/GW motifs. PLoS Pathog. 2010, 6, e1000996. [Google Scholar] [CrossRef]
- Zhuo, T.; Li, Y.Y.; Xiang, H.Y.; Wu, Z.Y.; Wang, X.B.; Wang, Y.; Zhang, Y.L.; Li, D.W.; Yu, J.L.; Han, C.G. Amino acid sequence motifs essential for P0-mediated suppression of RNA silencing in an isolate of potato leafroll virus from Inner Mongolia. Mol. Plant Microbe Interact. 2014, 27, 515–527. [Google Scholar] [CrossRef]
- Ye, K.; Patel, D.J. RNA silencing suppressor p21 of Beet yellows virus forms an RNA binding octameric ring structure. Structure 2005, 13, 1375–1384. [Google Scholar] [CrossRef][Green Version]
- Li, M.; Zhang, J.; Feng, M.; Wang, X.; Luo, C.; Wang, Q.; Cheng, Y. Characterization of silencing suppressor p24 of Grapevine leafroll-associated virus 2. Mol. Plant Pathol. 2018, 19, 355–368. [Google Scholar] [CrossRef]
- Sahana, N.; Kaur, H.; Jain, R.K.; Palukaitis, P.; Canto, T.; Praveen, S. The asparagine residue in the FRNK box of potyviral helper-component protease is critical for its small RNA binding and subcellular localization. J. Gen. Virol. 2014, 95, 1167–1177. [Google Scholar] [CrossRef]
- Zhou, X. Advances in understanding begomovirus satellites. Annu. Rev. Phytopathol. 2013, 51, 357–381. [Google Scholar] [CrossRef] [PubMed]
- Zerbini, F.M.; Briddon, R.W.; Idris, A.; Martin, D.P.; Moriones, E.; Navas-Castillo, J.; Rivera-Bustamante, R.; Roumagnac, P.; Varsani, A.; Ictv Report, C. ICTV Virus Taxonomy Profile: Geminiviridae. J. Gen. Virol. 2017, 98, 131–133. [Google Scholar] [CrossRef] [PubMed]
- Harrison, B.; Robinson, D. Natural genomic and antigenic variation in whitefly-transmitted geminiviruses (Begomoviruses). Annu. Rev. Phytopathol. 1999, 37, 369–398. [Google Scholar] [CrossRef] [PubMed]
- Harrison, B.D. Advances in Geminivirus Research. Annu. Rev. Phytopathol. 1985, 23, 55–82. [Google Scholar] [CrossRef]
- Hu, T.; Song, Y.; Wang, Y.; Zhou, X. Functional analysis of a novel βV1 gene identified in a geminivirus betasatellite. Sci. China Life Sci. 2020, 63, 688–696. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Yang, X.; Huang, C.; Zhang, X.; Zhou, X. Tomato leaf curl Yunnan virus-encoded C4 induces cell division through enhancing stability of Cyclin D 1.1 via impairing NbSKeta -mediated phosphorylation in Nicotiana benthamiana. PLoS Pathog. 2018, 14, e1006789. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Guo, W.; Li, F.; Sunter, G.; Zhou, X. Geminivirus-associated betasatellites: Exploiting chinks in the antiviral arsenal of Plants. Trends Plant Sci. 2019, 24, 519–529. [Google Scholar] [CrossRef]
- Jia, Q.; Liu, N.; Xie, K.; Dai, Y.; Han, S.; Zhao, X.; Qian, L.; Wang, Y.; Zhao, J.; Gorovits, R.; et al. CLCuMuB βC1 subverts ubiquitination by interacting with NbSKP1s to enhance geminivirus infection in Nicotiana benthamiana. PLoS Pathog. 2016, 12, e1005668. [Google Scholar] [CrossRef]
- Ismayil, A.; Yang, M.; Haxim, Y.; Wang, Y.; Li, J.; Han, L.; Wang, Y.; Zheng, X.; Wei, X.; Nagalakshmi, U.; et al. Cotton leaf curl Multan virus βC1 protein induces autophagy by disrupting the interaction of autophagy-related protein 3 with glyceraldehyde-3-phosphate dehydrogenases. Plant Cell 2020, 32, 1124–1135. [Google Scholar] [CrossRef]
- Rosas-Diaz, T.; Zhang, D.; Fan, P.; Wang, L.; Ding, X.; Jiang, Y.; Jimenez-Gongora, T.; Medina-Puche, L.; Zhao, X.; Feng, Z.; et al. A virus-targeted plant receptor-like kinase promotes cell-to-cell spread of RNAi. Proc. Natl. Acad. Sci. USA 2018, 115, 1388–1393. [Google Scholar] [CrossRef]
- Cui, X.; Li, G.; Wang, D.; Hu, D.; Zhou, X. A Begomovirus DNAβ-encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J. Virol. 2005, 79, 10764–10775. [Google Scholar] [CrossRef]
- Cui, X.; Zhou, X. AC2 and AC4 proteins of Tomato yellow leaf curl China virus and Tobacco curly shoot virus mediate suppression of RNA silencing. Chin. Sci. Bull. 2004, 49, 2607–2612. [Google Scholar] [CrossRef]
- Sun, M.; Jiang, K.; Li, C.; Du, J.; Li, M.; Ghanem, H.; Wu, G.; Qing, L. Tobacco curly shoot virus C3 protein enhances viral replication and gene expression in Nicotiana benthamiana plants. Virus Res. 2020, 281, 197939. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yang, X.; Wang, Y.; Xie, Y.; Zhou, X. Tomato yellow leaf curl virus V2 interacts with host HDA6 to suppress methylation-mediated transcriptional gene silencing in plants. J. Virol. 2018. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y.; Gong, Q.; Ismayil, A.; Yuan, Y.; Lian, B.; Jia, Q.; Han, M.; Deng, H.; Hong, Y.; et al. Geminiviral V2 protein suppresses transcriptional gene silencing through interaction with AGO4. J. Virol. 2019, 92, e00036. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Negrete, E.; Lozano-Duran, R.; Piedra-Aguilera, A.; Cruzado, L.; Bejarano, E.R.; Castillo, A.G. Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. New Phytol. 2013, 199, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, H.; Huang, X.; Xia, R.; Zhao, Q.; Lai, J.; Teng, K.; Li, Y.; Liang, L.; Du, Q.; et al. BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell 2011, 23, 273–288. [Google Scholar] [CrossRef]
- Jing, C.; Li, P.; Zhang, J.; Wang, R.; Wu, G.; Li, M.; Xie, L.; Qing, L. The Malvastrum yellow vein virus C4 protein promotes disease symptom development and enhances virus accumulation in plants. Front. Microbiol. 2019, 10, 2425. [Google Scholar] [CrossRef]
- Castillo-Gonzalez, C.; Liu, X.; Huang, C.; Zhao, C.; Ma, Z.; Hu, T.; Sun, F.; Zhou, Y.; Zhou, X.; Wang, X.J.; et al. Geminivirus-encoded TrAP suppressor inhibits the histone methyltransferase SUVH4/KYP to counter host defense. Elife 2015, 4, e06671. [Google Scholar] [CrossRef]
- Rishishwar, R.; Dasgupta, I. Suppressors of RNA silencing encoded by geminiviruses and associated DNA satellites. Virusdisease 2019, 30, 58–65. [Google Scholar] [CrossRef]
- Padidam, M.; Beachy, R.N.; Fauquet, C.M. The role of AV2 (“precoat”) and coat protein in viral replication and movement in tomato leaf curl geminivirus. Virology 1996, 224, 390–404. [Google Scholar] [CrossRef] [PubMed]
- Poornima Priyadarshini, C.G.; Ambika, M.V.; Tippeswamy, R.; Savithri, H.S. Functional characterization of coat protein and V2 involved in cell to cell movement of Cotton leaf curl Kokhran virus-Dabawali. PLoS ONE 2011, 6, e26929. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, Z.; Sattar, M.N.; Kvarnheden, A.; Mansoor, S.; Briddon, R.W. Effects of the mutation of selected genes of cotton leaf curl Kokhran virus on infectivity, symptoms and the maintenance of cotton leaf curl Multan betasatellite. Virus Res. 2012, 169, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Ikegami, M. Tomato leaf curl Java virus V2 protein is a determinant of virulence, hypersensitive response and suppression of posttranscriptional gene silencing. Virology 2010, 396, 85–93. [Google Scholar] [CrossRef]
- Mubin, M.; Amin, I.; Amrao, L.; Briddon, R.W.; Mansoor, S. The hypersensitive response induced by the V2 protein of a monopartite begomovirus is countered by the C2 protein. Mol. Plant Pathol. 2010, 11, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Luna, A.P.; Morilla, G.; Voinnet, O.; Bejarano, E.R. Functional analysis of gene-silencing suppressors from tomato yellow leaf curl disease viruses. Mol. Plant Microbe Interact. 2012, 25, 1294–1306. [Google Scholar] [CrossRef]
- Wang, B.; Li, F.; Huang, C.; Yang, X.; Qian, Y.; Xie, Y.; Zhou, X. V2 of tomato yellow leaf curl virus can suppress methylation-mediated transcriptional gene silencing in plants. J. Gen. Virol. 2014, 95, 225–230. [Google Scholar] [CrossRef]
- Zrachya, A.; Glick, E.; Levy, Y.; Arazi, T.; Citovsky, V.; Gafni, Y. Suppressor of RNA silencing encoded by Tomato yellow leaf curl virus-Israel. Virology 2007, 358, 159–165. [Google Scholar] [CrossRef]
- Sharma, P.; Ikegami, M.; Kon, T. Identification of the virulence factors and suppressors of posttranscriptional gene silencing encoded by Ageratum yellow vein virus, a monopartite begomovirus. Virus Res. 2010, 149, 19–27. [Google Scholar] [CrossRef]
- Amin, I.; Hussain, K.; Akbergenov, R.; Yadav, J.S.; Qazi, J.; Mansoor, S.; Hohn, T.; Fauquet, C.M.; Briddon, R.W. Suppressors of RNA silencing encoded by the components of the cotton leaf curl begomovirus-betasatellite complex. Mol. Plant Microbe Interact. 2011, 24, 973–983. [Google Scholar] [CrossRef]
- Zhang, J.; Dong, J.; Xu, Y.; Wu, J. V2 protein encoded by Tomato yellow leaf curl China virus is an RNA silencing suppressor. Virus Res. 2012, 163, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Luna, A.P.; Rodriguez-Negrete, E.A.; Morilla, G.; Wang, L.; Lozano-Duran, R.; Castillo, A.G.; Bejarano, E.R. V2 from a curtovirus is a suppressor of post-transcriptional gene silencing. J. Gen. Virol. 2017, 98, 2607–2614. [Google Scholar] [CrossRef] [PubMed]
- Goodin, M.M.; Dietzgen, R.G.; Schichnes, D.; Ruzin, S.; Jackson, A.O. pGD vectors: Versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 2002, 31, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.H.; Xiang, H.Y.; Wang, Q.; Li, Y.Y.; Wu, W.Q.; Han, C.G.; Li, D.W.; Yu, J.L. Ring structure amino acids affect the suppressor activity of melon aphid-borne yellows virus P0 protein. Virology 2010, 406, 21–27. [Google Scholar] [CrossRef]
- Zhao, W.; Ji, Y.; Wu, S.; Ma, X.; Li, S.; Sun, F.; Cheng, Z.; Zhou, Y.; Fan, Y. Single amino acid in V2 encoded by TYLCV is responsible for its self-interaction, aggregates and pathogenicity. Sci. Rep. 2018, 8, 3561. [Google Scholar] [CrossRef]
- Liu, Q.; Guo, R.; Li, M.; Feng, M.; Wang, X.; Wang, Q.; Cheng, Y. Critical regions and residues for self-interaction of grapevine leafroll-associated virus 2 protein p24. Virus Res. 2016, 220, 57–63. [Google Scholar] [CrossRef]
- Chowda-Reddy, R.V.; Achenjang, F.; Felton, C.; Etarock, M.T.; Anangfac, M.T.; Nugent, P.; Fondong, V.N. Role of a geminivirus AV2 protein putative protein kinase C motif on subcellular localization and pathogenicity. Virus Res. 2008, 135, 115–124. [Google Scholar] [CrossRef]
- Saeed, F.; Sattar, M.N.; Hameed, U.; Ilyas, M.; Haider, M.S.; Hamza, M.; Mansoor, S.; Amin, I. Infectivity of okra enation leaf curl virus and the role of its V2 protein in pathogenicity. Virus Res. 2018, 255, 90–94. [Google Scholar] [CrossRef]
- Wartig, L.; Kheyr-Pour, A.; Noris, E.; De Kouchkovsky, F.; Jouanneau, F.; Gronenborn, B.; Jupin, I. Genetic analysis of the monopartite tomato yellow leaf curl geminivirus: Roles of V1, V2, and C2 ORFs in viral pathogenesis. Virology 1997, 228, 132–140. [Google Scholar] [CrossRef]
- Aguilar, E.; Almendral, D.; Allende, L.; Pacheco, R.; Chung, B.N.; Canto, T.; Tenllado, F. The P25 protein of Potato virus X is the main pathogenicity determinant responsible for systemic necrosis in PVX-associated synergisms. J. Virol. 2014, 89, 2090–2103. [Google Scholar] [CrossRef]
- Li, F.; Ding, S.W. Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 2006, 60, 503–531. [Google Scholar] [CrossRef]
- Yang, X.; Xie, Y.; Raja, P.; Li, S.; Wolf, J.N.; Shen, Q.; Bisaro, D.M.; Zhou, X. Suppression of methylation-mediated transcriptional gene silencing by βC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog. 2011, 7, e1002329. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.; Krczal, G.; Wassenegger, M. Three gene products of a begomovirus-betasatellite complex restore expression of a transcriptionally silenced green fluorescent protein transgene in Nicotiana benthamiana. Virus Genes 2015, 50, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Lakatos, L.; Csorba, T.; Pantaleo, V.; Chapman, E.J.; Carrington, J.C.; Liu, Y.P.; Dolja, V.V.; Calvino, L.F.; Lopez-Moya, J.J.; Burgyan, J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006, 25, 2768–2780. [Google Scholar] [CrossRef]
- Kontra, L.; Csorba, T. Distinct effects of p19 RNA silencing suppressor on small RNA mediated pathways in plants. PLoS Pathog. 2016, 12, e1005935. [Google Scholar] [CrossRef]
- Merai, Z.; Kerenyi, Z.; Kertesz, S.; Magna, M.; Lakatos, L.; Silhavy, D. Double-stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J. Virol. 2006, 80, 5747–5756. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, R.; Doudna, J.A. dsRNA with 5’ overhangs contributes to endogenous and antiviral RNA silencing pathways in plants. EMBO J. 2009, 28, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Yang, J.; Lin, C.; Yuan, Y.A. Structural basis for RNA-silencing suppression by Tomato aspermy virus protein 2b. EMBO Rep. 2008, 9, 754–760. [Google Scholar] [CrossRef]
- Schwach, F.; Vaistij, F.E.; Jones, L.; Baulcombe, D.C. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 2005, 138, 1842–1852. [Google Scholar] [CrossRef]
- Di Serio, F.; Martinez de Alba, A.E.; Navarro, B.; Gisel, A.; Flores, R. RNA-dependent RNA polymerase 6 delays accumulation and precludes meristem invasion of a viroid that replicates in the nucleus. J. Virol. 2010, 84, 2477–2489. [Google Scholar] [CrossRef]
- Ghoshal, B.; Sanfacon, H. Symptom recovery in virus-infected plants: Revisiting the role of RNA silencing mechanisms. Virology 2015, 479–480, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.P.; Liu, D.S.; Yan, T.; Fang, X.D.; Dong, K.; Xu, J.; Wang, Y.; Yu, J.L.; Wang, X.B. Cucumber mosaic virus coat protein modulates the accumulation of 2b protein and antiviral silencing that causes symptom recovery in planta. PLoS Pathog. 2017, 13, e1006522. [Google Scholar] [CrossRef] [PubMed]
- Sunpapao, A.; Nakai, T.; Dong, F.; Mochizuki, T.; Ohki, S.T. The 2b protein of cucumber mosaic virus is essential for viral infection of the shoot apical meristem and for efficient invasion of leaf primordia in infected tobacco plants. J. Gen. Virol. 2009, 90, 3015–3021. [Google Scholar] [CrossRef] [PubMed]
- Martin-Hernandez, A.M.; Baulcombe, D.C. Tobacco rattle virus 16-kilodalton protein encodes a suppressor of RNA silencing that allows transient viral entry in meristems. J. Virol. 2008, 82, 4064–4071. [Google Scholar] [CrossRef]
- Hormuzdi, S.G.; Bisaro, D.M. Genetic analysis of beet curly top virus: Examination of the roles of L2 and L3 genes in viral pathogenesis. Virology 1995, 206, 1044–1054. [Google Scholar] [CrossRef][Green Version]
- Raja, P.; Sanville, B.C.; Buchmann, R.C.; Bisaro, D.M. Viral genome methylation as an epigenetic defense against geminiviruses. J. Virol. 2008, 82, 8997–9007. [Google Scholar] [CrossRef]
- Raja, P.; Jackel, J.N.; Li, S.; Heard, I.M.; Bisaro, D.M. Arabidopsis double-stranded RNA binding protein DRB3 participates in methylation-mediated defense against geminiviruses. J. Virol. 2014, 88, 2611–2622. [Google Scholar] [CrossRef]
- Coursey, T.; Regedanz, E.; Bisaro, D.M. Arabidopsis RNA polymerase V mediates enhanced compaction and silencing of geminivirus and transposon chromatin during host recovery from infection. J. Virol. 2018, 92, e01320-17. [Google Scholar] [CrossRef]
- Cao, X.; Zhou, P.; Zhang, X.; Zhu, S.; Zhong, X.; Xiao, Q.; Ding, B.; Li, Y. Identification of an RNA silencing suppressor from a plant double-stranded RNA virus. J. Virol. 2005, 79, 13018–13027. [Google Scholar] [CrossRef]
- Li, P.; Jing, C.; Ren, H.; Jia, Z.; Ghanem, H.; Wu, G.; Li, M.; Qing, L. Analysis of Pathogenicity and Virulence Factors of Ageratum leaf curl Sichuan virus. Front. Plant Sci. 2020, 11, 527787. [Google Scholar] [CrossRef]
- Mason, G.; Caciagli, P.; Accotto, G.P.; Noris, E. Real-time PCR for the quantitation of Tomato yellow leaf curl Sardinia virus in tomato plants and in Bemisia tabaci. J. Virol. Methods 2008, 147, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−∆∆C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Li, C.; Jiang, K.; Li, K.; Zhang, J.; Sun, M.; Wu, G.; Qing, L. Characterization of Pathogenicity-Associated V2 Protein of Tobacco Curly Shoot Virus. Int. J. Mol. Sci. 2021, 22, 923. https://doi.org/10.3390/ijms22020923
Li M, Li C, Jiang K, Li K, Zhang J, Sun M, Wu G, Qing L. Characterization of Pathogenicity-Associated V2 Protein of Tobacco Curly Shoot Virus. International Journal of Molecular Sciences. 2021; 22(2):923. https://doi.org/10.3390/ijms22020923
Chicago/Turabian StyleLi, Mingjun, Changchang Li, Kairong Jiang, Ke Li, Junlei Zhang, Miao Sun, Gentu Wu, and Ling Qing. 2021. "Characterization of Pathogenicity-Associated V2 Protein of Tobacco Curly Shoot Virus" International Journal of Molecular Sciences 22, no. 2: 923. https://doi.org/10.3390/ijms22020923
APA StyleLi, M., Li, C., Jiang, K., Li, K., Zhang, J., Sun, M., Wu, G., & Qing, L. (2021). Characterization of Pathogenicity-Associated V2 Protein of Tobacco Curly Shoot Virus. International Journal of Molecular Sciences, 22(2), 923. https://doi.org/10.3390/ijms22020923



