Transcriptome Analysis Revealed GhWOX4 Intercedes Myriad Regulatory Pathways to Modulate Drought Tolerance and Vascular Growth in Cotton
Abstract
1. Introduction
2. Results
2.1. Identification and Structure Analysis of GhWOX4
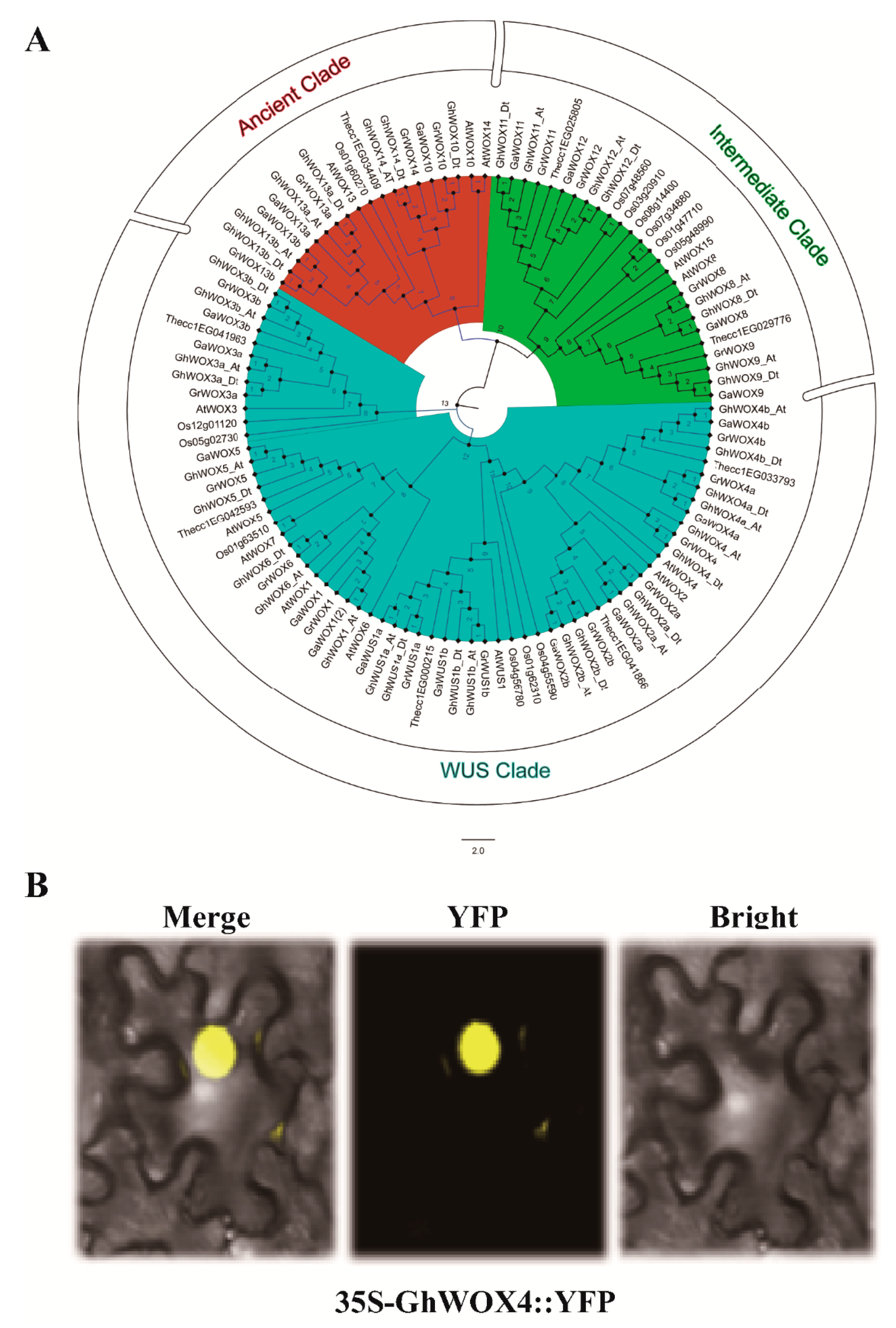
2.2. Sub-Cellular Localization and Phylogenetic Analysis of GhWOX4
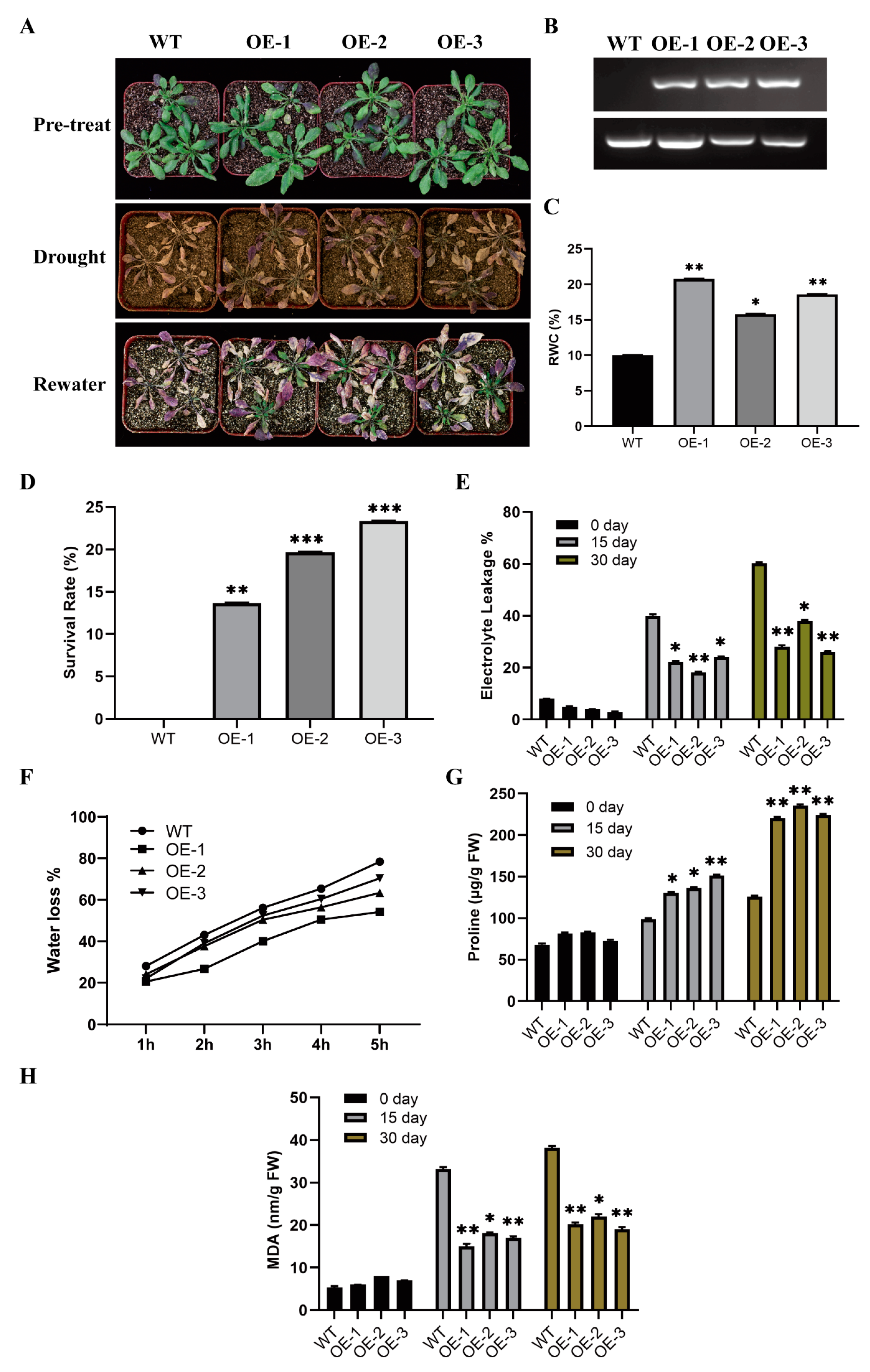
2.3. Overexpression of GhWOX4 Enhanced Drought Tolerance
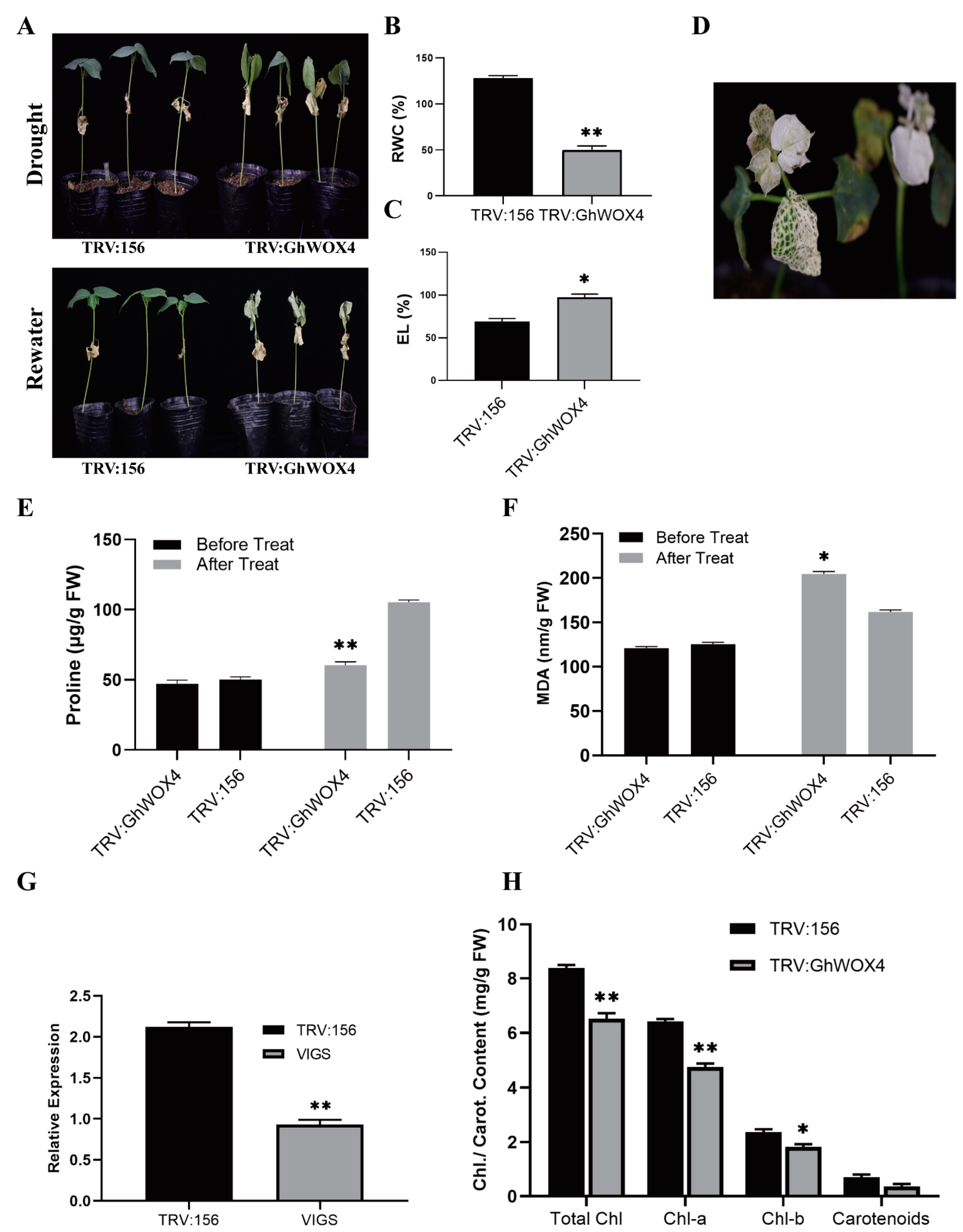
2.4. Knock-Down of GhWOX4 Bottled-Up Tolerance to Drought Stress
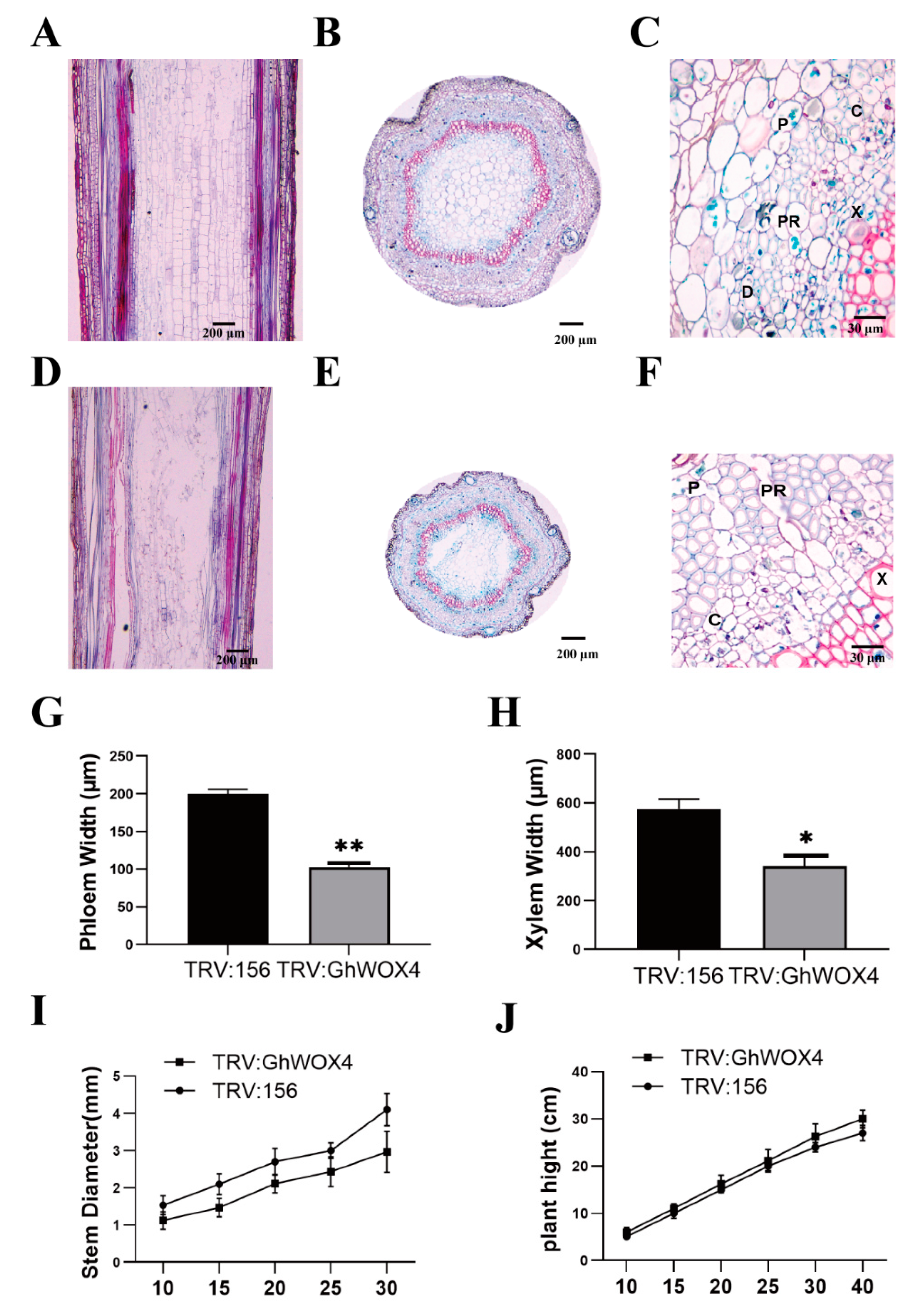
2.5. Knock-Down of GhWOX4 Inhibits the Vascular Cell Growth
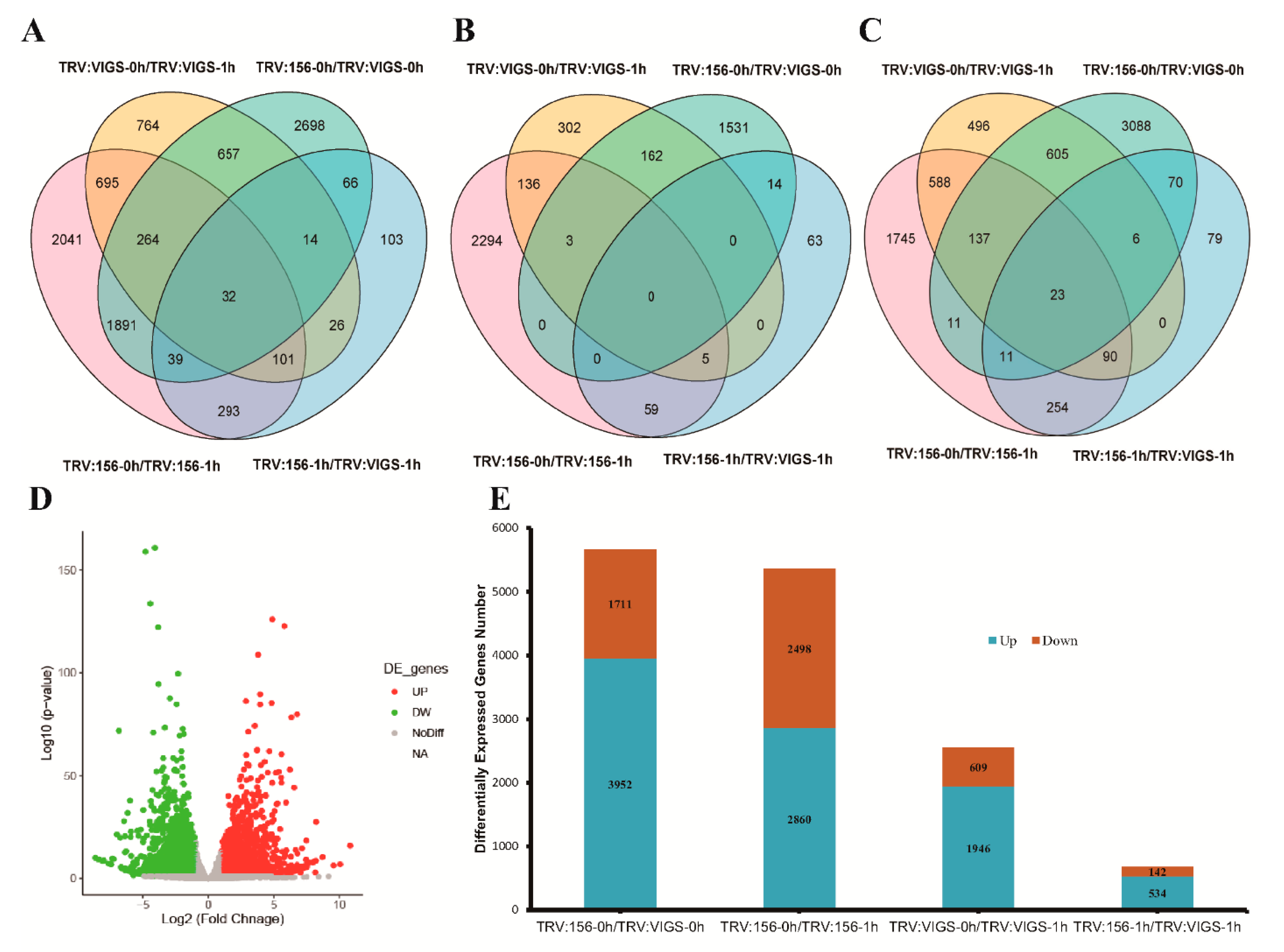
2.6. Transcriptome Analysis and Result Summary
2.7. mRNA-Seq Revealed Differentially Expressed Genes (DEGs)
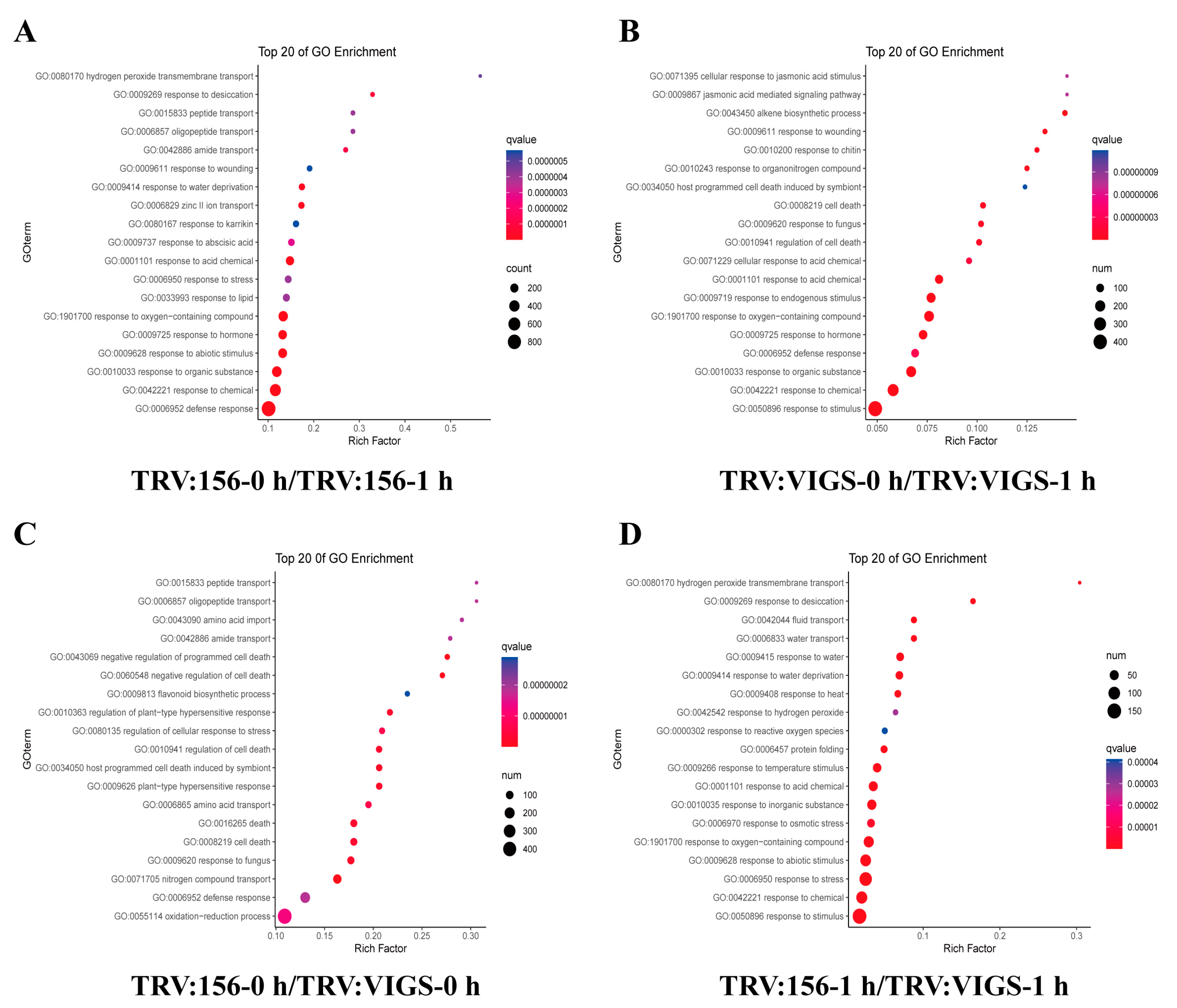
2.8. GO Analysis Revealed GhWOX4 Biological Functions
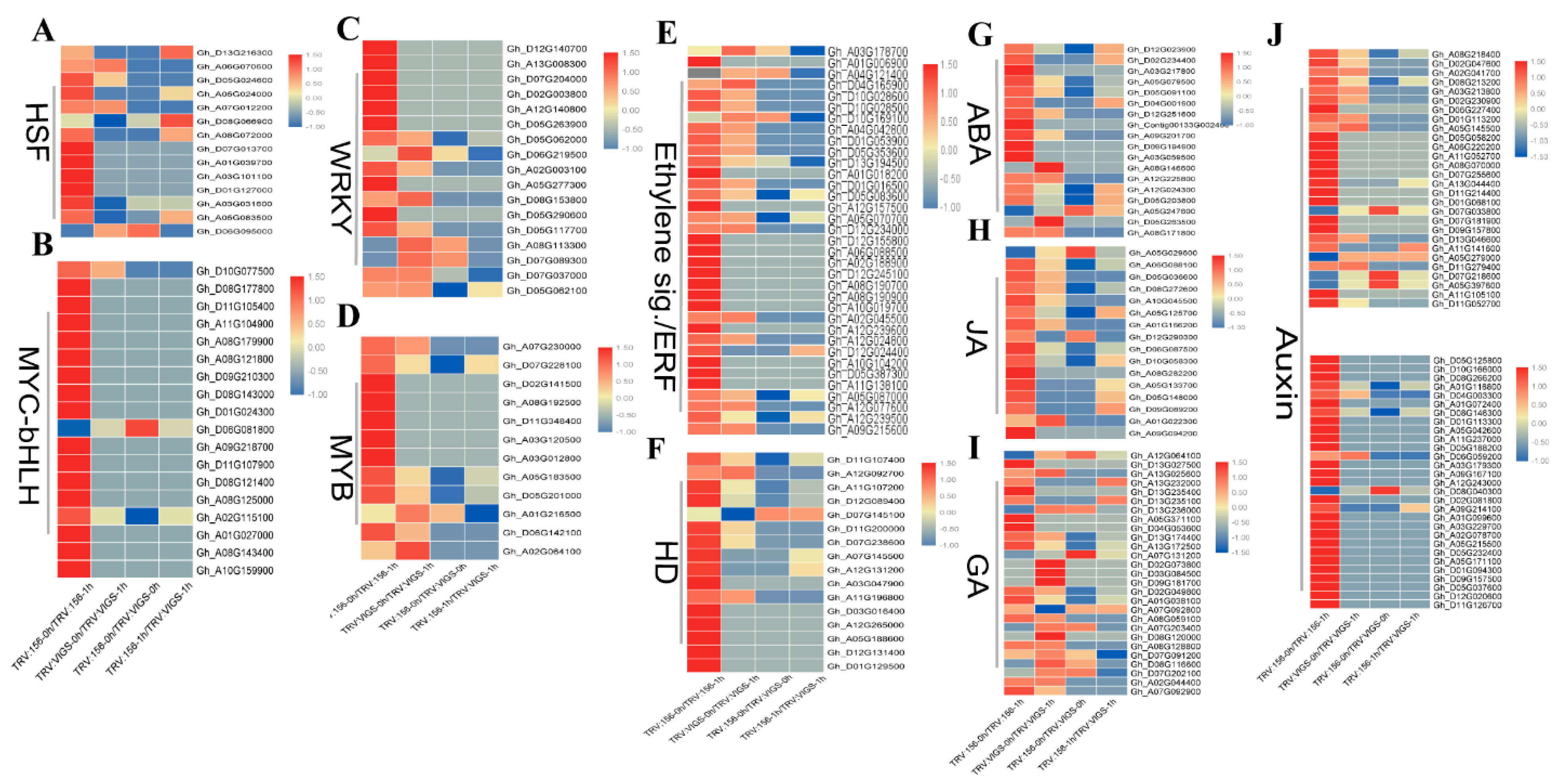
2.9. TFs and Hormones Play Key Roles in GhWOX4 Mediated Plant Growth and Drought Tolerance
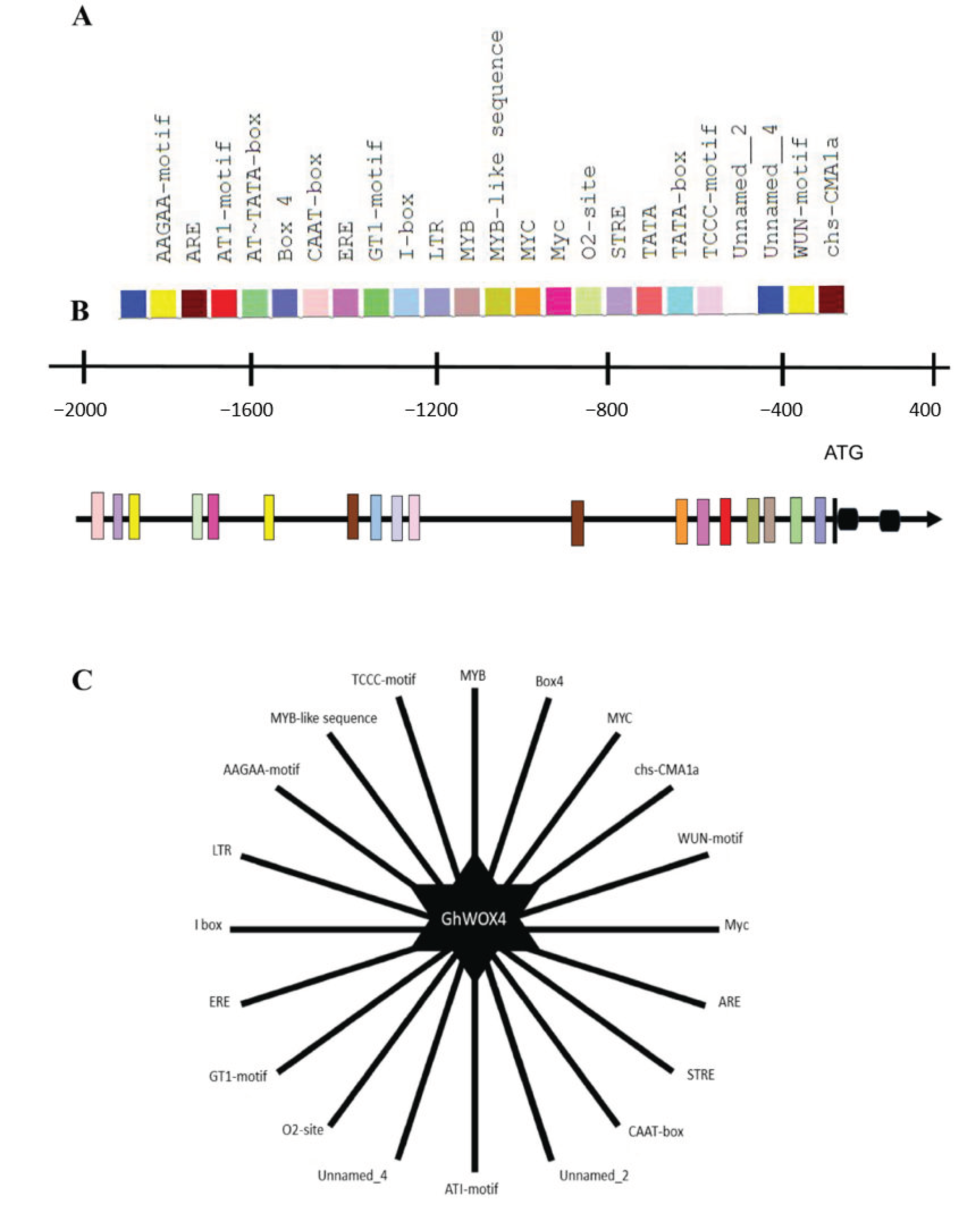
2.10. Insilico Promotor Analysis Identified the Important Cis-Acting Regulatory Elements (CAREs)
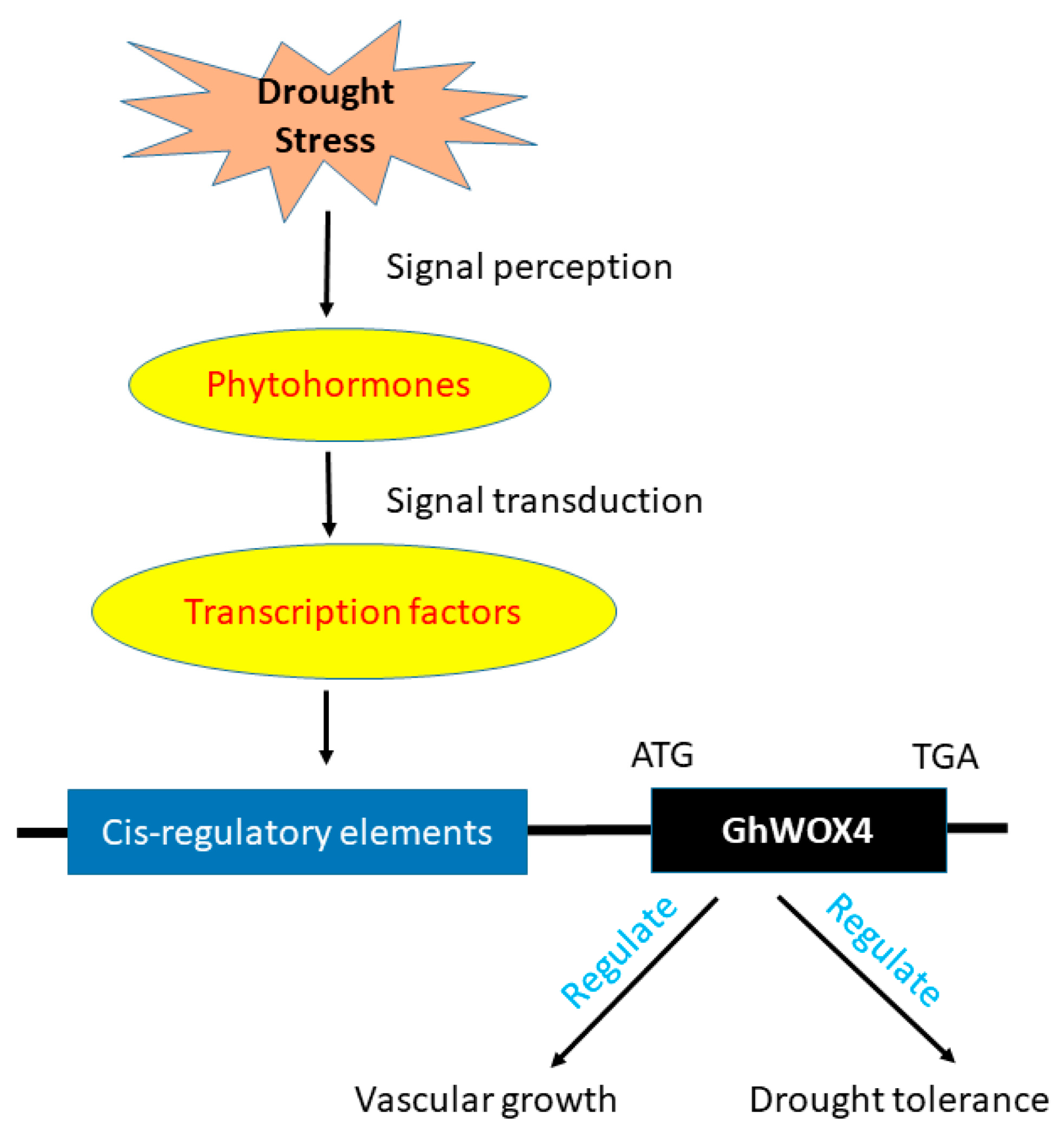
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Cloning and Ectopic Expression of GhWOX4
4.3. RNA Extraction and Expression Analysis by qRT-PCR
4.4. TRV-Mediated Gene Silencing and Drought Assay
4.5. GhWOX4 Subcellular Localization
4.6. Physiological Indices
4.7. Chlorophyll and Carotenoid Contents
4.8. Anatomical Analysis
4.9. RNA-Seq Library Construction and Data Analysis
4.10. Insilico Promotor Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GhWOX | Gossypium hirsutum WUSCHEL-Related homeobox |
| AUX/1AA | Auxin/Indole-acetic-acid |
| bHLH | Basic-helix-loop-helix protein |
| WRKY | Transcription factor (TF), partake WRKYGQK heptapeptide at N-terminal |
| MYB | Myeloblastosis (Avian Myeloblastosis Viral Oncogene Homolog) TF |
| myc | Melocytomatosis (isolated from an avian myelocytomatosis virus) TF |
| TRV | Tobacco rattle virus |
| ERF | Ethylene response factor |
| YFP | Yellow-fluorescent protein |
| PDS | Phytoene-desaturase |
| ROS | Reactive oxygen species |
| FDR | False discovery rate |
| PYR/PYL | Pyrabactin resistance1/PYR1; PYR-like (PYL) |
| CBL | Calcineurin B-like protein |
| AP2/EREBP | Apetala2/Ethylene response element binding protein |
| AGD1 | ADP-Ribosylation factor GTPase-activating protein domain |
| ARF | Auxin response factor |
| ARE | Auxin response element |
| SAUR | Small-auxin upregulated RNA |
| ABRE/ABF | ABA-responsive element/ABRE-binding factor |
| AIP1 | Actin-interacting protein1 |
| CYPs | Cytochrome 450 proteins |
| GA2OX8/GASA4 | Gibbrellin 2-Oxidase; Gibbrellic acid stimulated Arabidopsis |
| HB/HD | Homeobox/Homeobox-domain |
| GRAS | GIBBERELLIC ACID INSENSITIVE (GAI), REPRESSOR OF GA1 (RGA), and SCARECROW (SCR) |
| LOB | Lateral-organ-boundaries domain |
| bZIP | Bacic-leucine zipper |
| ATHB | Arabidopsis thaliana-Homeobox |
| ANL | Anthocyanin-Less |
| LHW | Lonesome-highway |
| RF2B | Fertility restorer-B |
| RVE7 | Protein REVEILLE7 |
| HAT | HD-Zip protein |
| STRE | StuA-response element |
| MES | Morpholinoethanesulfonic acid |
| pEG101 | pEarleyGate101 (vector) |
| ATP | Adenosine tri-phosphate |
References
- Gao, X.; Wheeler, T.; Li, Z.; Kenerley, C.M.; He, P.; Shan, L. Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 2011, 66, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Li, S.; Yang, S.; Wang, L.; Guo, W. RETRACTED ARTICLE: Overexpression of a cotton annexin gene, GhAnn1, enhances drought and salt stress tolerance in transgenic cotton. Plant Mol. Biol. 2014, 87, 47–67. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Feng, L.; Wei, N.; Liu, Z.-H.; Hu, S.; Li, X.-B. Overexpression of cotton PYL genes in Arabidopsis enhances the transgenic plant tolerance to drought stress. Plant Physiol. Biochem. 2017, 115, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.K.; Agarwal, P.; Reddy, M.K.; Sopory, S.K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006, 25, 1263–1274. [Google Scholar] [CrossRef]
- Tuteja, N. Abscisic Acid and abiotic stress signaling. Plant Signal Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef]
- Osakabe, Y.; Osakabe, K.; Shinozaki, K.; Tran, L.-S.P. Response of plants to water stress. Front. Plant Sci. 2014, 5, 86. [Google Scholar] [CrossRef]
- Campo, S.; Baldrich, P.; Messeguer, J.; Lalanne, E.; Coca, M.; San Segundo, B. Overexpression of a Calcium-Dependent Protein Kinase Confers Salt and Drought Tolerance in Rice by Preventing Membrane Lipid Peroxidation. Plant Physiol. 2014, 165, 688–704. [Google Scholar] [CrossRef]
- Lucas, W.J.; Groover, A.; Lichtenberger, R.; Furuta, K.; Yadav, S.-R.; Helariutta, Y.; He, X.-Q.; Fukuda, H.; Kang, J.; Brady, S.M.; et al. The Plant Vascular System: Evolution, Development and FunctionsF. J. Integr. Plant Biol. 2013, 55, 294–388. [Google Scholar] [CrossRef]
- Kehr, J.; Buhtz, A. Long distance transport and movement of RNA through the phloem. J. Exp. Bot. 2007, 59, 85–92. [Google Scholar] [CrossRef]
- Notaguchi, M.; Okamoto, S. Dynamics of long-distance signaling via plant vascular tissues. Front. Plant Sci. 2015, 6, 161. [Google Scholar] [CrossRef]
- Suer, S.; Agusti, J.; Sanchez, P.; Schwarz, M.; Greb, T. WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis. Plant Cell 2011, 23, 3247–3259. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Strable, J.; Shimizu, R.; Koenig, D.; Sinha, N.; Scanlon, M.J. WOX4 promotes procambial development. Plant Physiol. 2010, 152, 1346–1356. [Google Scholar] [CrossRef] [PubMed]
- Etchells, J.P.; Provost, C.M.; Mishra, L.; Turner, S.R. WOX4 and WOX14 act downstream of the PXY receptor kinase to regulate plant vascular proliferation independently of any role in vascular organisation. Development 2013, 140, 2224–2234. [Google Scholar] [CrossRef] [PubMed]
- Yasui, Y.; Ohmori, Y.; Takebayashi, Y.; Sakakibara, H.; Hirano, H.-Y. WUSCHEL-RELATED HOMEOBOX4 acts as a key regulator in early leaf development in rice. PLoS Genet. 2018, 14, e1007365. [Google Scholar] [CrossRef] [PubMed]
- Kucukoglu, M.; Nilsson, J.; Zheng, B.; Chaabouni, S.; Nilsson, O. WUSCHEL-RELATED HOMEOBOX4 (WOX4)-like genes regulate cambial cell division activity and secondary growth in Populus trees. New Phytol. 2017, 215, 642–657. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Tan, F.; Lu, Y.; Liu, X.; Li, T.; Yuan, W.; Zhao, Y.; Zhou, D.-X. WOX11 recruits a histone H3K27me3 demethylase to promote gene expression during shoot development in rice. Nucleic Acids Res. 2018, 46, 2356–2369. [Google Scholar] [CrossRef]
- Cheng, S.; Huang, Y.; Zhu, N.; Zhao, Y. The rice WUSCHEL-related homeobox genes are involved in reproductive organ development, hormone signaling and abiotic stress response. Gene 2014, 549, 266–274. [Google Scholar] [CrossRef]
- Minh-Thu, P.-H.; Kim, J.S.; Chae, S.; Jun, K.M.; Lee, G.; Kim, D.; Cheong, J.; Song, S.I.; Nahm, B.H.; Kim, Y. A WUSCHEL Homeobox Transcription Factor, OsWOX13, Enhances Drought Tolerance and Triggers Early Flowering in Rice. Mol. Cells 2018, 41, 781–798. [Google Scholar] [CrossRef]
- Qiao, Y.; Liu, L.; Xiong, Q.; Flores, C.; Wong, J.; Shi, J.; Wang, X.; Liu, X.; Xiang, Q.; Jiang, S.; et al. Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 2013, 45, 330–333. [Google Scholar] [CrossRef]
- Smetana, O.; Mäkilä, R.; Lyu, M.; Amiryousefi, A.; Sánchez Rodríguez, F.; Wu, M.-F.; Solé-Gil, A.; Leal Gavarrón, M.; Siligato, R.; Miyashima, S.; et al. High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature 2019, 565, 485–489. [Google Scholar] [CrossRef]
- Shariatipour, N.; Heidari, B. Meta-Analysis of Expression of the Stress Tolerance Associated Genes and Uncover their Cis-Regulatory Elements in Rice (Oryza sativa L.). Open Bioinform. J. 2020, 13, 39–49. [Google Scholar] [CrossRef]
- Yang, Z.; Chi, X.; Guo, F.; Jin, X.; Luo, H.; Hawar, A.; Chen, Y.; Feng, K.; Wang, B.; Qi, J.; et al. SbWRKY30 enhances the drought tolerance of plants and regulates a drought stress-responsive gene, SbRD19, in sorghum. J. Plant Physiol. 2020, 246–247, 153142. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Yang, X.; Wang, L.; Zhu, L.; Zhou, T.; Deng, J.; Zhang, X. Molecular cloning and functional characterization of a novel cotton CBL-interacting protein kinase gene (GhCIPK6) reveals its involvement in multiple abiotic stress tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 2013, 435, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Ogden, A.J.; Bhatt, J.J.; Brewer, H.M.; Kintigh, J.; Kariuki, S.M.; Rudrabhatla, S.; Adkins, J.N.; Curtis, W.R. Phloem Exudate Protein Profiles during Drought and Recovery Reveal Abiotic Stress Responses in Tomato Vasculature. Int. J. Mol. Sci. 2020, 21, 4461. [Google Scholar] [CrossRef]
- You, J.; Zhang, Y.; Liu, A.; Li, D.; Wang, X.; Dossa, K.; Zhou, R.; Yu, J.; Zhang, Y.; Wang, L.; et al. Transcriptomic and metabolomic profiling of drought-tolerant and susceptible sesame genotypes in response to drought stress. BMC Plant Biol. 2019, 19, 267. [Google Scholar] [CrossRef]
- Zhang, Z.-F.; Li, Y.-Y.; Xiao, B.-Z. Comparative transcriptome analysis highlights the crucial roles of photosynthetic system in drought stress adaptation in upland rice. Sci. Rep. 2016, 6, 19349. [Google Scholar] [CrossRef]
- Xie, Z.; Li, D.; Wang, L.; Sack, F.D.; Grotewold, E. Role of the stomatal development regulators FLP/MYB88 in abiotic stress responses. Plant J. 2010, 64, 731–739. [Google Scholar] [CrossRef]
- Huang, D.; Wu, W.; Abrams, S.R.; Cutler, A.J. The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 2008, 59, 2991–3007. [Google Scholar] [CrossRef]
- Oshima, Y.; Shikata, M.; Koyama, T.; Ohtsubo, N.; Mitsuda, N.; Ohme-Takagi, M. MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell 2013, 25, 1609–1624. [Google Scholar] [CrossRef]
- Moreno, A.A.; Mukhtar, M.S.; Blanco, F.; Boatwright, J.L.; Moreno, I.; Jordan, M.R.; Chen, Y.; Brandizzi, F.; Dong, X.; Orellana, A.; et al. IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS ONE 2012, 7, e31944. [Google Scholar] [CrossRef]
- Brady, S.M.; Zhang, L.; Megraw, M.; Martinez, N.J.; Jiang, E.; Yi, C.S.; Liu, W.; Zeng, A.; Taylor-Teeples, M.; Kim, D.; et al. A stele-enriched gene regulatory network in the Arabidopsis root. Mol. Syst. Biol. 2011, 7, 459. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Zhang, Z.; Chen, S.; Beachy, R.N. RF2b, a rice bZIP transcription activator, interacts with RF2a and is involved in symptom development of rice tungro disease. Proc. Natl. Acad. Sci. USA 2004, 101, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kang, J.-y.; Cho, D.-I.; Park, J.H.; Kim, S.Y. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 2004, 40, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Yamaguchi-Shinozaki, K.; Urao, T.; Iwasaki, T.; Hosokawa, D.; Shinozaki, K. Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar] [CrossRef] [PubMed]
- Rawat, R.; Schwartz, J.; Jones, M.A.; Sairanen, I.; Cheng, Y.; Andersson, C.R.; Zhao, Y.; Ljung, K.; Harmer, S.L. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc. Natl. Acad. Sci. USA 2009, 106, 16883–16888. [Google Scholar] [CrossRef]
- Kwon, Y.; Kim, J.H.; Nguyen, H.N.; Jikumaru, Y.; Kamiya, Y.; Hong, S.-W.; Lee, H. A novel Arabidopsis MYB-like transcription factor, MYBH, regulates hypocotyl elongation by enhancing auxin accumulation. J. Exp. Bot. 2013, 64, 3911–3922. [Google Scholar] [CrossRef]
- Chen, J.; Nolan, T.M.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yin, Y. Arabidopsis WRKY46, WRKY54, and WRKY70 Transcription Factors Are Involved in Brassinosteroid-Regulated Plant Growth and Drought Responses. Plant Cell 2017, 29, 1425–1439. [Google Scholar] [CrossRef]
- Ueda, M.; Zhang, Z.; Laux, T. Transcriptional Activation of Arabidopsis Axis Patterning Genes WOX8/9 Links Zygote Polarity to Embryo Development. Dev. Cell 2011, 20, 264–270. [Google Scholar] [CrossRef]
- Ma, Q.; Xia, Z.; Cai, Z.; Li, L.; Cheng, Y.; Liu, J.; Nian, H. GmWRKY16 Enhances Drought and Salt Tolerance Through an ABA-Mediated Pathway in Arabidopsis thaliana. Front. Plant Sci. 2019, 9, 1979. [Google Scholar] [CrossRef]
- Soderman, E.; Mattsson, J.; Engstrom, P. The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. Plant J. 1996, 10, 375–381. [Google Scholar] [CrossRef]
- Shi, W.; Cheng, J.; Wen, X.; Wang, J.; Shi, G.; Yao, J.; Hou, L.; Sun, Q.; Xiang, P.; Yuan, X.; et al. Transcriptomic studies reveal a key metabolic pathway contributing to a well-maintained photosynthetic system under drought stress in foxtail millet (Setaria italica L.). PeerJ 2018, 6, e4752. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [PubMed]
- Bouchez, O.; Huard, C.; Lorrain, S.; Roby, D.; Balagué, C. Ethylene is one of the key elements for cell death and defense response control in the Arabidopsis lesion mimic mutant vad1. Plant Physiol. 2007, 145, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Barra-Jiménez, A.; Ragni, L. Secondary development in the stem: When Arabidopsis and trees are closer than it seems. Curr. Opin. Plant Biol. 2017, 35, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Oparka, K.J.; Turgeon, R. Sieve elements and companion cells-traffic control centers of the phloem. Plant Cell 1999, 11, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Gujas, B.; Rodriguez-Villalon, A. Plant Phosphoglycerolipids: The Gatekeepers of Vascular Cell Differentiation. Front. Plant Sci. 2016, 7, 103. [Google Scholar] [CrossRef]
- Liu, Y.; Patra, B.; Pattanaik, S.; Wang, Y.; Yuan, L. GATA and Phytochrome Interacting Factor Transcription Factors Regulate Light-Induced Vindoline Biosynthesis in Catharanthus roseus. Plant Physiol. 2019, 180, 1336–1350. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Exposito-Rodriguez, M.; Laissue, P.P.; Yvon-Durocher, G.; Smirnoff, N.; Mullineaux, P.M. Photosynthesis-dependent H2O2 transfer from chloroplasts to nuclei provides a high-light signalling mechanism. Nat. Commun. 2017, 8, 49. [Google Scholar] [CrossRef]
- Zhang, C.; Fan, L.; Le, B.H.; Ye, P.; Mo, B.; Chen, X. Regulation of ARGONAUTE10 Expression Enables Temporal and Spatial Precision in Axillary Meristem Initiation in Arabidopsis. Dev. Cell 2020. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method forAgrobacterium-mediated transformation ofArabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, M.; Wang, P.; Cox, K.L.; Duan, L.; Dever, J.K.; Shan, L.; Li, Z.; He, P. Regulation of cotton (Gossypium hirsutum) drought responses by mitogen-activated protein (MAP) kinase cascade-mediated phosphorylation of GhWRKY59. New Phytol. 2017, 215, 1462–1475. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, L.; Zhou, K.; Zhang, Y.; Han, X.; Din, Y.; Ge, X.; Qin, W.; Wang, P.; Li, F.; et al. GhWRKY6 Acts as a Negative Regulator in Both Transgenic Arabidopsis and Cotton During Drought and Salt Stress. Front. Genet. 2019, 10, 392. [Google Scholar] [CrossRef] [PubMed]
- Parsons, S.A. A Practical Handbook of Seawater Analysis. Fisheries Research Board of Canada Bulletin 167. J. D. H. Strickland, T. R. Parsons. Q. Rev. Biol. 1969, 44, 327. [Google Scholar] [CrossRef]
- Porra, R.J.; Thompson, W.A.; Kriedemann, P.E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta (BBA) Bioenergy 1989, 975, 384–394. [Google Scholar] [CrossRef]
- Tuominen, H.; Puech, L.; Fink, S.; Sundberg, B. A Radial Concentration Gradient of Indole-3-Acetic Acid Is Related to Secondary Xylem Development in Hybrid Aspen. Plant Physiol. 1997, 115, 577–585. [Google Scholar] [CrossRef]
- Nilsson, J.; Karlberg, A.; Antti, H.; Lopez-Vernaza, M.; Mellerowicz, E.; Perrot-Rechenmann, C.; Sandberg, G.; Bhalerao, R.P. Dissecting the molecular basis of the regulation of wood formation by auxin in hybrid aspen. Plant Cell 2008, 20, 843–855. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y.; Wang, X.; Wang, W.; Li, Z.; Wu, J.; Wang, G.; Wu, L.; Zhang, G.; Ma, Z. HyPRP1 performs a role in negatively regulating cotton resistance to V. dahliae via the thickening of cell walls and ROS accumulation. BMC Plant Biol. 2018, 18, 339. [Google Scholar] [CrossRef]

| Sample ID | Read Sum | Mapped Read Sum | Map Rate (%) | GC (%) |
|---|---|---|---|---|
| * PYL_VIGS-0 h_R1 | 46,162,896 | 44,371,337 | 96.12 | 45.59 |
| PYL_VIGS-0 h_R2 | 43,242,842 | 41,587,275 | 96.17 | 45.22 |
| PYL_VIGS-0 h_R3 | 42,443,503 | 40,570,587 | 95.59 | 45.35 |
| PYL_VIGS-1 h_R1 | 56,984,332 | 54,501,470 | 95.64 | 45.14 |
| PYL_VIGS-1 h_R2 | 47,557,755 | 45,492,367 | 95.66 | 45.11 |
| PYL_VIGS-1 h_R3 | 44,546,148 | 42,544,884 | 95.51 | 45.29 |
| * PYL_156-0 h_R1 | 51,385,206 | 49,241,433 | 95.83 | 45.39 |
| PYL_156-0 h_R2 | 45,698,425 | 43,788,845 | 95.82 | 45.45 |
| PYL_156-0 h_R3 | 49,625,416 | 47,602,859 | 95.92 | 45.39 |
| PYL_156-1 h_R1 | 43,066,450 | 41,246,640 | 95.77 | 45.05 |
| PYL_156-1 h_R2 | 41,804,713 | 39,678,862 | 94.91 | 45 |
| PYL_156-1 h_R3 | 47,323,891 | 45,363,867 | 95.86 | 45.32 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sajjad, M.; Wei, X.; Liu, L.; Li, F.; Ge, X. Transcriptome Analysis Revealed GhWOX4 Intercedes Myriad Regulatory Pathways to Modulate Drought Tolerance and Vascular Growth in Cotton. Int. J. Mol. Sci. 2021, 22, 898. https://doi.org/10.3390/ijms22020898
Sajjad M, Wei X, Liu L, Li F, Ge X. Transcriptome Analysis Revealed GhWOX4 Intercedes Myriad Regulatory Pathways to Modulate Drought Tolerance and Vascular Growth in Cotton. International Journal of Molecular Sciences. 2021; 22(2):898. https://doi.org/10.3390/ijms22020898
Chicago/Turabian StyleSajjad, Muhammad, Xi Wei, Lisen Liu, Fuguang Li, and Xiaoyang Ge. 2021. "Transcriptome Analysis Revealed GhWOX4 Intercedes Myriad Regulatory Pathways to Modulate Drought Tolerance and Vascular Growth in Cotton" International Journal of Molecular Sciences 22, no. 2: 898. https://doi.org/10.3390/ijms22020898
APA StyleSajjad, M., Wei, X., Liu, L., Li, F., & Ge, X. (2021). Transcriptome Analysis Revealed GhWOX4 Intercedes Myriad Regulatory Pathways to Modulate Drought Tolerance and Vascular Growth in Cotton. International Journal of Molecular Sciences, 22(2), 898. https://doi.org/10.3390/ijms22020898






