NMR-Based Structural Characterization of a Two-Disulfide-Bonded Analogue of the FXIIIa Inhibitor Tridegin: New Insights into Structure–Activity Relationships
Abstract
1. Introduction
2. Results and Discussion
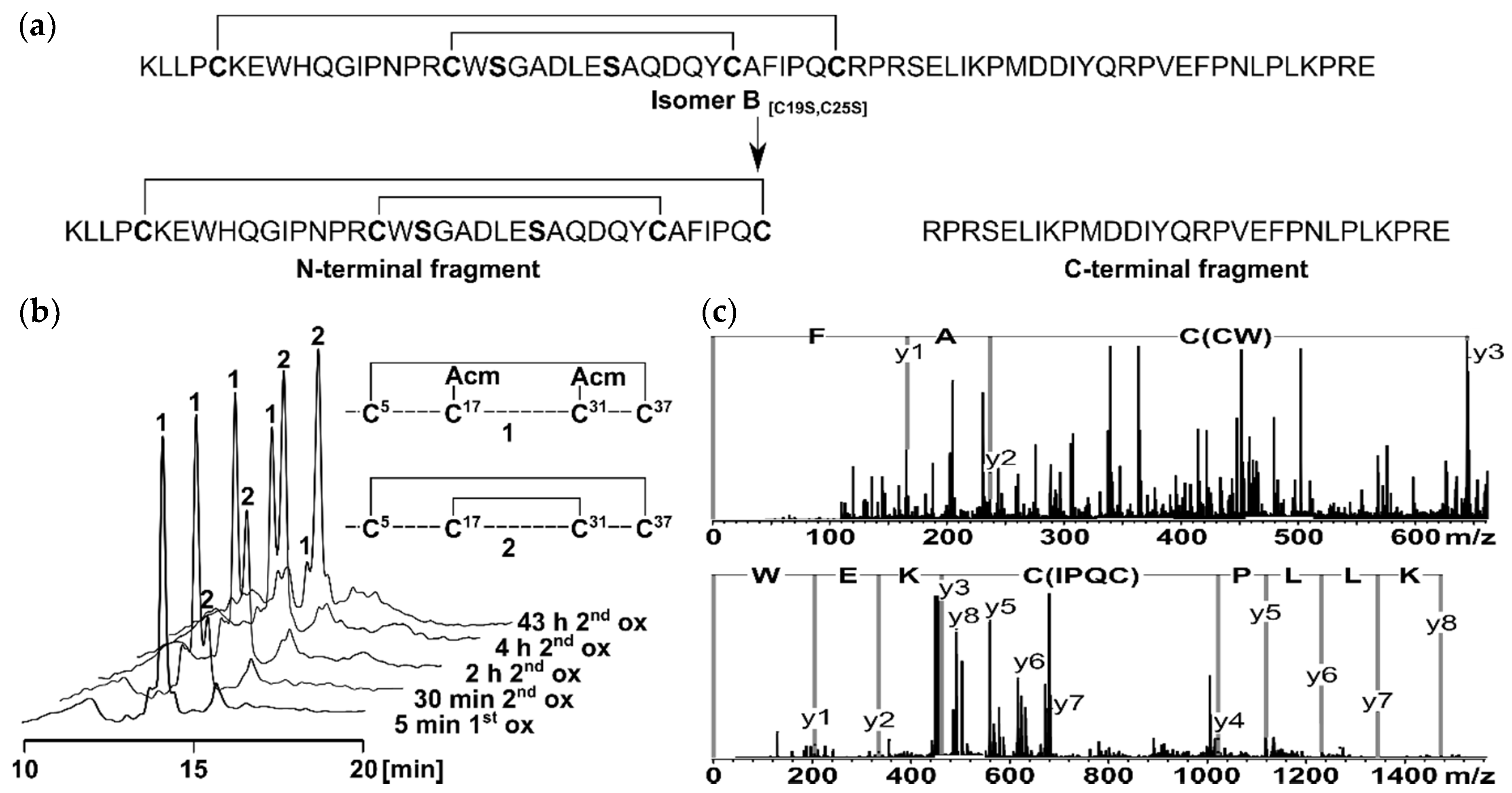
2.1. Peptide Synthesis and Characterization
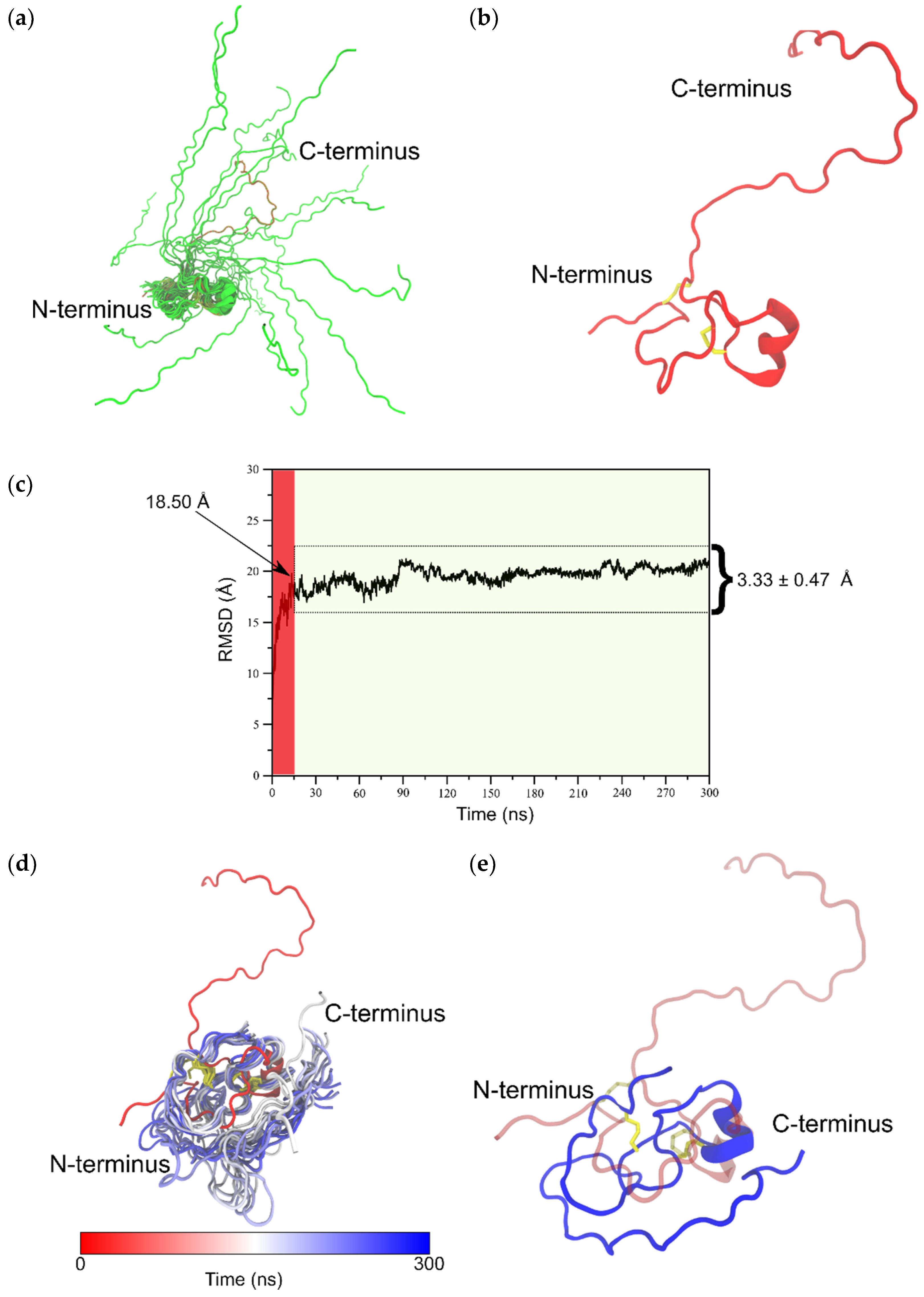
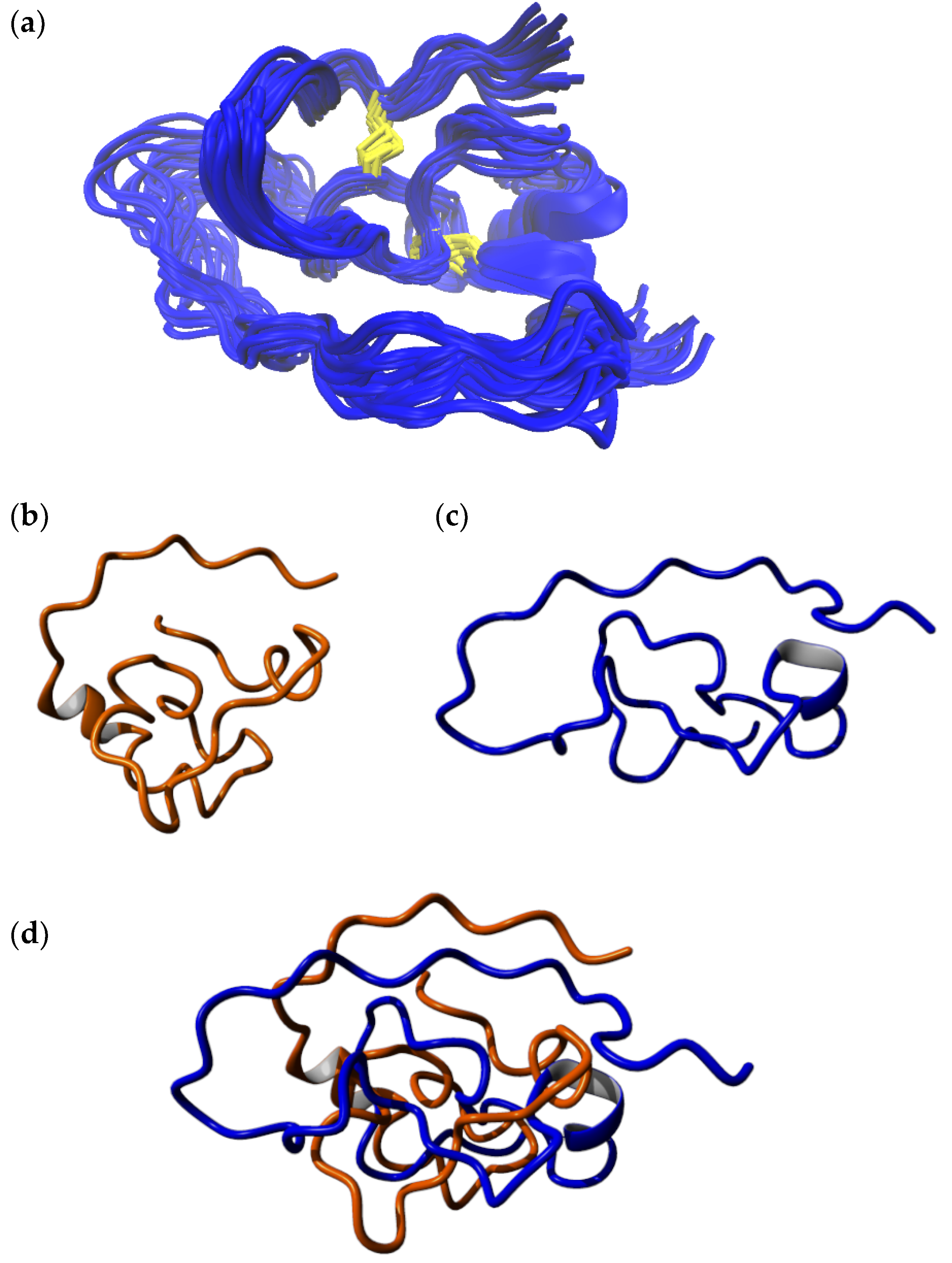
2.2. Structure Elucidation by NMR Spectroscopy
2.3. MD-Based Analysis of Isomer B[C19S,C25S]
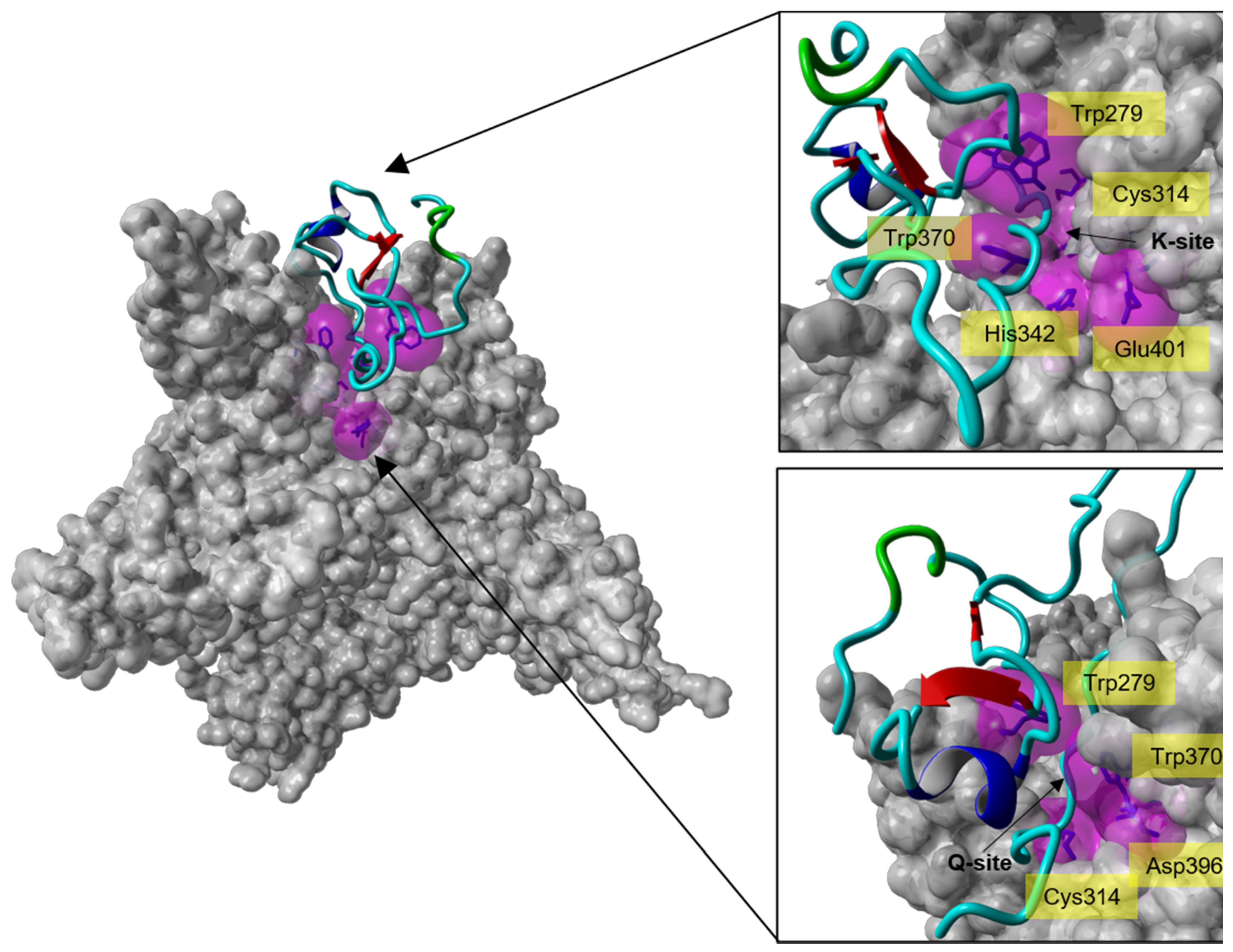
2.4. Molecular Docking of Full-Length Tridegin Variant on FXIIIa
3. Materials and Methods
3.1. Materials
3.2. Peptide Synthesis and Purification
3.3. Selective Oxidation of the N-Terminal Fragment
3.4. Peptide Characterization and Determination of Disulfide Connectivity
3.5. Enzyme Activity Assay
3.6. NMR Spectroscopy and Structure Prediction
3.7. Molecular Dynamics (MD) Simulations
3.8. Blind Docking Studies on the FXIIIa Crystall Structure (PDB ID: 4KTY)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Acm | Acetamidomethyl |
| AcOH | Acetic acid |
| Arg | Arginine |
| Asp | Aspartic acid |
| BPTI | Bovine pancreatic trypsin inhibitor |
| CASP | critical assessment of protein structure prediction |
| COSY | Correlated spectroscopy |
| Cys | Cysteine |
| 2,5-DHAP | 2,5-Dihydroxyacetophenone |
| DMF | Dimethylformamide |
| ESI | Electrospray ionization |
| Fmoc | Fluorenylmethoxycarbonyl |
| FXIII | Blood coagulation factor XIII |
| FXIIIa | Activated blood coagulation factor XIII |
| Glu | Glutamic acid |
| HBTU | 2-(1H-Benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate |
| HSQC | Heteronuclear single quantum coherence |
| His | Histidine |
| Ile | Isoleucine |
| Lys | Lysine |
| MALDI | Matrix-assisted laser desorption/ionization |
| MD | Molecular dynamics |
| MS | Mass spectrometry |
| MS/MS | Tandem mass spectrometry |
| NMR | Nuclear magnetic resonance |
| NOESY | Nuclear Overhauser enhancement spectroscopy |
| NPT | constant number of atoms, pressure, temperature |
| NSGM | Non-saccharide glucosaminoglycan mimetics |
| NVT | constant number of atoms, volume, temperature |
| Phe | Phenylalanine |
| Pro | Proline |
| RMSD | Root mean square deviation |
| RP-HPLC | Reversed-phase high-performance liquid chromatography |
| Ser | Serine |
| SPPS | Solid phase peptide synthesis |
| TFA | Trifluoroacetic acid |
| TOCSY | Total correlation spectroscopy |
| Trt | Trityl |
| Trp | Tryptophane |
| Tyr | Tyrosine |
| VMD | Visual molecular dynamics |
References
- Whitaker, I.S.; Rao, J.; Izadi, D.; Butler, P.E. Historical Article: Hirudo medicinalis: Ancient origins of, and trends in the use of medicinal leeches throughout history. Br. J. Oral Maxillofac. Surg. 2004, 42, 133–137. [Google Scholar] [CrossRef]
- Kvist, S.; Manzano-Marín, A.; de Carle, D.; Trontelj, P.; Siddall, M.E. Draft genome of the European medicinal leech Hirudo medicinalis (Annelida, Clitellata, Hirudiniformes) with emphasis on anticoagulants. Sci. Rep. 2020, 10, 9885. [Google Scholar] [CrossRef] [PubMed]
- Salzet, M. Anticoagulants and inhibitors of platelet aggregation derived from leeches. FEBS Lett. 2001, 492, 187–192. [Google Scholar] [CrossRef]
- Greinacher, A.; Warkentin, T.E. The direct thrombin inhibitor hirudin. Thromb. Haemost. 2008, 99, 819–829. [Google Scholar] [CrossRef]
- Markwardt, F. Hirudin as alternative anticoagulant—A historical review. Semin. Thromb. Hemost. 2002, 28, 405–414. [Google Scholar] [CrossRef]
- Callas, D.D.; Hoppensteadt, D.; Fareed, J. Comparative studies on the anticoagulant and protease generation inhibitory actions of newly developed site-directed thrombin inhibitory drugs. Efegatran, argatroban, hirulog, and hirudin. Semin. Thromb. Hemost. 1995, 21, 177–183. [Google Scholar] [CrossRef]
- Arsenault, K.A.; Hirsh, J.; Whitlock, R.P.; Eikelboom, J.W. Direct thrombin inhibitors in cardiovascular disease. Nat. Rev. Cardiol. 2012, 9, 402–414. [Google Scholar] [CrossRef]
- Lee, C.J.; Ansell, J.E. Direct thrombin inhibitors. Br. J. Clin. Pharmacol. 2011, 72, 581–592. [Google Scholar] [CrossRef]
- Griffin, J.H. Blood coagulation. The thrombin paradox. Nature 1995, 378, 337–338. [Google Scholar] [CrossRef]
- Shaw, C.M.; O’Hanlon, D.M.; McEntee, G.P. Venous thrombosis. Am. J. Surg. 2003, 186, 167–168. [Google Scholar] [CrossRef]
- Lane, D.A.; Philippou, H.; Huntington, J.A. Directing thrombin. Blood 2005, 106, 2605–2612. [Google Scholar] [CrossRef] [PubMed]
- Muszbek, L.; Bereczky, Z.; Bagoly, Z.; Komáromi, I.; Katona, É. Factor XIII: A coagulation factor with multiple plasmatic and cellular functions. Physiol. Rev. 2011, 91, 931–972. [Google Scholar] [CrossRef] [PubMed]
- Weisel, J.W.; Litvinov, R.I. Fibrin Formation, Structure and Properties. Subcell. Biochem. 2017, 82, 405–456. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Biswas, A.; Akhter, M.S.; Krettler, C.; Reinhart, C.; Dodt, J.; Reuter, A.; Philippou, H.; Ivaskevicius, V.; Oldenburg, J. Revisiting the mechanism of coagulation factor XIII activation and regulation from a structure/functional perspective. Sci. Rep. 2016, 6, 30105. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Nazabal, A.; Kaniyappan, S.; Pellequer, J.-L.; Wolberg, A.S.; Imhof, D.; Oldenburg, J.; Biswas, A. The Plasma Factor XIII Heterotetrameric Complex Structure: Unexpected Unequal Pairing within a Symmetric Complex. Biomolecules 2019, 9, 765. [Google Scholar] [CrossRef]
- Stieler, M.; Weber, J.; Hils, M.; Kolb, P.; Heine, A.; Büchold, C.; Pasternack, R.; Klebe, G. Structure of active coagulation factor XIII triggered by calcium binding: Basis for the design of next-generation anticoagulants. Angew. Chem. Int. Ed. Engl. 2013, 52, 11930–11934. [Google Scholar] [CrossRef]
- Aleman, M.M.; Byrnes, J.R.; Wang, J.-G.; Tran, R.; Lam, W.A.; Di Paola, J.; Mackman, N.; Degen, J.L.; Flick, M.J.; Wolberg, A.S. Factor XIII activity mediates red blood cell retention in venous thrombi. J. Clin. Investig. 2014, 124, 3590–3600. [Google Scholar] [CrossRef]
- Byrnes, J.R.; Wolberg, A.S. Newly-Recognized Roles of Factor XIII in Thrombosis. Semin. Thromb. Hemost. 2016, 42, 445–454. [Google Scholar] [CrossRef]
- Duval, C.; Allan, P.; Connell, S.D.A.; Ridger, V.C.; Philippou, H.; Ariëns, R.A.S. Roles of fibrin α- and γ-chain specific cross-linking by FXIIIa in fibrin structure and function. Thromb. Haemost. 2014, 111, 842–850. [Google Scholar] [CrossRef]
- Wolberg, A.S. Fibrinogen and factor XIII: Newly recognized roles in venous thrombus formation and composition. Curr. Opin. Hematol. 2018, 25, 358–364. [Google Scholar] [CrossRef]
- Avery, C.A.; Pease, R.J.; Smith, K.; Boothby, M.; Buckley, H.M.; Grant, P.J.; Fishwick, C.W.G. (±) cis-Bisamido epoxides: A novel series of potent FXIII-A inhibitors. Eur. J. Med. Chem. 2015, 98, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Iwata, Y.; Tago, K.; Kiho, T.; Kogen, H.; Fujioka, T.; Otsuka, N.; Suzuki-Konagai, K.; Ogita, T.; Miyamoto, S. Conformational analysis and docking study of potent factor XIIIa inhibitors having a cyclopropenone ring. J. Mol. Graph. Model. 2000, 18, 591–599. [Google Scholar] [CrossRef]
- Schmitz, T.; Bäuml, C.A.; Imhof, D. Inhibitors of blood coagulation factor XIII. Anal. Biochem. 2020, 605, 113708. [Google Scholar] [CrossRef] [PubMed]
- Al-Horani, R.A.; Kar, S. Factor XIIIa inhibitors as potential novel drugs for venous thromboembolism. Eur. J. Med. Chem. 2020, 200, 112442. [Google Scholar] [CrossRef] [PubMed]
- Pasternack, R.; Büchold, C.; Jähnig, R.; Pelzer, C.; Sommer, M.; Heil, A.; Florian, P.; Nowak, G.; Gerlach, U.; Hils, M. Novel inhibitor ZED3197 as potential drug candidate in anticoagulation targeting coagulation FXIIIa (F13a). J. Thromb. Haemost. 2020, 18, 191–200. [Google Scholar] [CrossRef]
- Al-Horani, R.A.; Karuturi, R.; Lee, M.; Afosah, D.K.; Desai, U.R. Allosteric Inhibition of Factor XIIIa. Non-Saccharide Glycosaminoglycan Mimetics, but Not Glycosaminoglycans, Exhibit Promising Inhibition Profile. PLoS ONE 2016, 11, e0160189. [Google Scholar] [CrossRef]
- Finney, S.; Seale, L.; Sawyer, R.T.; Wallis, R.B. Tridegin, a new peptidic inhibitor of factor XIIIa, from the blood-sucking leech Haementeria ghilianii. Biochem. J. 1997, 324 Pt 3, 797–805. [Google Scholar] [CrossRef][Green Version]
- Bäuml, C.A.; Paul George, A.A.; Schmitz, T.; Sommerfeld, P.; Pietsch, M.; Podsiadlowski, L.; Steinmetzer, T.; Biswas, A.; Imhof, D. Distinct 3-disulfide-bonded isomers of tridegin differentially inhibit coagulation factor XIIIa: The influence of structural stability on bioactivity. Eur. J. Med. Chem. 2020, 201, 112474. [Google Scholar] [CrossRef]
- Bäuml, C.A.; Schmitz, T.; Paul George, A.A.; Sudarsanam, M.; Hardes, K.; Steinmetzer, T.; Holle, L.A.; Wolberg, A.S.; Pötzsch, B.; Oldenburg, J.; et al. Coagulation Factor XIIIa Inhibitor Tridegin: On the Role of Disulfide Bonds for Folding, Stability, and Function. J. Med. Chem. 2019, 62, 3513–3523. [Google Scholar] [CrossRef]
- Böhm, M.; Bäuml, C.A.; Hardes, K.; Steinmetzer, T.; Roeser, D.; Schaub, Y.; Than, M.E.; Biswas, A.; Imhof, D. Novel insights into structure and function of factor XIIIa-inhibitor tridegin. J. Med. Chem. 2014, 57, 10355–10365. [Google Scholar] [CrossRef]
- Böhm, M.; Kühl, T.; Hardes, K.; Coch, R.; Arkona, C.; Schlott, B.; Steinmetzer, T.; Imhof, D. Synthesis and functional characterization of tridegin and its analogues: Inhibitors and substrates of factor XIIIa. ChemMedChem 2012, 7, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.-Y. Diverse pathways of oxidative folding of disulfide proteins: Underlying causes and folding models. Biochemistry 2011, 50, 3414–3431. [Google Scholar] [CrossRef] [PubMed]
- Paul George, A.A.; Heimer, P.; Maaß, A.; Hamaekers, J.; Hofmann-Apitius, M.; Biswas, A.; Imhof, D. Insights into the Folding of Disulfide-Rich μ-Conotoxins. ACS Omega 2018, 3, 12330–12340. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Boudreau, M.A.; Zheng, J.; Whittal, R.M.; Austin, P.; Roskelley, C.D.; Roberge, M.; Andersen, R.J.; Vederas, J.C. Chemical synthesis and biological activity of the neopetrosiamides and their analogues: Revision of disulfide bond connectivity. J. Am. Chem. Soc. 2010, 132, 1486–1487. [Google Scholar] [CrossRef]
- Günther, P.; Buchner, L. Combined automated NOE assignment and structure calculation with CYANA. J. Biomol. NMR 2015, 62, 453–471. [Google Scholar] [CrossRef]
- Konagurthu, A.S.; Whisstock, J.C.; Stuckey, P.J.; Lesk, A.M. MUSTANG: A multiple structural alignment algorithm. Proteins 2006, 64, 559–574. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Van der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 14101. [Google Scholar] [CrossRef] [PubMed]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Nosé, S.; Klein, M.L. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983, 50, 1055–1076. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. The HDOCK server for integrated protein-protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Moult, J.; Fidelis, K.; Kryshtafovych, A.; Schwede, T.; Tramontano, A. Critical assessment of methods of protein structure prediction (CASP)--round x. Proteins 2014, 82 (Suppl. 2), 1–6. [Google Scholar] [CrossRef]
- Van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef]
- Tina, K.G.; Bhadra, R.; Srinivasan, N. PIC: Protein Interactions Calculator. Nucleic Acids Res. 2007, 35, W473–W476. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmitz, T.; Paul George, A.A.; Nubbemeyer, B.; Bäuml, C.A.; Steinmetzer, T.; Ohlenschläger, O.; Biswas, A.; Imhof, D. NMR-Based Structural Characterization of a Two-Disulfide-Bonded Analogue of the FXIIIa Inhibitor Tridegin: New Insights into Structure–Activity Relationships. Int. J. Mol. Sci. 2021, 22, 880. https://doi.org/10.3390/ijms22020880
Schmitz T, Paul George AA, Nubbemeyer B, Bäuml CA, Steinmetzer T, Ohlenschläger O, Biswas A, Imhof D. NMR-Based Structural Characterization of a Two-Disulfide-Bonded Analogue of the FXIIIa Inhibitor Tridegin: New Insights into Structure–Activity Relationships. International Journal of Molecular Sciences. 2021; 22(2):880. https://doi.org/10.3390/ijms22020880
Chicago/Turabian StyleSchmitz, Thomas, Ajay Abisheck Paul George, Britta Nubbemeyer, Charlotte A. Bäuml, Torsten Steinmetzer, Oliver Ohlenschläger, Arijit Biswas, and Diana Imhof. 2021. "NMR-Based Structural Characterization of a Two-Disulfide-Bonded Analogue of the FXIIIa Inhibitor Tridegin: New Insights into Structure–Activity Relationships" International Journal of Molecular Sciences 22, no. 2: 880. https://doi.org/10.3390/ijms22020880
APA StyleSchmitz, T., Paul George, A. A., Nubbemeyer, B., Bäuml, C. A., Steinmetzer, T., Ohlenschläger, O., Biswas, A., & Imhof, D. (2021). NMR-Based Structural Characterization of a Two-Disulfide-Bonded Analogue of the FXIIIa Inhibitor Tridegin: New Insights into Structure–Activity Relationships. International Journal of Molecular Sciences, 22(2), 880. https://doi.org/10.3390/ijms22020880






