Description of AtCAX4 in Response to Abiotic Stress in Arabidopsis
Abstract
1. Introduction
2. Results
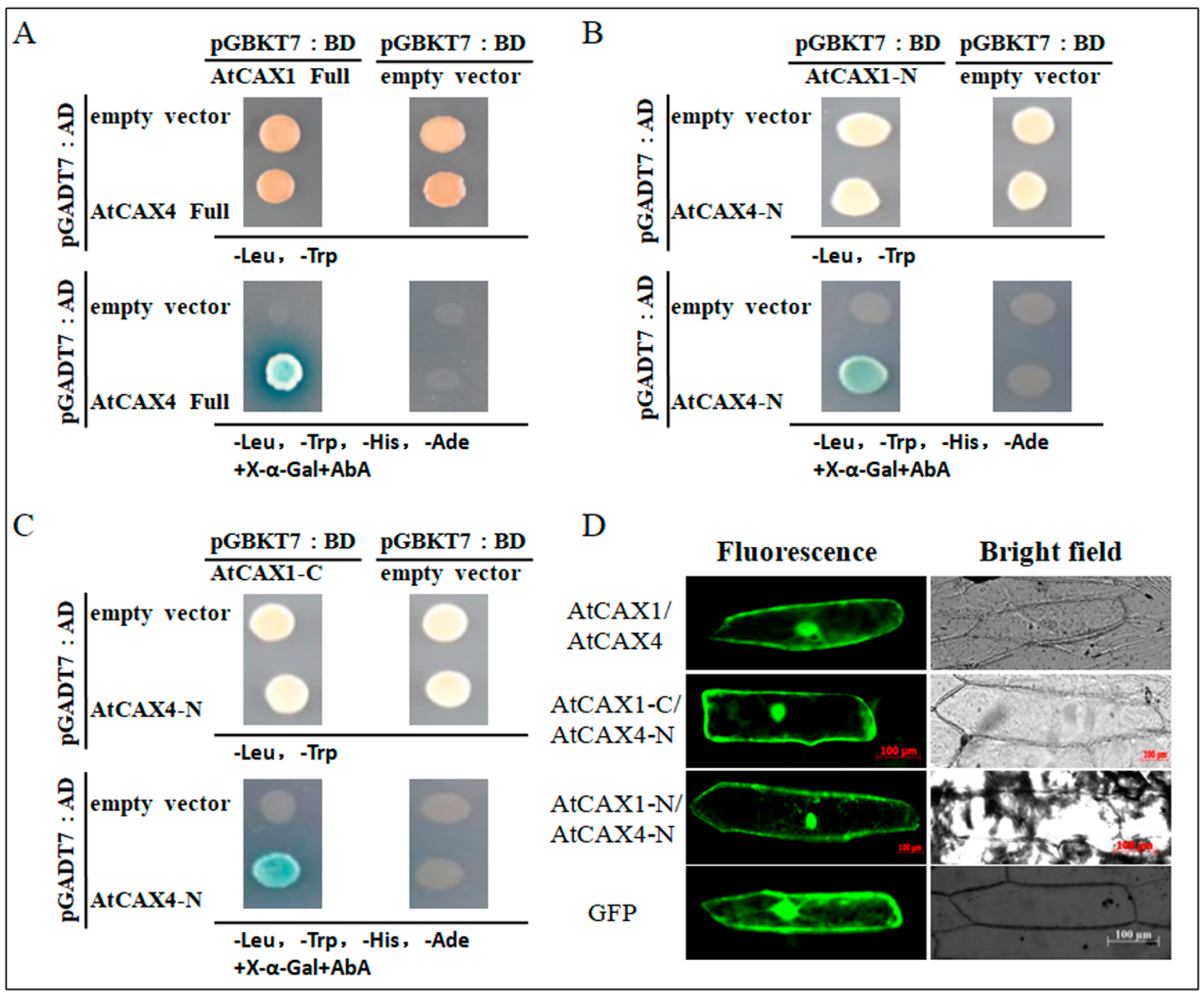
2.1. AtCAX1 Interacts with the N-Terminus of AtCAX4
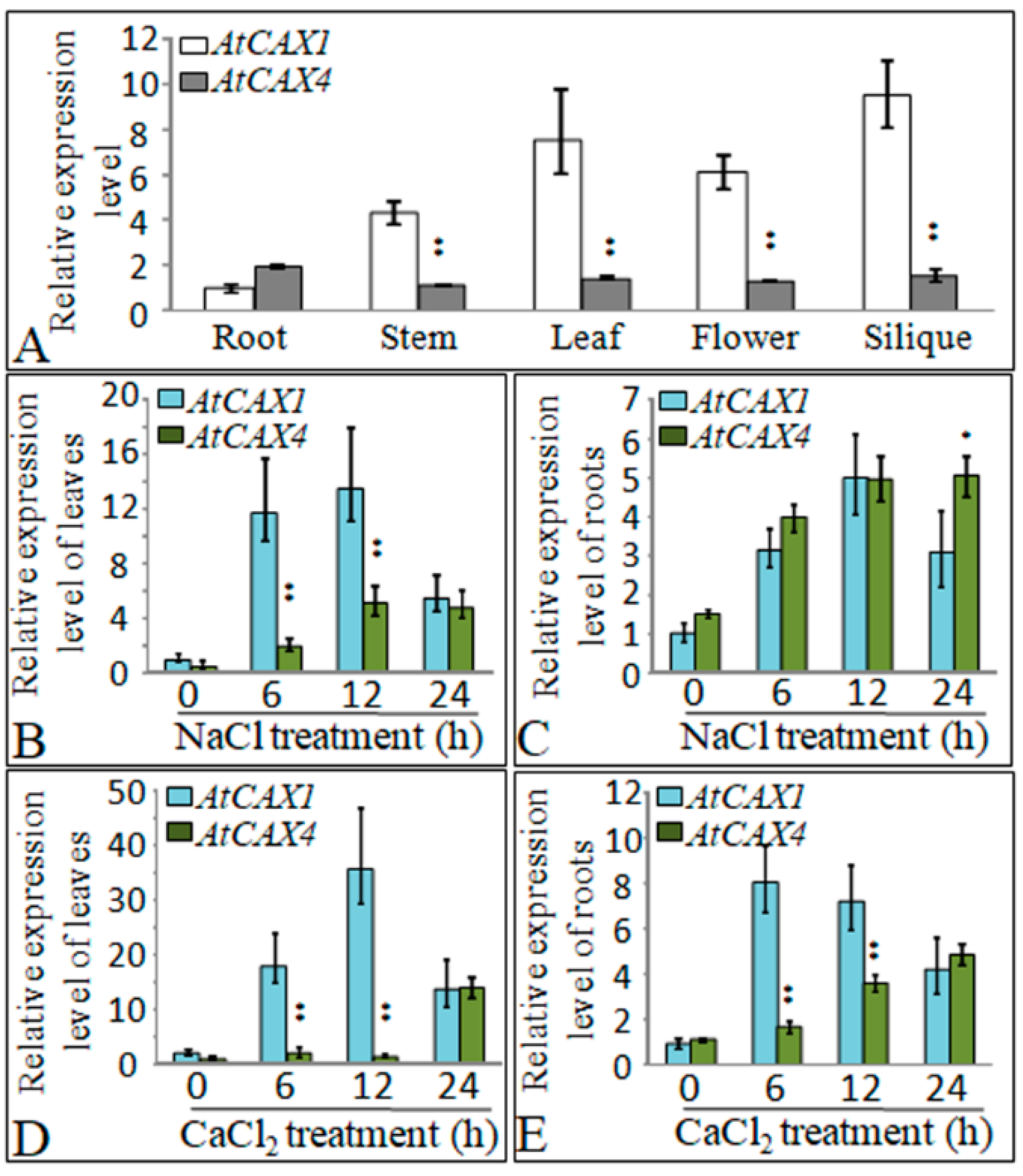
2.2. Expression of AtCAX1 and AtCAX4 in Response to Salt and Ion Stress
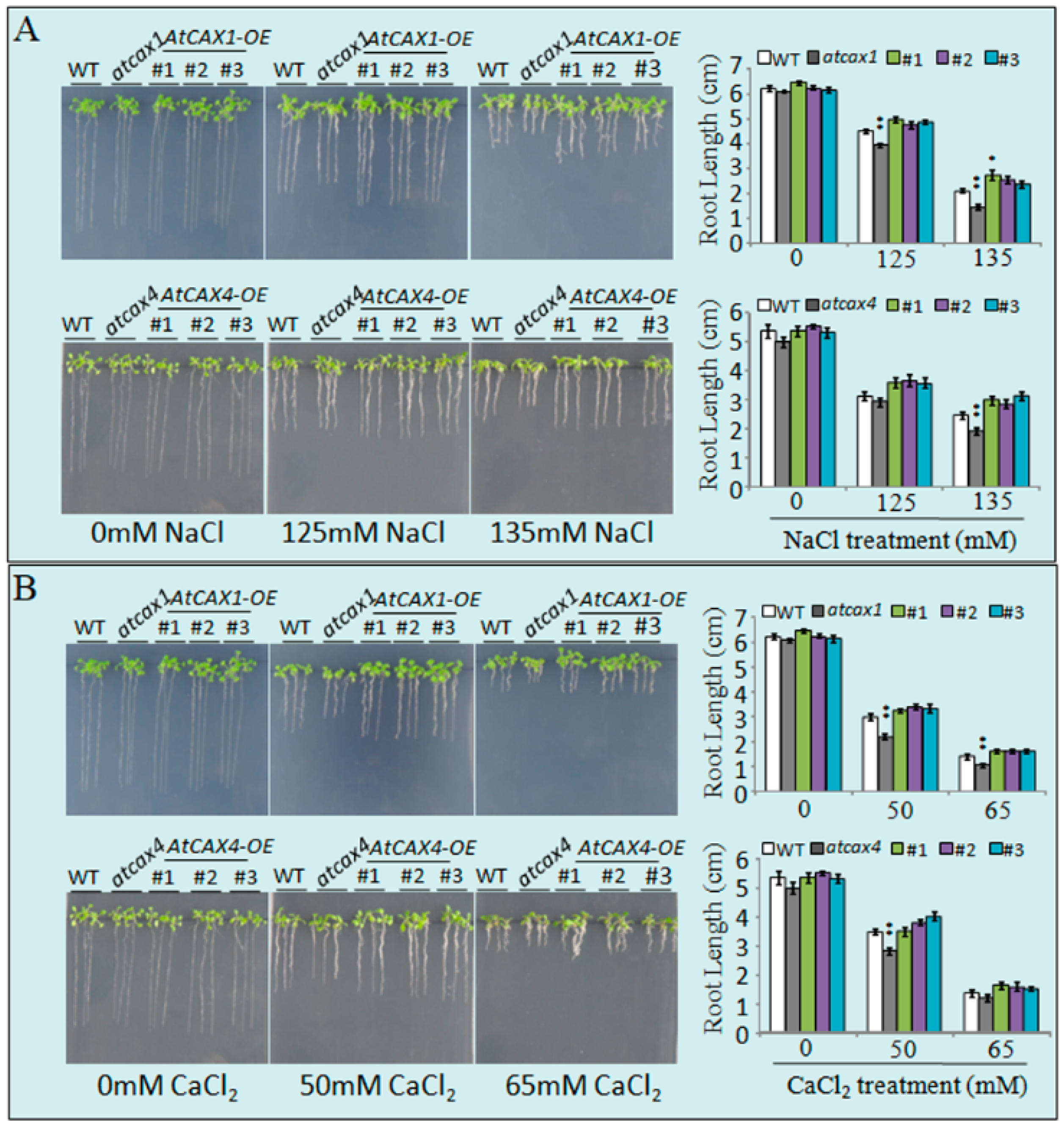
2.3. Loss of Function Mutants of AtCAX1 and AtCAX4 Render Arabidopsis Plants Sensitive to Salt and Ion Stress
3. Discussion
3.1. AtCAX1 and AtCAX4 form A Heterodimer
3.2. AtCAX1 and AtCAX4 Interact in Plants in Response to Environmental Stress
4. Materials and Methods
4.1. Yeast Two-Hybrid (Y2H) Screening
4.2. Bimolecular Fluorescence Complementation (BiFC) Assay
4.3. Plant Material and Transformation
4.4. RNA Isolation and Northern Blotting
4.5. Analysis of Gene Expression Using Reverse Transcription–Quantitative PCR (RT-qPCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CAXs | Ca2+/H+ antiporters |
| BiFC | Bimolecular fluorescence complementation |
| Ca2+ | Calcium ion |
| Ba2+ | Barium ion |
| AbA | Aureobasidin A |
| MS | Murashige and Skoog |
References
- Reddy, A.S.N.; Ali, G.S.; Celesnik, H.; Day, I.S. Coping with stresses: Roles of calcium-and calcium/calmodulin-regulated gene expression. Plant Cell 2011, 23, 2010–2032. [Google Scholar] [CrossRef]
- Steinhorst, L.; Kudla, J. Signaling in cells and organisms-calcium holds the line. Curr. Opin. Plant Biol. 2014, 22, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.Y.; Dunand, C.; Snedden, W.; Galaud, J.P. CaM and CML emergence in the green lineage. Trends Plant Sci. 2015, 20, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Clapham, D.E. Calcium signaling. Cell 2007, 13, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Bose, J.; Pottosin, I.I.; Shabala, S.S.; Palmgren, M.G.; Shabala, S. Calcium efflux systems in stress signaling and adaptation in plants. Front. Plant Sci. 2011, 2, 85. [Google Scholar] [CrossRef]
- Shabala, S. Physiological and cellular aspects of phytotoxicity tolerance in plants: The role of membrane transporters and implications for crop breeding for waterlogging tolerance. New Phytol. 2011, 190, 289–298. [Google Scholar] [CrossRef]
- Shigaki, T.; Hirschi, K.D. Diverse functions and molecular properties emerging for CAX cation/H+ exchangers in plants. Plant Biol. 2006, 8, 419–429. [Google Scholar] [CrossRef]
- Pittman, J.K.; Bonza, M.C.; De Michelis, M.I. Ca2+ pumps and Ca2+ antiporters in plant development. Transp. Pumps Plant Signal. Signal. Commun. Plants 2011, 7, 133–161. [Google Scholar]
- Kamiya, T.; Maeshima, M. Residues in internal repeats of the rice cation/H+ exchanger are involved in the transport and selection of cations. J. Biol. Chem. 2004, 279, 812–819. [Google Scholar] [CrossRef]
- Shigaki, T.; Barkla, B.J.; Miranda-Vergara, M.C.; Zhao, J.; Pantoja, O.; Hirschi, K.D. Identification of a crucial histidine involved in metal transport activity in the Arabidopsis cation/H+ exchanger CAX1. J. Biol. Chem. 2005, 280, 30136–30142. [Google Scholar] [CrossRef]
- Shigaki, T.; Rees, I.; Nakhleh, L.; Hirschi, K.D. Identification of three distinct phylogenetic groups of CAX cation ⁄proton antiporters. J. Mol. Evol. 2006, 63, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Hirschi, K.D.; Zhen, R.G.; Cunningham, K.W.; Rea, P.A.; Fink, G.R. CAX1, an H+/Ca2+ antiporter from Arabidopsis. Proc. Nat. Acad. Sci. USA 1996, 93, 8782–8786. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, X.; Takano, T.; Liu, S. Characterization of a PutCAX1 gene from Puccinellia tenuiflora that confers Ca2+ and Ba2+ tolerance in yeast. Biochem. Biophys. Res. Commun. 2009, 383, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Shigaki, T.; Mei, H.; Marshall, J.; Li, X.; Manohar, M.; Hirschi, K.D. The expression of the open reading frame of Arabidopsis CAX1, but not its cDNA, confers metal tolerance in yeast. Plant Biol 2010, 12, 935–939. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, T.; Akahori, T.; Maeshima, M. Expression profile of the genes for rice cation⁄H+ exchanger family and functional analysis in yeast. Plant Cell Physiol. 2005, 46, 1735–1740. [Google Scholar] [CrossRef] [PubMed]
- Korenkov, V.; Hirschi, K.; Crutchfield, J.D.; Wagner, G.J. Enhancing tonoplast Cd/H antiport activity increases Cd, Zn, and Mn tolerance, and impacts root/shoot Cd partitioning in Nicotiana tabacum L. Planta 2007, 226, 1379–1387. [Google Scholar] [CrossRef]
- Mei, H.; Cheng, N.H.; Zhao, J.; Park, S.; Escareno, R.A.; Pittman, J.K.; Hirschi, K.D. Root development under metal stress in Arabidopsis thaliana requires the H+/cation antiporter CAX4. New Phytol. 2009, 183, 95–105. [Google Scholar] [CrossRef]
- Conn, S.J.; Gilliham, M.; Athman, A.; Schreiber, A.W.; Baumann, U.; Moller, I.; Cheng, N.H.; Stancombe, M.A.; Hirschi, K.D.; Webb, A.A.R.; et al. Cell-specific vacuolar calcium storage mediated by CAX1 regulates apoplastic calcium concentration, gas exchange, and plant productivity in Arabidopsis. Plant Cell 2011, 23, 240–257. [Google Scholar] [CrossRef]
- Pittman, J.K.; Sreevidya, C.S.; Shigaki, T.; Ueoka-Nakanishi, H.; Hirschi, K.D. Distinct N-terminal regulatory domains of Ca2+/H+ antiporters. Plant Physiol 2002, 130, 1054–1062. [Google Scholar] [CrossRef]
- Pittman, J.K.; Shigaki, T.; Marshall, J.L.; Morris, J.L.; Cheng, N.H.; Hirschi, K.D. Functional and regulatory analysis of the Arabidopsis thaliana CAX2 cation transporter. Plant. Mol. Biol. 2004, 56, 959–971. [Google Scholar] [CrossRef]
- Schaaf, G.; Catoni, E.; Fitz, M.; Schwacke, R.; Schneider, A.; von Wiren, N.; Frommer, W.B. A putative role for the vacuolar calcium⁄manganese proton antiporter AtCAX2 in heavy metal detoxification. Plant Biol. 2002, 4, 612–618. [Google Scholar] [CrossRef]
- Mei, H.; Zhao, J.; Pittman, J.K.; Lachmansingh, J.; Park, S.; Hirschi, K.D. In planta regulation of the Arabidopsis Ca2+/H+ antiporter CAX1. J. Exp. Bot. 2007, 58, 3419–3427. [Google Scholar] [CrossRef] [PubMed]
- Pittman, J.K.; Hirschi, K.D. Regulation of CAX1, an Arabidopsis Ca2+/H+ antiporter. Identification of an N-terminal autoinhibitory domain. Plant Physiol. 2001, 127, 1020–1029. [Google Scholar] [CrossRef] [PubMed]
- Pittman, J.K.; Shigaki, T.; Cheng, N.H.; Hirschi, K.D. Mechanism of N-terminal autoinhibition in the Arabidopsis Ca2+/H+ antiporter CAX1. J. Biol. Chem. 2002, 277, 26452–26459. [Google Scholar] [CrossRef]
- Cheng, N.H.; Pittman, J.K.; Barkla, B.J.; Shigaki, T.; Hirschi, K.D. The Arabidopsis cax1 mutant exhibits impaired ion homeostasis, development, and hormonal responses and reveals interplay among vacuolar transporters. Plant Cell 2003, 15, 347–364. [Google Scholar] [CrossRef]
- Cheng, N.H.; Pittman, J.K.; Shigaki, L.J.; LeClere, S.; Lahner, B.; Salt, D.E.; Hirschi, K.D. Functional association of Arabidopsis CAX1 and CAX3 is required for normal growth and ion homeostasis. Plant Physiol 2005, 138, 2048–2060. [Google Scholar] [CrossRef]
- Shigaki, T.; Hirschi, K. Characterization of CAX-like genes in plants: Implications for functional diversity. Gene 2000, 31, 291–298. [Google Scholar] [CrossRef]
- Cheng, N.H.; Pittman, J.K.; Shigaki, T.; Hirschi, K.D. Characterization of CAX4, an Arabidopsis H+/cation antiporter. Plant Physiol 2002, 128, 1245–1254. [Google Scholar] [CrossRef]
- Zhao, J.; Shigaki, T.; Mei, H.; Guo, Y.; Cheng, N.H.; Hirschi, K.D. Interaction between Arabidopsis Ca2+/H+ Exchangers CAX1 and CAX3. J. Biol. Chem. 2009, 284, 4605–4615. [Google Scholar] [CrossRef]
- Ma, Y.; Berkowitz, G.A. Multimeric CAX complexes and Ca2+ signaling-beyond humdrum housekeeping. J. Exp. Bot. 2017, 68, 3997–3999. [Google Scholar] [CrossRef]
- Loqué, D.; Lalonde, S.; Looger, L.L.; von Wire’n, N.; Frommer, W.B. A cytosolic trans-activation domain essential for ammonium uptake. Nature 2007, 446, 195–198. [Google Scholar] [CrossRef] [PubMed]
- Hocking, B.; Conn, S.J.; Manohar, M.; Xu, B.; Athman, A.; Stancombe, M.A.; Webb, A.R.; Hirschi, K.D.; Gilliham, M. Heterodimerization of Arabidopsis calcium/proton exchangers contributes to regulation of guard cell dynamics and plant defense responses. J. Exp. Bot. 2017, 68, 4171–4183. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N.; Liub, J.; Nelsonb, R.S.; Hirschi, K.D. Characterization of CXIP4, a novel Arabidopsis protein that activates the H+/Ca2+ antiporter, CAX1. FEBS Lett. 2004, 559, 99–106. [Google Scholar] [CrossRef]
- Gebreselassie, D.; Rajarathnam, K.; Fliegel, L. Expression, purification, and characterization of the carboxyl-terminal region of the Na+/H+ exchanger. Biochem. Cell Biol. 1998, 76, 837–842. [Google Scholar] [CrossRef]
- Li, X.; Liu, Y.; Kay, C.M.; Muller-Esterl, W.; Fliegel, L. The Na+/H+ exchanger cytoplastic tail: Structure, function, and interactions with tescalcin. Biochemistry 2003, 42, 7448–7456. [Google Scholar] [CrossRef]
- Waditee, R.; Hibino, T.; Tanaka, Y.; Nakamura, T.; Incharoensakdi, A.; Takabe, T. Halotolerant cyanobacterium Aphanothece halophytica contains an Na+/H+ antiporter, homologous to eukaryotic ones, with novel ion specificity affected by C-terminal tail. J. Biol. Chem. 2001, 276, 36931–36938. [Google Scholar] [CrossRef]
- Bickerton, P.D.; Pittman, J.K. Role of cation/proton exchangers in abiotic stress signaling and stress tolerance in plants. In Elucidation of Abiotic Stress Signaling in Plants: Functional Genomics Perspectives, 1st ed.; Pandey, K.G., Ed.; Springer: New York, NY, USA, 2015; pp. 95–117. [Google Scholar]
- Pittman, J.K.; Hirschi, K.D. CAX-ing a wide net: Cation/H+ transporters in metal remediation and abiotic stress signaling. Plant Biol. 2016, 18, 741–749. [Google Scholar] [CrossRef]
- Manohar, M.; Shigaki, T.; Hirschi, K.D. Plant cation/H+ exchangers (CAXs): Biological functions and genetic manipulations. Plant Biol. 2011, 13, 561–569. [Google Scholar] [CrossRef]
- Guo, K.; Bu, Y.; Takano, T.; Liu, S.; Zhang, X. Arabidopsis cysteine proteinase inhibitor AtCYSb interacts with a Ca2+-dependent nuclease, AtCaN2. FEBS Lett. 2013, 587, 3417–3421. [Google Scholar] [CrossRef]
- Tsugama, D.; Liu, S.; Takano, T. A putative myristoylated 2C-type protein phosphatase, PP2C74, interacts with SnRK1 in Arabidopsis. FEBS Lett. 2012, 586, 693–698. [Google Scholar] [CrossRef]
- Zhang, C.Q.; Nishiuchi, S.; Liu, S.; Takano, T. Characterization of two plasma membrane protein 3 genes (PutPMP3) from the alkali grass, Puccinellia tenuiflora, and functional comparison of the rice homologues, OsLti6a/b from rice. BMB Rep. 2008, 41, 448–454. [Google Scholar]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bu, Y.; Fu, W.; Chen, J.; Takano, T.; Liu, S. Description of AtCAX4 in Response to Abiotic Stress in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 856. https://doi.org/10.3390/ijms22020856
Bu Y, Fu W, Chen J, Takano T, Liu S. Description of AtCAX4 in Response to Abiotic Stress in Arabidopsis. International Journal of Molecular Sciences. 2021; 22(2):856. https://doi.org/10.3390/ijms22020856
Chicago/Turabian StyleBu, Yuanyuan, Weichao Fu, Jiangpo Chen, Tetsuo Takano, and Shenkui Liu. 2021. "Description of AtCAX4 in Response to Abiotic Stress in Arabidopsis" International Journal of Molecular Sciences 22, no. 2: 856. https://doi.org/10.3390/ijms22020856
APA StyleBu, Y., Fu, W., Chen, J., Takano, T., & Liu, S. (2021). Description of AtCAX4 in Response to Abiotic Stress in Arabidopsis. International Journal of Molecular Sciences, 22(2), 856. https://doi.org/10.3390/ijms22020856




