Comparative Proteomic Analysis of the Mesenchymal Stem Cells Secretome from Adipose, Bone Marrow, Placenta and Wharton’s Jelly
Abstract
:1. Introduction
2. Results
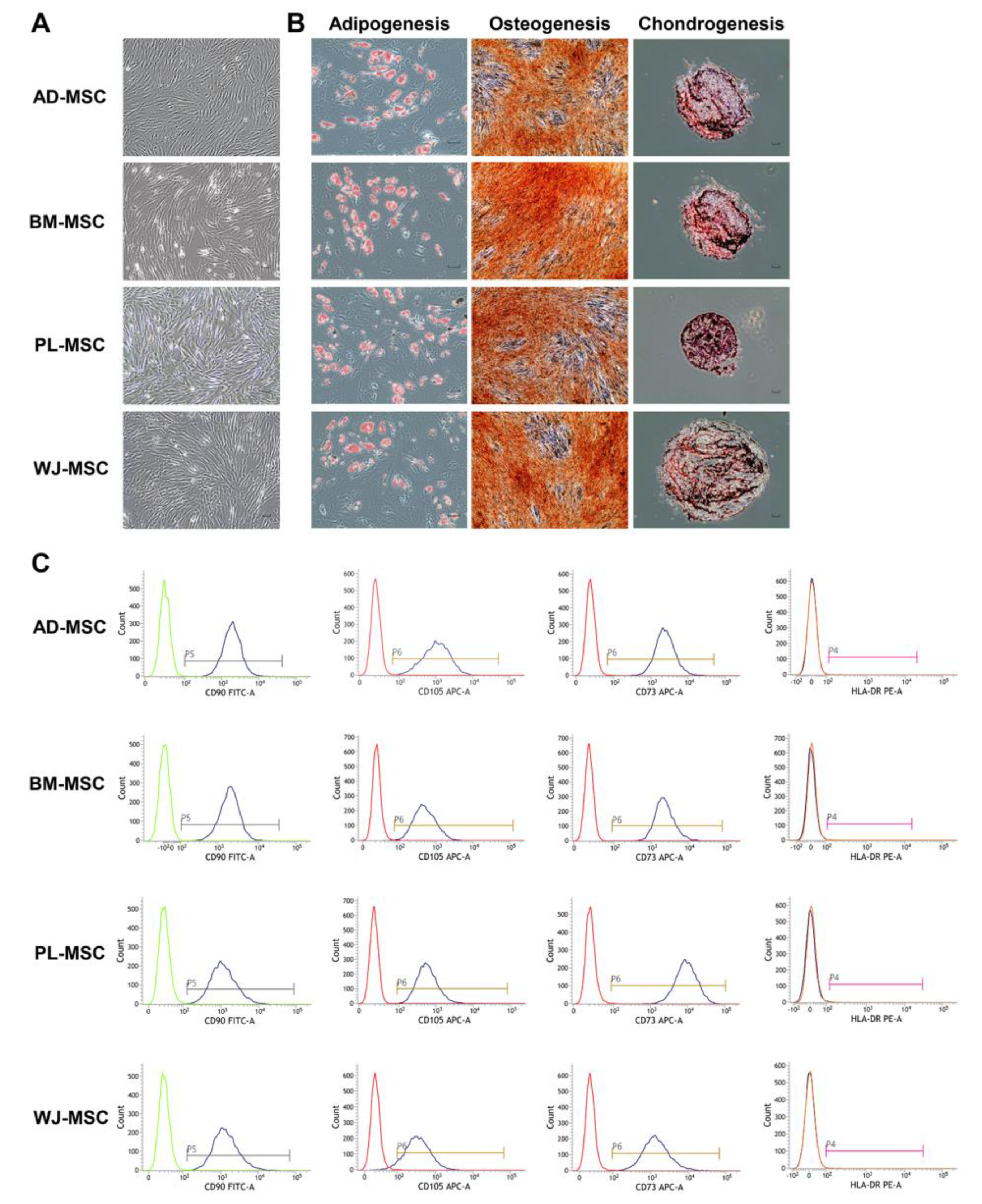
2.1. Characterization of MSCs from Different Sources
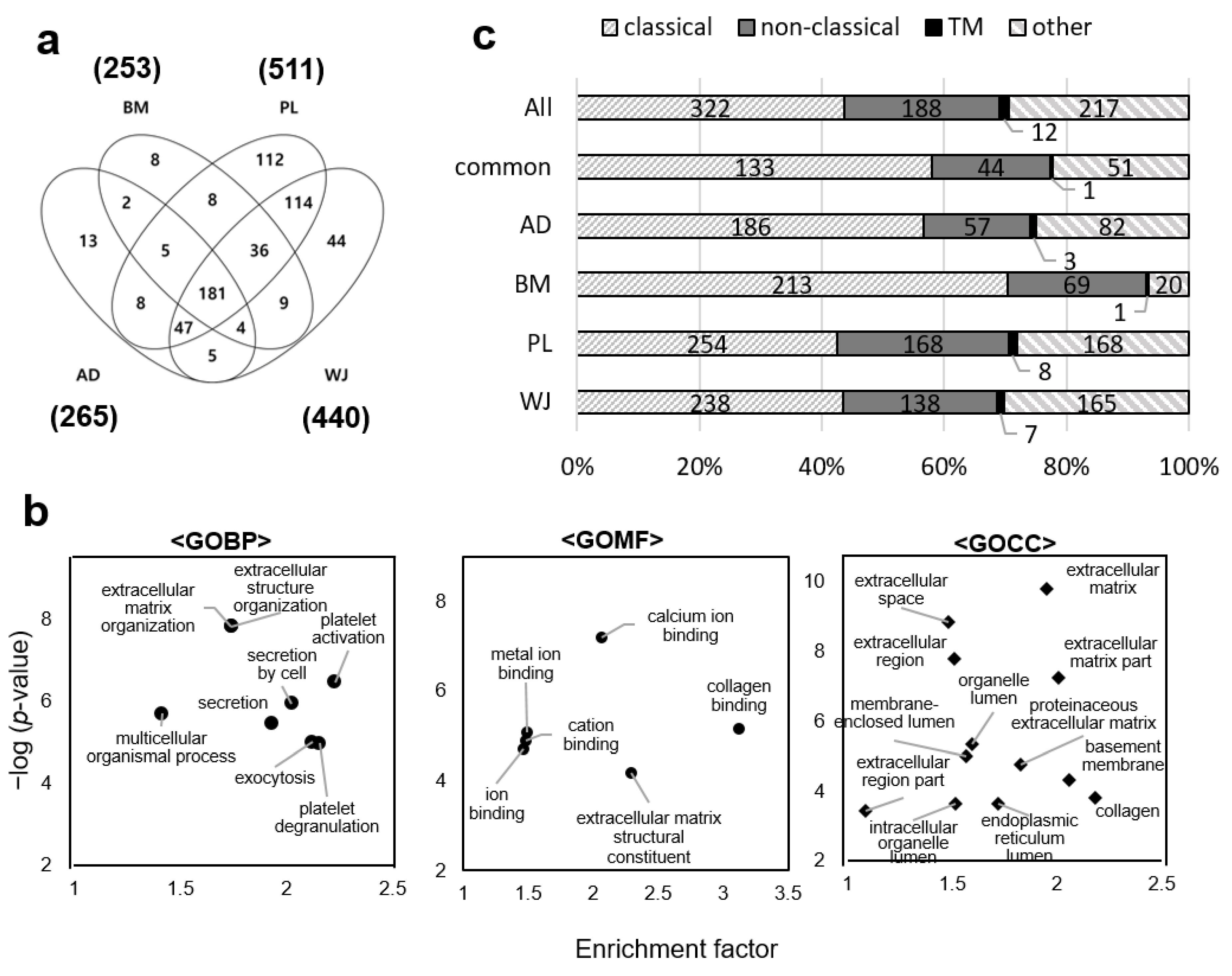
2.2. Profiling of MSCs Secretome by Mass Spectrometry
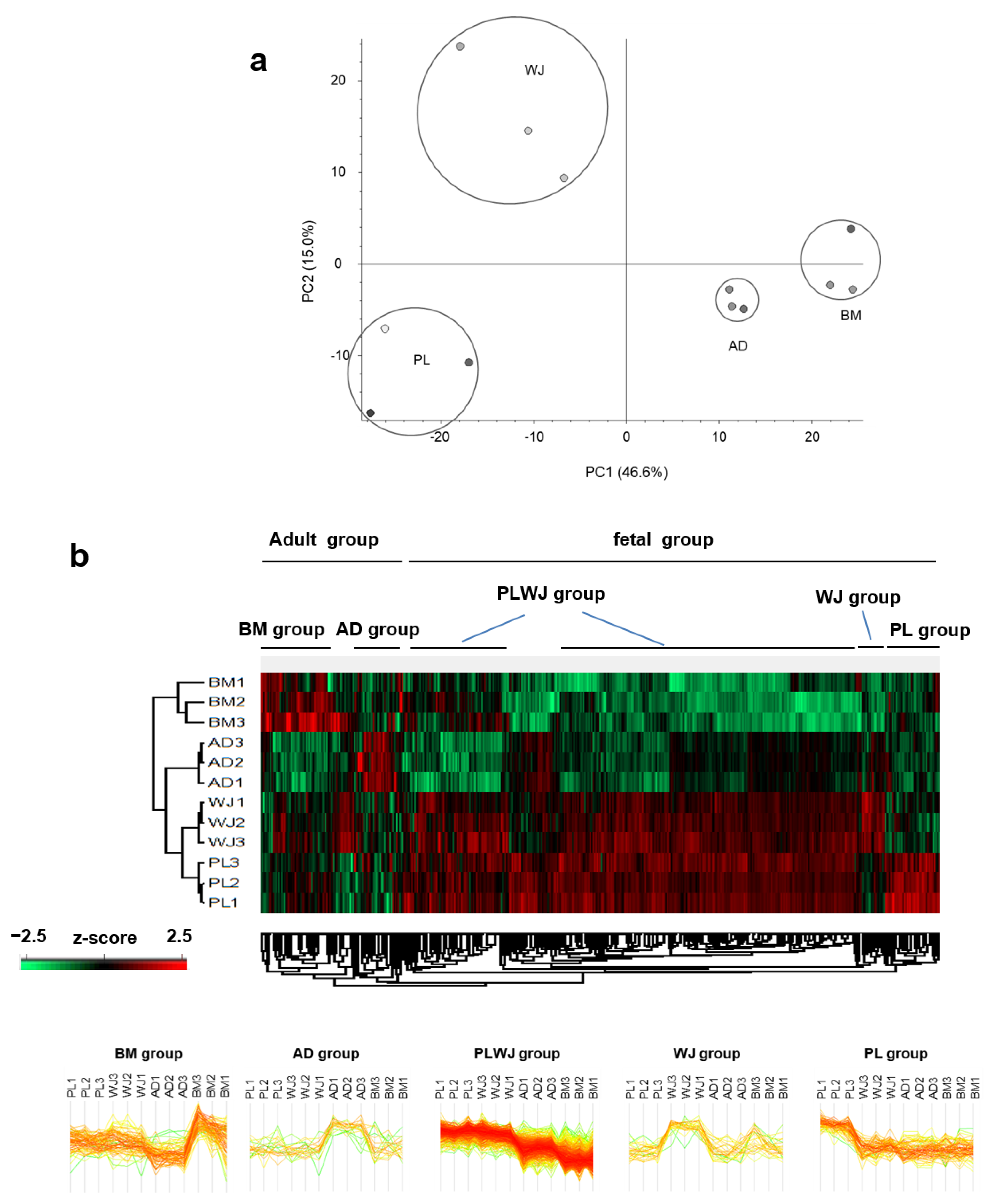
2.3. Gene Ontology Analysis of MSCs Secretome and Clustering of Differential Expressed Proteins
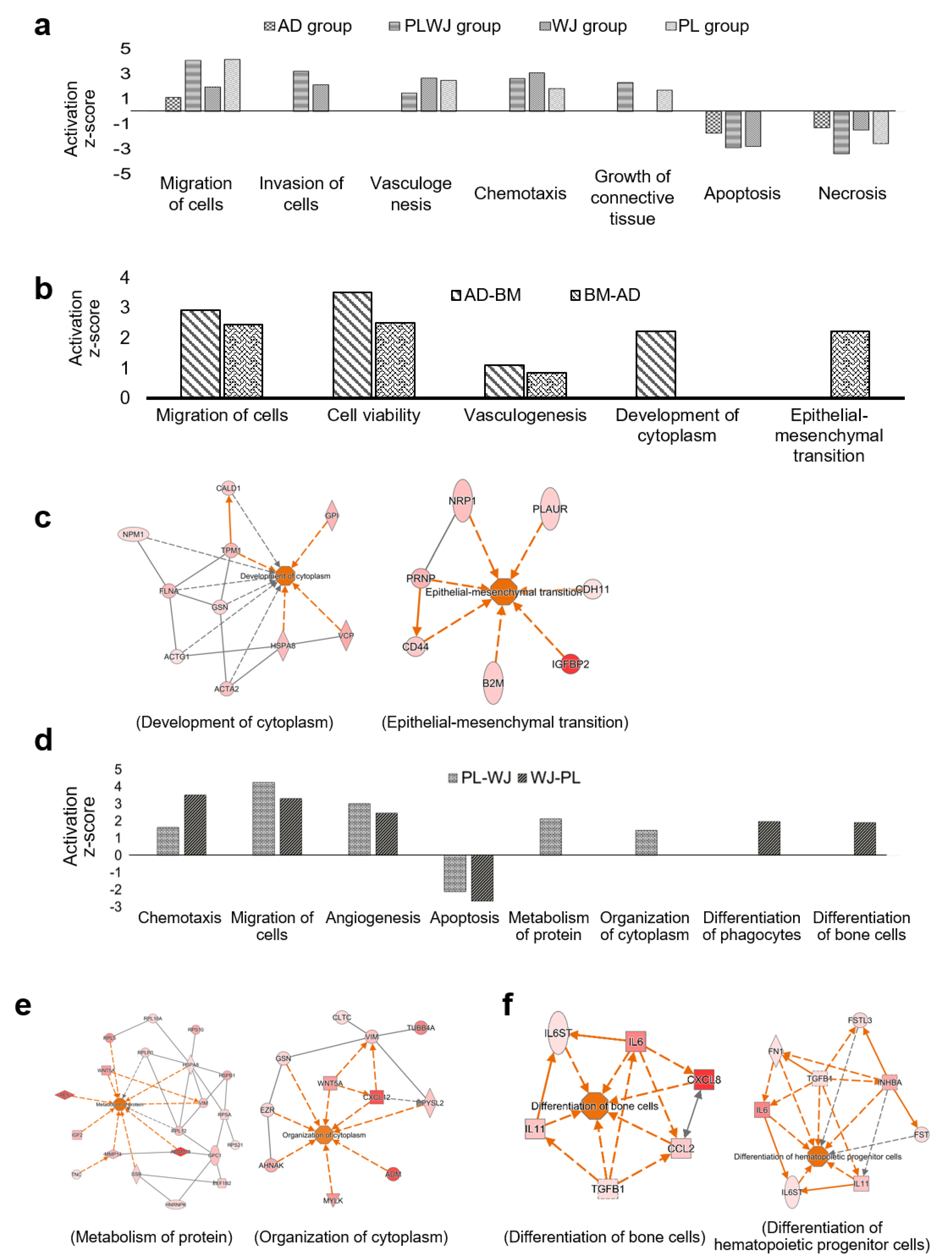
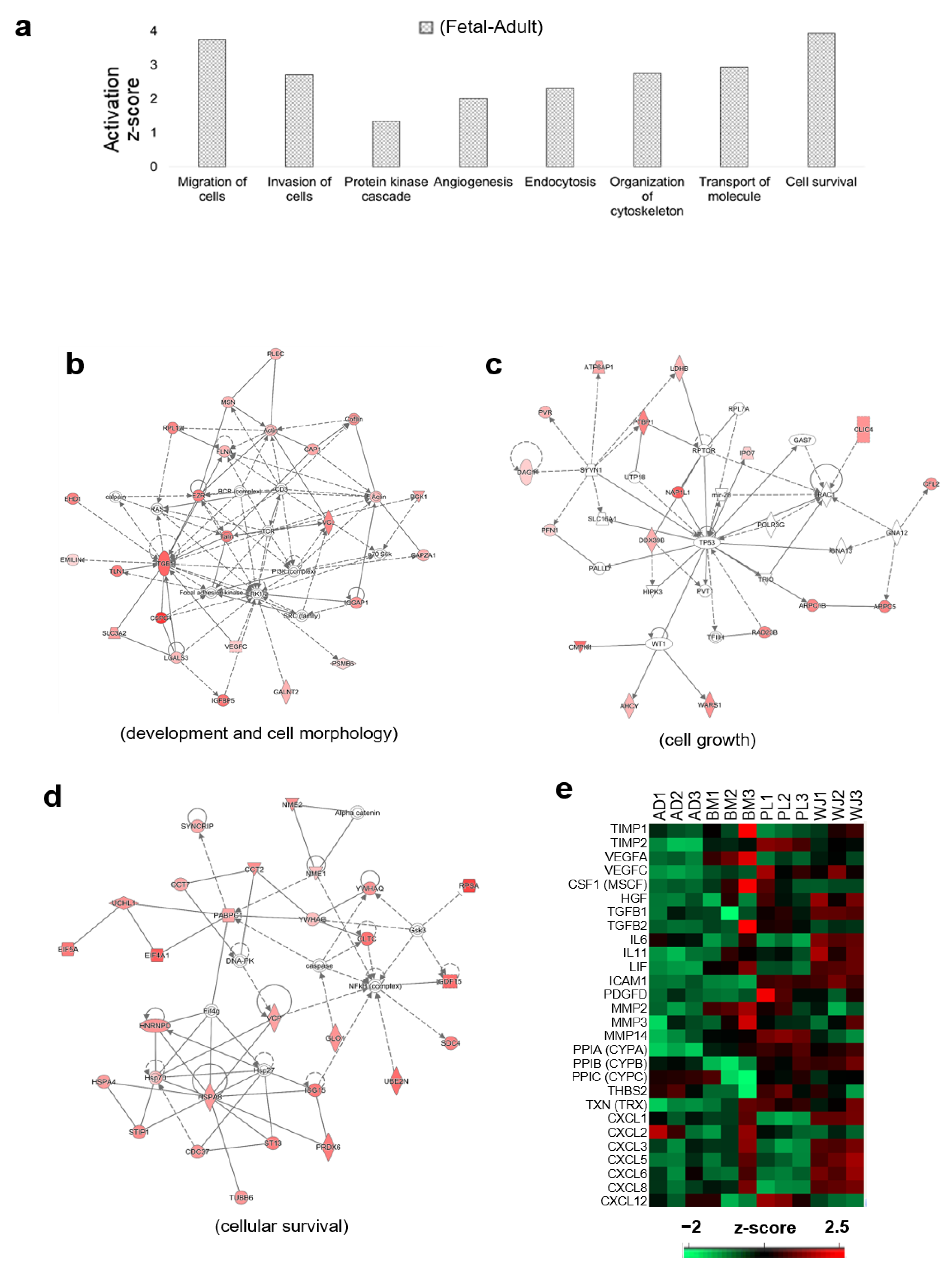
2.4. Functional Analyses of Secretome from Different Sources of MSCs
3. Discussion
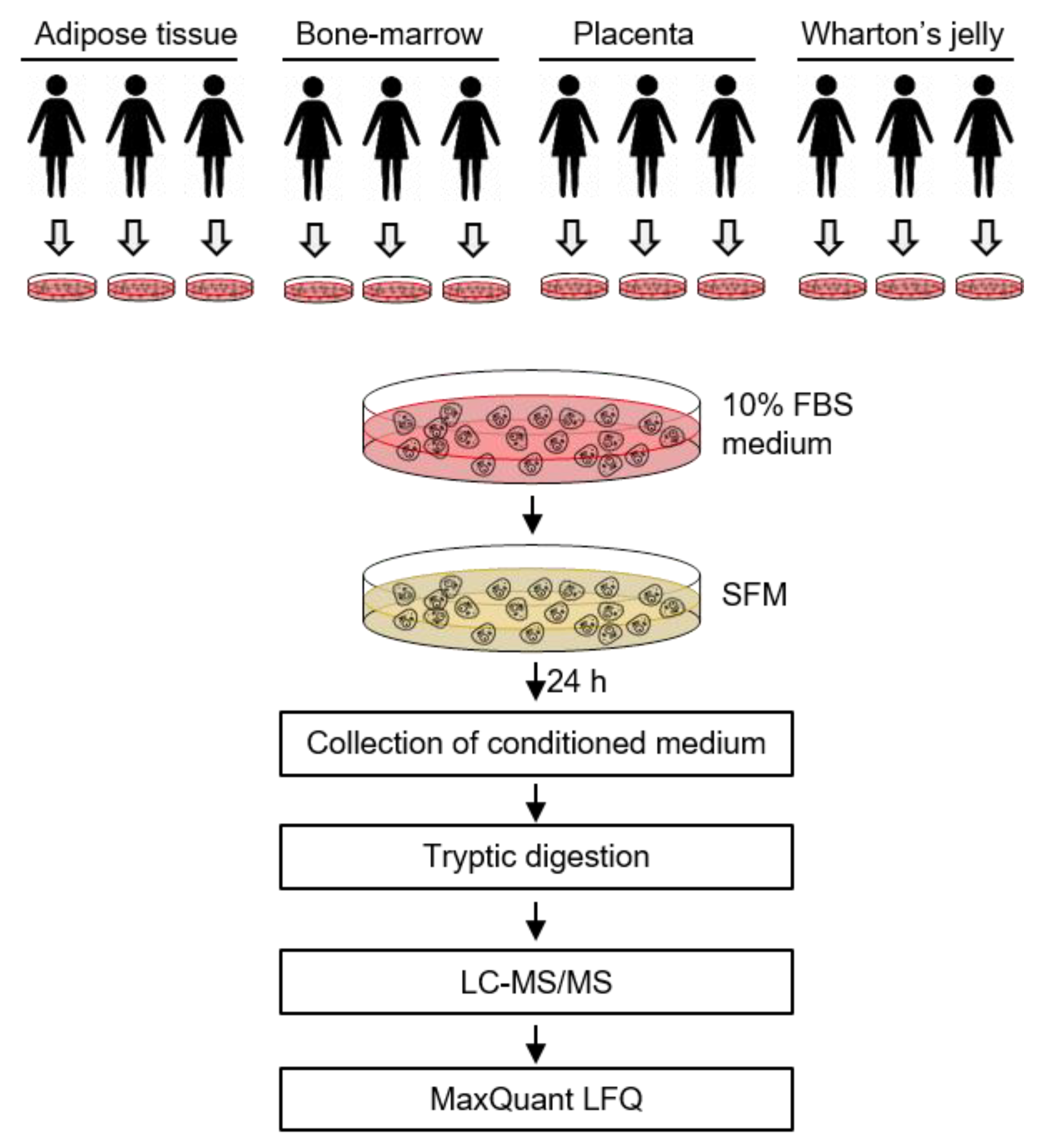
4. Materials and Methods
4.1. Isolation and Culture of MSCs from four Different Sources
4.2. Stemness Evaluation of MSCs
4.3. Preparation of the Secretome
4.4. In-Solution Digestion
4.5. Liquid Chromatography and Tandem Mass Spectrometry (LC–MS/MS)
4.6. Analysis of Mass Spectrometric Data
4.7. Bioinformatics Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Caplan, A.I. Mesenchymal stem cells. J. Orthop. Res. 1991, 9, 641–650. [Google Scholar] [PubMed]
- Friedenstein, A.J.; Piatetzky, S., II; Petrakova, K.V. Osteogenesis in transplants of bone marrow cells. J. Embryol. Exp. Morphol. 1966, 16, 381–390. [Google Scholar] [PubMed]
- Tavassoli, M.; Crosby, W.H. Transplantation of marrow to extramedullary sites. Science 1968, 161, 54–56. [Google Scholar] [CrossRef] [PubMed]
- Friedenstein, A.; Kuralesova, A.I. Osteogenic precursor cells of bone marrow in radiation chimeras. Transplantation 1971, 12, 99–108. [Google Scholar] [CrossRef]
- Friedenstein, A.J.; Chailakhyan, R.K.; Gerasimov, U.V. Bone marrow osteogenic stem cells: In vitro cultivation and transplantation in diffusion chambers. Cell Prolif. 1987, 20, 263–272. [Google Scholar] [CrossRef]
- Owen, M.; Friedenstein, A.J. Stromal stem cells: Marrow-derived osteogenic precursors. Ciba Found. Symp. 1988, 136, 42–60. [Google Scholar]
- Caplan, A.I.; Correa, D. The MSC: An injury drugstore. Cell Stem Cell 2011, 9, 11–15. [Google Scholar] [CrossRef] [Green Version]
- Gao, F.; Chiu, S.M.; Motan, D.A.; Zhang, Z.; Chen, L.; Ji, H.L.; Tse, H.F.; Fu, Q.L.; Lian, Q. Mesenchymal stem cells and immunomodulation: Current status and future prospects. Cell Death Dis. 2016, 7, e2062. [Google Scholar] [CrossRef] [Green Version]
- Squillaro, T.; Peluso, G.; Galderisi, U. Clinical Trials with Mesenchymal Stem Cells: An Update. Cell Transplant. 2016, 25, 829–848. [Google Scholar] [CrossRef] [Green Version]
- Galipeau, J.; Sensebe, L. Mesenchymal Stromal Cells: Clinical Challenges and Therapeutic Opportunities. Cell Stem Cell 2018, 22, 824–833. [Google Scholar] [CrossRef] [Green Version]
- da Silva Meirelles, L.; Chagastelles, P.C.; Nardi, N.B. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J. Cell Sci. 2006, 119, 2204–2213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crisan, M.; Yap, S.; Casteilla, L.; Chen, C.W.; Corselli, M.; Park, T.S.; Andriolo, G.; Sun, B.; Zheng, B.; Zhang, L.; et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 2008, 3, 301–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kern, S.; Eichler, H.; Stoeve, J.; Kluter, H.; Bieback, K. Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells 2006, 24, 1294–1301. [Google Scholar] [CrossRef] [PubMed]
- Hass, R.; Kasper, C.; Bohm, S.; Jacobs, R. Different populations and sources of human mesenchymal stem cells (MSC): A comparison of adult and neonatal tissue-derived MSC. Cell Commun. Signal. 2011, 9, 12. [Google Scholar] [CrossRef] [Green Version]
- Maleki, M.; Ghanbarvand, F.; Reza Behvarz, M.; Ejtemaei, M.; Ghadirkhomi, E. Comparison of mesenchymal stem cell markers in multiple human adult stem cells. Int. J. Stem Cells 2014, 7, 118–126. [Google Scholar] [CrossRef] [Green Version]
- Roth, A.; Schleyer, E.; Schoppmeyer, K.; Kluge, R.; Wittekind, C.; Mossner, J.; Wiedmann, M. Imatinib mesylate for palliative second-line treatment of advanced biliary tract cancer: A bicentric phase II study. Onkologie 2011, 34, 469–470. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Ponten, F.; Gry, M.; Fagerberg, L.; Lundberg, E.; Asplund, A.; Berglund, L.; Oksvold, P.; Bjorling, E.; Hober, S.; Kampf, C.; et al. A global view of protein expression in human cells, tissues, and organs. Mol. Syst. Biol. 2009, 5, 337. [Google Scholar] [CrossRef]
- Sonawane, A.R.; Platig, J.; Fagny, M.; Chen, C.Y.; Paulson, J.N.; Lopes-Ramos, C.M.; DeMeo, D.L.; Quackenbush, J.; Glass, K.; Kuijjer, M.L. Understanding Tissue-Specific Gene Regulation. Cell Rep. 2017, 21, 1077–1088. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Smith, J.W.; Huang, C.M. Mass spectrometry-based label-free quantitative proteomics. J. Biomed. Biotechnol. 2010, 2010, 840518. [Google Scholar] [CrossRef]
- Kinnaird, T.; Stabile, E.; Burnett, M.S.; Lee, C.W.; Barr, S.; Fuchs, S.; Epstein, S.E. Marrow-derived stromal cells express genes encoding a broad spectrum of arteriogenic cytokines and promote in vitro and in vivo arteriogenesis through paracrine mechanisms. Circ. Res. 2004, 94, 678–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehman, J.; Traktuev, D.; Li, J.; Merfeld-Clauss, S.; Temm-Grove, C.J.; Bovenkerk, J.E.; Pell, C.L.; Johnstone, B.H.; Considine, R.V.; March, K.L. Secretion of angiogenic and antiapoptotic factors by human adipose stromal cells. Circulation 2004, 109, 1292–1298. [Google Scholar] [CrossRef] [PubMed]
- Meirelles, L.D.S.; Fontes, A.M.; Covas, D.T.; Caplan, A.I. Mechanisms involved in the therapeutic properties of mesenchymal stem cells. Cytokine Growth Factor Rev. 2009, 20, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Elsner, T.; Botella, L.M.; Velasco, B.; Corbi, A.; Attisano, L.; Bernabeu, C. Synergistic cooperation between hypoxia and transforming growth factor-beta pathways on human vascular endothelial growth factor gene expression. J. Biol. Chem. 2001, 276, 38527–38535. [Google Scholar] [CrossRef] [Green Version]
- Lozito, T.P.; Tuan, R.S. Mesenchymal stem cells inhibit both endogenous and exogenous MMPs via secreted TIMPs. J. Cell. Physiol. 2011, 226, 385–396. [Google Scholar] [CrossRef]
- Pu, C.M.; Liu, C.W.; Liang, C.J.; Yen, Y.H.; Chen, S.H.; Jiang-Shieh, Y.F.; Chien, C.L.; Chen, Y.C.; Chen, Y.L. Adipose-Derived Stem Cells Protect Skin Flaps against Ischemia/Reperfusion Injury via IL-6 Expression. J. Investig. Dermatol. 2017, 137, 1353–1362. [Google Scholar] [CrossRef] [Green Version]
- Mauney, J.; Olsen, B.R.; Volloch, V. Matrix remodeling as stem cell recruitment event: A novel in vitro model for homing of human bone marrow stromal cells to the site of injury shows crucial role of extracellular collagen matrix. Matrix Biol. 2010, 29, 657–663. [Google Scholar] [CrossRef]
- Wei, Y.; Jinchuan, Y.; Yi, L.; Jun, W.; Zhongqun, W.; Cuiping, W. Antiapoptotic and proapoptotic signaling of cyclophilin A in endothelial cells. Inflammation 2013, 36, 567–572. [Google Scholar] [CrossRef]
- Kim, J.; Choi, T.G.; Ding, Y.; Kim, Y.; Ha, K.S.; Lee, K.H.; Kang, I.; Ha, J.; Kaufman, R.J.; Lee, J.; et al. Overexpressed cyclophilin B suppresses apoptosis associated with ROS and Ca2+ homeostasis after ER stress. J. Cell Sci. 2008, 121, 3636–3648. [Google Scholar] [CrossRef] [Green Version]
- Shoji, M.; Koba, S.; Kobayashi, Y. Roles of bone-marrow-derived cells and inflammatory cytokines in neointimal hyperplasia after vascular injury. Biomed. Res. Int. 2014, 2014, 945127. [Google Scholar] [CrossRef] [Green Version]
- Forte, A.; Rinaldi, B.; Sodano, L.; Berrino, L.; Rossi, F.; Finicelli, M.; Grossi, M.; Cobellis, G.; Botti, C.; De Feo, M.; et al. Stem cell therapy for arterial restenosis: Potential parameters contributing to the success of bone marrow-derived mesenchymal stromal cells. Cardiovasc. Drugs Ther. 2012, 26, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Gabrielyan, A.; Quade, M.; Gelinsky, M.; Rosen-Wolff, A. IL-11 and soluble VCAM-1 are important components of Hypoxia Conditioned Media and crucial for Mesenchymal Stromal Cells attraction. Stem Cell Res. 2020, 45, 101814. [Google Scholar] [CrossRef] [PubMed]
- Uccelli, A.; Moretta, L.; Pistoia, V. Mesenchymal stem cells in health and disease. Nat. Rev. Immunol. 2008, 8, 726–736. [Google Scholar] [CrossRef] [PubMed]
- Ankrum, J.; Karp, J.M. Mesenchymal stem cell therapy: Two steps forward, one step back. Trends Mol. Med. 2010, 16, 203–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ullah, I.; Subbarao, R.B.; Rho, G.J. Human mesenchymal stem cells—Current trends and future prospective. Biosci. Rep. 2015, 35, e00191. [Google Scholar] [CrossRef]
- Trounson, A.; McDonald, C. Stem Cell Therapies in Clinical Trials: Progress and Challenges. Cell Stem Cell 2015, 17, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Lazarus, H.M.; Haynesworth, S.E.; Gerson, S.L.; Rosenthal, N.S.; Caplan, A.I. Ex vivo expansion and subsequent infusion of human bone marrow-derived stromal progenitor cells (mesenchymal progenitor cells): Implications for therapeutic use. Bone Marrow Transpl. 1995, 16, 557–564. [Google Scholar]
- Lalu, M.M.; McIntyre, L.; Pugliese, C.; Fergusson, D.; Winston, B.W.; Marshall, J.C.; Granton, J.; Stewart, D.J.; Canadian Critical Care Trials, G. Safety of cell therapy with mesenchymal stromal cells (SafeCell): A systematic review and meta-analysis of clinical trials. PLoS ONE 2012, 7, e47559. [Google Scholar] [CrossRef]
- Bianco, P.; Cao, X.; Frenette, P.S.; Mao, J.J.; Robey, P.G.; Simmons, P.J.; Wang, C.Y. The meaning, the sense and the significance: Translating the science of mesenchymal stem cells into medicine. Nat. Med. 2013, 19, 35–42. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.S.; Lee, N.K.; Yoo, D.; Lee, J.; Choi, S.J.; Oh, W.; Chang, J.W.; Na, D.L. Lowering the concentration affects the migration and viability of intracerebroventricular-delivered human mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2017, 493, 751–757. [Google Scholar] [CrossRef]
- Vizoso, F.J.; Eiro, N.; Cid, S.; Schneider, J.; Perez-Fernandez, R. Mesenchymal Stem Cell Secretome: Toward Cell-Free Therapeutic Strategies in Regenerative Medicine. Int. J. Mol. Sci. 2017, 18, 1852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mead, B.; Logan, A.; Berry, M.; Leadbeater, W.; Scheven, B.A. Paracrine-mediated neuroprotection and neuritogenesis of axotomised retinal ganglion cells by human dental pulp stem cells: Comparison with human bone marrow and adipose-derived mesenchymal stem cells. PLoS ONE 2014, 9, e109305. [Google Scholar] [CrossRef] [PubMed]
- Pires, A.O.; Mendes-Pinheiro, B.; Teixeira, F.G.; Anjo, S.I.; Ribeiro-Samy, S.; Gomes, E.D.; Serra, S.C.; Silva, N.A.; Manadas, B.; Sousa, N.; et al. Unveiling the Differences of Secretome of Human Bone Marrow Mesenchymal Stem Cells, Adipose Tissue-Derived Stem Cells, and Human Umbilical Cord Perivascular Cells: A Proteomic Analysis. Stem Cells Dev. 2016, 25, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Talwadekar, M.D.; Kale, V.P.; Limaye, L.S. Placenta-derived mesenchymal stem cells possess better immunoregulatory properties compared to their cord-derived counterparts-a paired sample study. Sci. Rep. 2015, 5, 15784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolbank, S.; Peterbauer, A.; Fahrner, M.; Hennerbichler, S.; van Griensven, M.; Stadler, G.; Redl, H.; Gabriel, C. Dose-dependent immunomodulatory effect of human stem cells from amniotic membrane: A comparison with human mesenchymal stem cells from adipose tissue. Tissue Eng. 2007, 13, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Tachida, Y.; Sakurai, H.; Okutsu, J.; Suda, K.; Sugita, R.; Yaginuma, Y.; Ogura, Y.; Shimada, K.; Isono, F.; Kubota, K.; et al. Proteomic Comparison of the Secreted Factors of Mesenchymal Stem Cells from Bone Marrow, Adipose Tissue and Dental Pulp. J. Proteom. Bioinform. 2015, 8, 266–273. [Google Scholar] [CrossRef]
- Chen, L.; Tredget, E.E.; Wu, P.Y.; Wu, Y. Paracrine factors of mesenchymal stem cells recruit macrophages and endothelial lineage cells and enhance wound healing. PLoS ONE 2008, 3, e1886. [Google Scholar] [CrossRef] [Green Version]
- Di Ianni, M.; Del Papa, B.; De Ioanni, M.; Moretti, L.; Bonifacio, E.; Cecchini, D.; Sportoletti, P.; Falzetti, F.; Tabilio, A. Mesenchymal cells recruit and regulate T regulatory cells. Exp. Hematol. 2008, 36, 309–318. [Google Scholar] [CrossRef]
- Tasso, R.; Augello, A.; Boccardo, S.; Salvi, S.; Carida, M.; Postiglione, F.; Fais, F.; Truini, M.; Cancedda, R.; Pennesi, G. Recruitment of a host’s osteoprogenitor cells using exogenous mesenchymal stem cells seeded on porous ceramic. Tissue Eng. Part A 2009, 15, 2203–2212. [Google Scholar] [CrossRef]
- Kwon, Y.W.; Heo, S.C.; Jeong, G.O.; Yoon, J.W.; Mo, W.M.; Lee, M.J.; Jang, I.H.; Kwon, S.M.; Lee, J.S.; Kim, J.H. Tumor necrosis factor-alpha-activated mesenchymal stem cells promote endothelial progenitor cell homing and angiogenesis. Biochim. Biophys. Acta 2013, 1832, 2136–2144. [Google Scholar] [CrossRef] [Green Version]
- Shin, L.; Peterson, D.A. Human mesenchymal stem cell grafts enhance normal and impaired wound healing by recruiting existing endogenous tissue stem/progenitor cells. Stem Cells Transl. Med. 2013, 2, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, S.; Gu, S.; Sang, L.; Dai, C. Mesenchymal stem cells recruit macrophages to alleviate experimental colitis through TGFbeta1. Cell. Physiol. Biochem. 2015, 35, 858–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.J.; Seo, D.H.; Lee, S.H.; Lee, S.H.; An, G.H.; Ahn, H.J.; Kwon, D.; Seo, K.W.; Kang, K.S. Conditioned media from human umbilical cord blood-derived mesenchymal stem cells stimulate rejuvenation function in human skin. Biochem. Biophys. Rep. 2018, 16, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Webb, T.L.; Ning, F.; Shiraishi, Y.; Regan, D.P.; Chow, L.; Smith, M.J.; Ashino, S.; Guth, A.M.; Hopkins, S.; et al. Mesenchymal Stem Cells Recruit CCR2(+) Monocytes to Suppress Allergic Airway Inflammation. J. Immunol. 2018, 200, 1261–1269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menezes, K.; Rosa, B.G.; Freitas, C.; da Cruz, A.S.; de Siqueira Santos, R.; Nascimento, M.A.; Alves, D.V.L.; Bonamino, M.; Rossi, M.I.; Borojevic, R.; et al. Human mesenchymal stromal/stem cells recruit resident pericytes and induce blood vessels maturation to repair experimental spinal cord injury in rats. Sci. Rep. 2020, 10, 19604. [Google Scholar] [CrossRef] [PubMed]
- Weiss, A.R.R.; Dahlke, M.H. Immunomodulation by Mesenchymal Stem Cells (MSCs): Mechanisms of Action of Living, Apoptotic, and Dead MSCs. Front. Immunol. 2019, 10, 1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yubo, M.; Yanyan, L.; Li, L.; Tao, S.; Bo, L.; Lin, C. Clinical efficacy and safety of mesenchymal stem cell transplantation for osteoarthritis treatment: A meta-analysis. PLoS ONE 2017, 12, e0175449. [Google Scholar] [CrossRef]
- Xu, P.; Yang, X. The Efficacy and Safety of Mesenchymal Stem Cell Transplantation for Spinal Cord Injury Patients: A Meta-Analysis and Systematic Review. Cell Transplant. 2019, 28, 36–46. [Google Scholar] [CrossRef] [Green Version]
- Ren, G.; Zhao, X.; Wang, Y.; Zhang, X.; Chen, X.; Xu, C.; Yuan, Z.R.; Roberts, A.I.; Zhang, L.; Zheng, B.; et al. CCR2-dependent recruitment of macrophages by tumor-educated mesenchymal stromal cells promotes tumor development and is mimicked by TNFalpha. Cell Stem Cell 2012, 11, 812–824. [Google Scholar] [CrossRef] [Green Version]
- Yu, P.F.; Huang, Y.; Han, Y.Y.; Lin, L.Y.; Sun, W.H.; Rabson, A.B.; Wang, Y.; Shi, Y.F. TNFalpha-activated mesenchymal stromal cells promote breast cancer metastasis by recruiting CXCR2(+) neutrophils. Oncogene 2017, 36, 482–490. [Google Scholar] [CrossRef]
- Lee, H.Y.; Hong, I.S. Double-edged sword of mesenchymal stem cells: Cancer-promoting versus therapeutic potential. Cancer Sci. 2017, 108, 1939–1946. [Google Scholar] [CrossRef] [PubMed]
- Amable, P.R.; Teixeira, M.V.; Carias, R.B.; Granjeiro, J.M.; Borojevic, R. Protein synthesis and secretion in human mesenchymal cells derived from bone marrow, adipose tissue and Wharton’s jelly. Stem Cell Res. Ther. 2014, 5, 53. [Google Scholar] [CrossRef] [Green Version]
- Nakanishi, C.; Nagaya, N.; Ohnishi, S.; Yamahara, K.; Takabatake, S.; Konno, T.; Hayashi, K.; Kawashiri, M.A.; Tsubokawa, T.; Yamagishi, M. Gene and protein expression analysis of mesenchymal stem cells derived from rat adipose tissue and bone marrow. Circ. J. 2011, 75, 2260–2268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, L.; Jia, Z.; Yin, X.; Zhang, X.; Liu, Y.; Chen, P.; Ma, K.; Zhou, C. Comparative analysis of mesenchymal stem cells from bone marrow, cartilage, and adipose tissue. Stem Cells Dev. 2008, 17, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.J.; Kim, J.; Cho, J.H.; Chung, H.M.; Chae, J.I. Comparative Analysis of Human Mesenchymal Stem Cells Derived from Bone Marrow, Placenta, and Adipose Tissue as Sources of Cell Therapy. J. Cell. Biochem. 2016, 117, 1112–1125. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.S.; Choi, Y.; Kim, H.S.; Kim, H.O. Comparison of molecular profiles of human mesenchymal stem cells derived from bone marrow, umbilical cord blood, placenta and adipose tissue. Int. J. Mol. Med. 2016, 37, 115–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nonnis, S.; Maffioli, E.; Zanotti, L.; Santagata, F.; Negri, A.; Viola, A.; Elliman, S.; Tedeschi, G. Effect of fetal bovine serum in culture media on MS analysis of mesenchymal stromal cells secretome. EuPA Open Proteom. 2016, 10, 28–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, J.; Kim, H.J.; Kim, G.; Song, M.; Woo, S.J.; Lee, S.T.; Kim, H.; Lee, C. Discovery of melanotransferrin as a serological marker of colorectal cancer by secretome analysis and quantitative proteomics. J. Proteome Res. 2014, 13, 4919–4931. [Google Scholar] [CrossRef]
- Shin, J.; Kim, G.; Kabir, M.H.; Park, S.J.; Lee, S.T.; Lee, C. Use of composite protein database including search result sequences for mass spectrometric analysis of cell secretome. PLoS ONE 2015, 10, e0121692. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Bendtsen, J.D.; Jensen, L.J.; Blom, N.; Von Heijne, G.; Brunak, S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 2004, 17, 349–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moller, S.; Croning, M.D.; Apweiler, R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 2001, 17, 646–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, S.; Lee, J.; Kwon, Y.; Park, K.-S.; Jeong, J.-H.; Choi, S.-J.; Bang, S.I.; Chang, J.W.; Lee, C. Comparative Proteomic Analysis of the Mesenchymal Stem Cells Secretome from Adipose, Bone Marrow, Placenta and Wharton’s Jelly. Int. J. Mol. Sci. 2021, 22, 845. https://doi.org/10.3390/ijms22020845
Shin S, Lee J, Kwon Y, Park K-S, Jeong J-H, Choi S-J, Bang SI, Chang JW, Lee C. Comparative Proteomic Analysis of the Mesenchymal Stem Cells Secretome from Adipose, Bone Marrow, Placenta and Wharton’s Jelly. International Journal of Molecular Sciences. 2021; 22(2):845. https://doi.org/10.3390/ijms22020845
Chicago/Turabian StyleShin, Sungho, Jeongmin Lee, Yumi Kwon, Kang-Sik Park, Jae-Hoon Jeong, Suk-Joo Choi, Sa Ik Bang, Jong Wook Chang, and Cheolju Lee. 2021. "Comparative Proteomic Analysis of the Mesenchymal Stem Cells Secretome from Adipose, Bone Marrow, Placenta and Wharton’s Jelly" International Journal of Molecular Sciences 22, no. 2: 845. https://doi.org/10.3390/ijms22020845






