Delivery of miR-424-5p via Extracellular Vesicles Promotes the Apoptosis of MDA-MB-231 TNBC Cells in the Tumor Microenvironment
Abstract
1. Introduction
2. Results
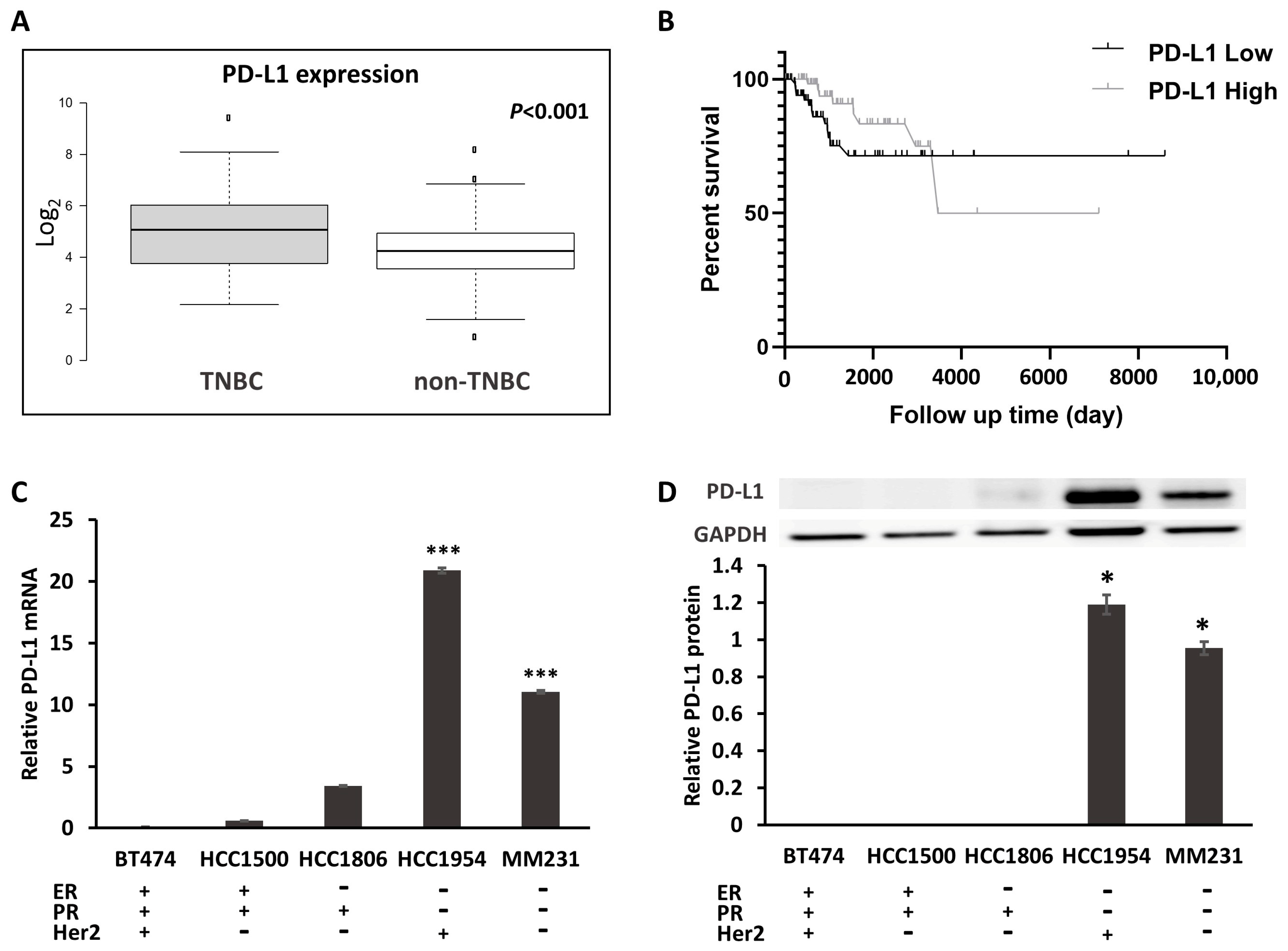
2.1. PD-L1 Expression Is Increased in TNBC
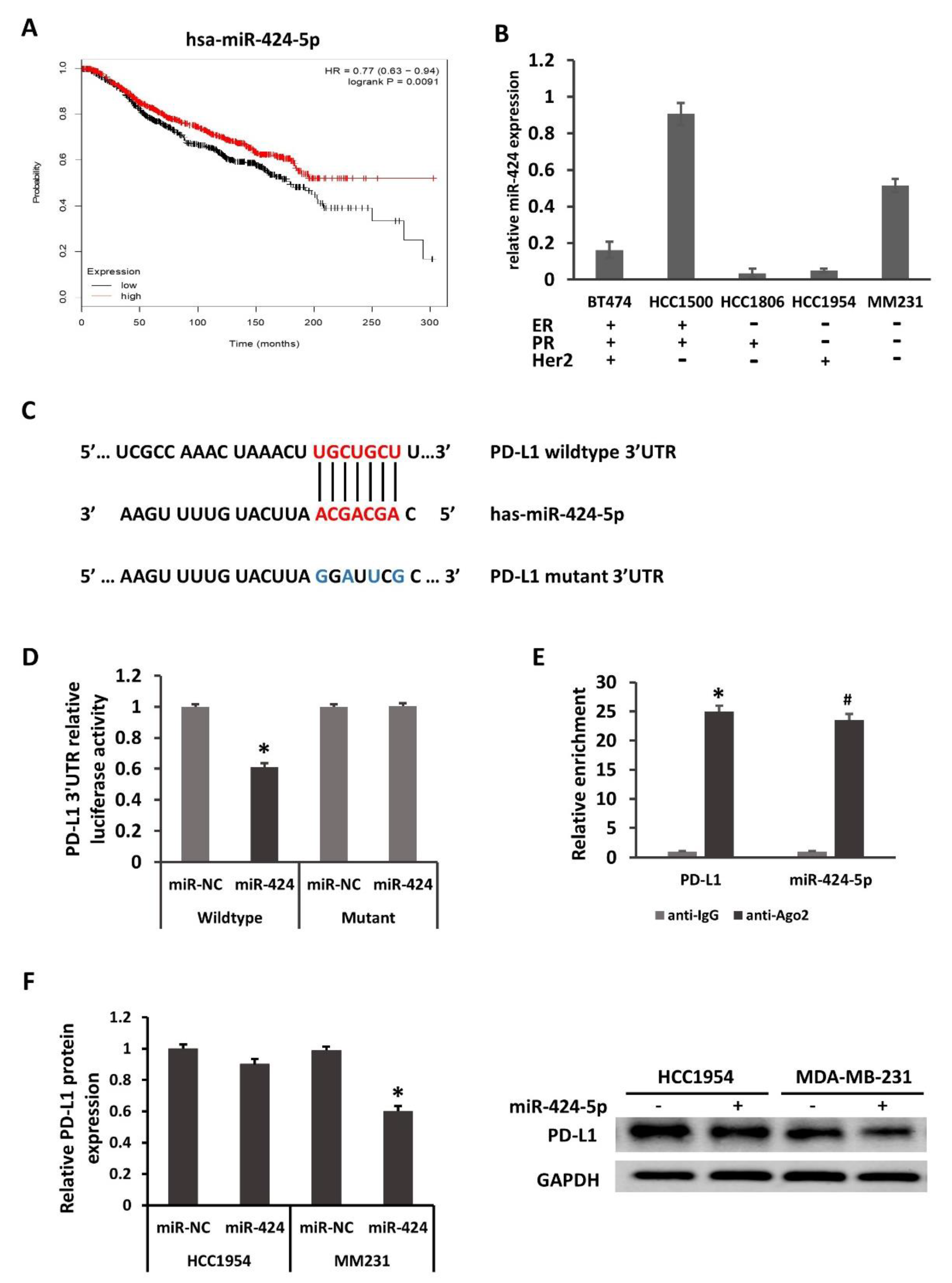
2.2. PD-L1 Is Directly Regulated by miR-424-5p
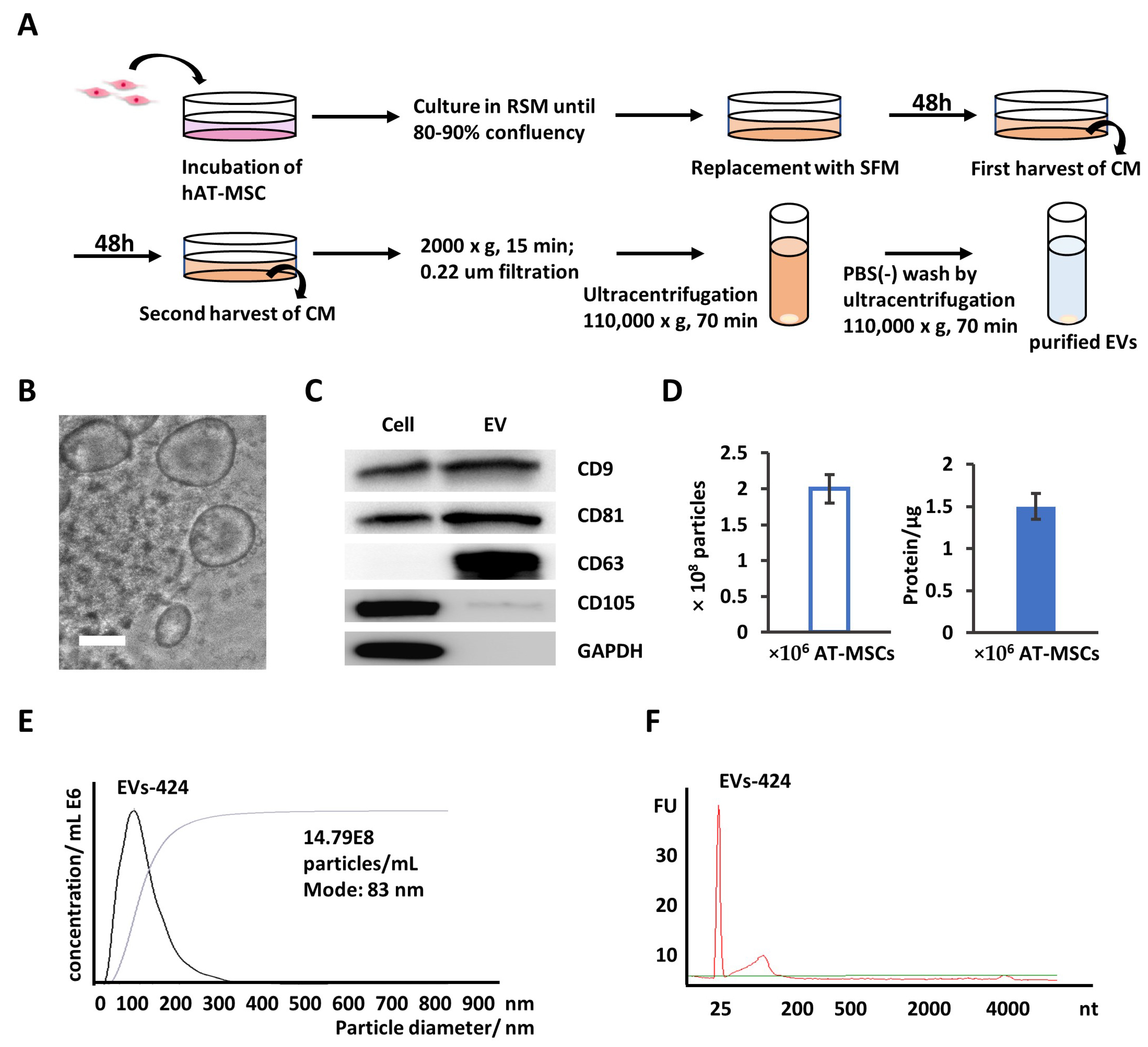
2.3. EVs Are Secreted into AT-MSC Culture Supernatant
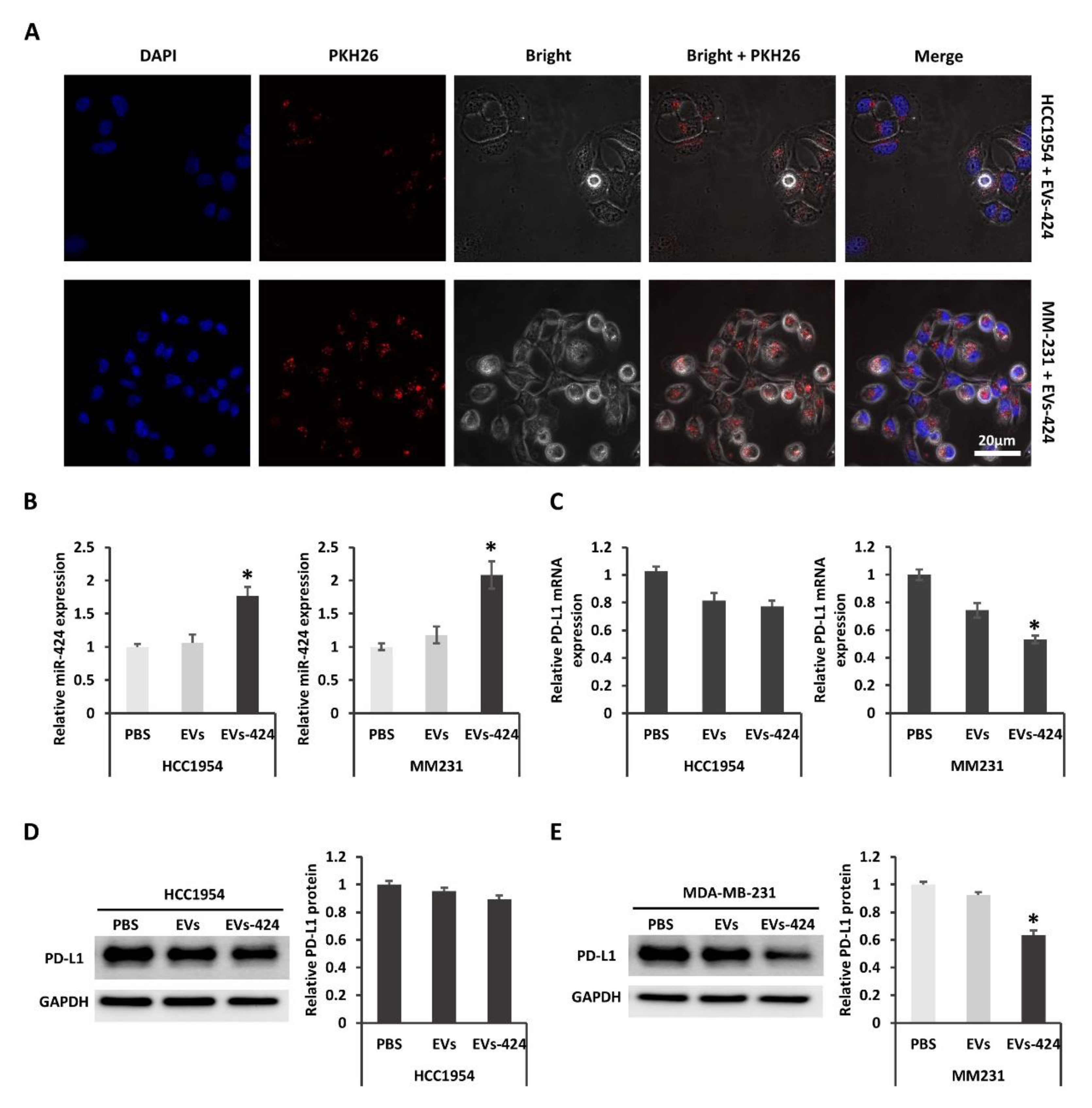
2.4. EVs-Mediated Delivery of miR-424 Decreases PD-L1 Expression in Recipient Cells
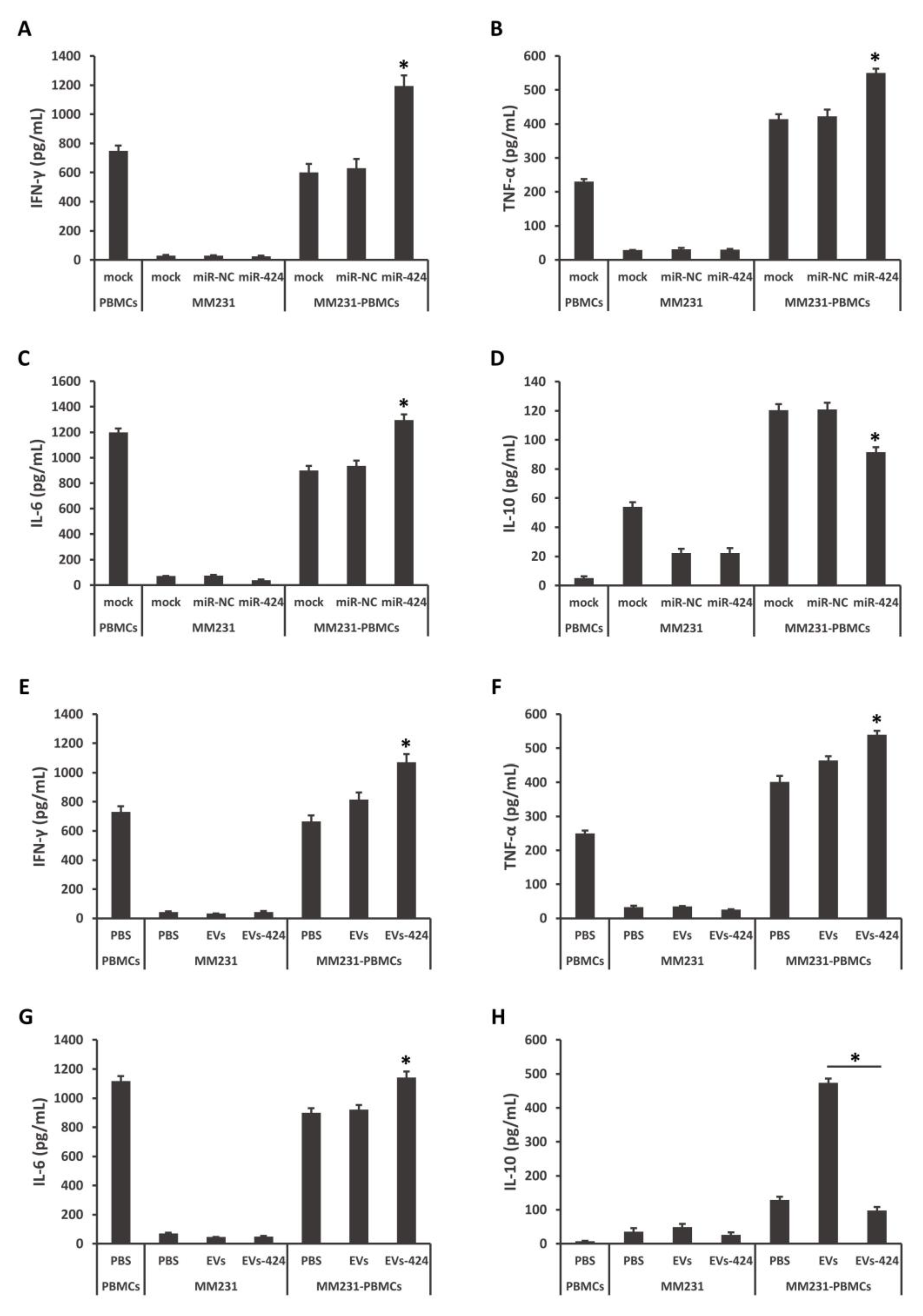
2.5. miR-424-5p Regulates Cytokine Secretion
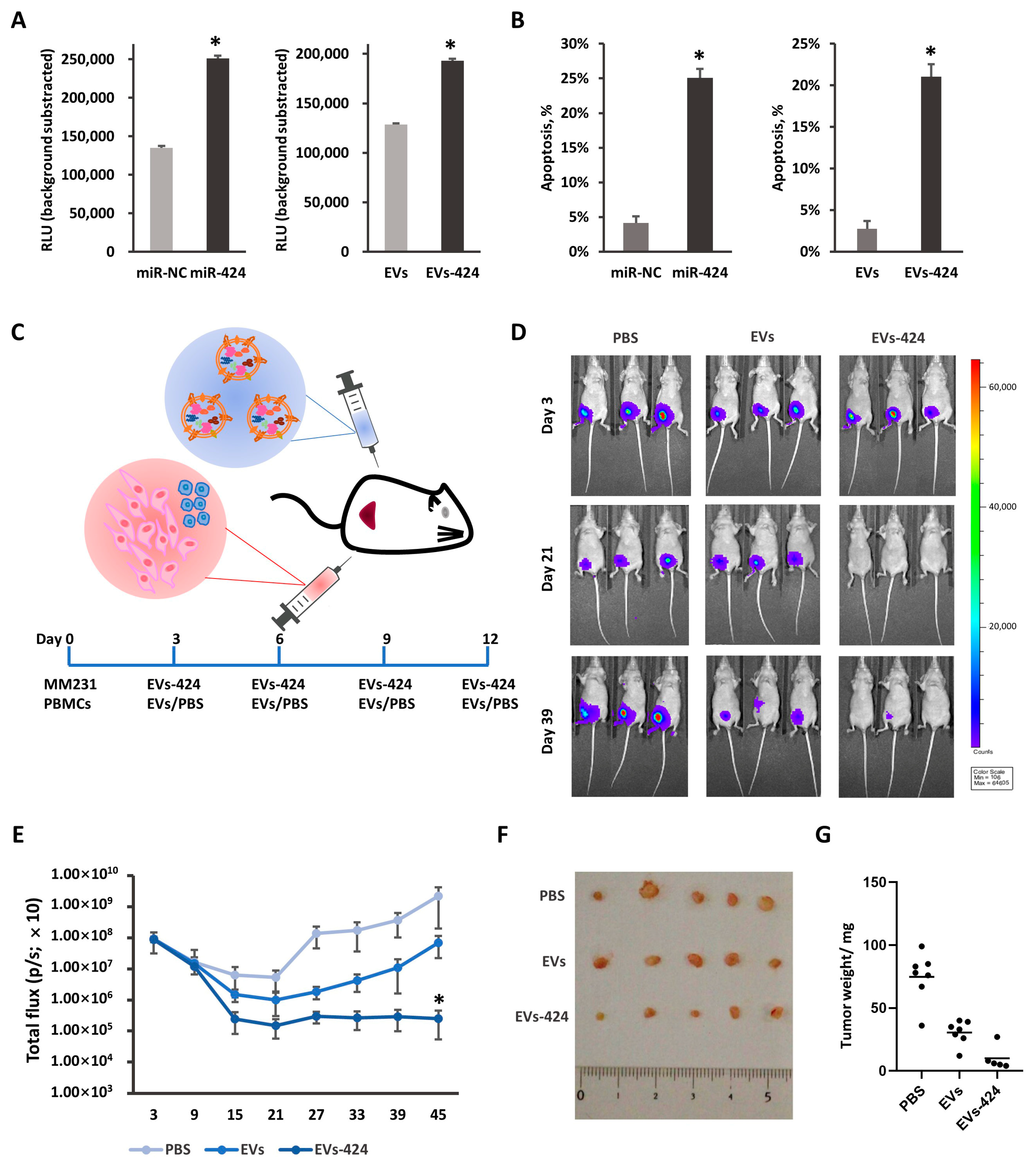
2.6. miR-424-5p Upregulation Promotes Cytotoxicity against Tumors
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Culture
4.2. Preparation of EVs
4.3. Electron Microscopy
4.4. Measurement of the Size Distribution and Particle Number by NTA
4.5. Real-Time Reverse Transcription-PCR
4.6. Cell Transfection
4.7. Dual-Luciferase Reporter Assay
4.8. AGO2-RNA Immunoprecipitation
4.9. Western Blotting
4.10. Tumor Cell/T Cell Coculture Model
4.11. Immunofluorescence
4.12. Apoptosis Assessment of Tumor Cells in the Coculture System
4.13. Caspase 3/7 Assay
4.14. LDH Release Assay
4.15. In Vivo Studies
4.16. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-Negative Breast Cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef]
- Shao, F.; Sun, H.; Deng, C.-X. Potential Therapeutic Targets of Triple-Negative Breast Cancer Based on Its Intrinsic Subtype. Oncotarget 2017, 8, 73329–73344. [Google Scholar] [CrossRef] [PubMed]
- Loi, S.; Sirtaine, N.; Piette, F.; Salgado, R.; Viale, G.; Van Eenoo, F.; Rouas, G.; Francis, P.; Crown, J.P.A.; Hitre, E.; et al. Prognostic and Predictive Value of Tumor-Infiltrating Lymphocytes in a Phase III Randomized Adjuvant Breast Cancer Trial in Node-Positive Breast Cancer Comparing the Addition of Docetaxel to Doxorubicin With Doxorubicin-Based Chemotherapy: BIG 02-98. JCO 2013, 31, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Sabatier, R.; Finetti, P.; Mamessier, E.; Adelaide, J.; Chaffanet, M.; Ali, H.R.; Viens, P.; Caldas, C.; Birnbaum, D.; Bertucci, F. Prognostic and Predictive Value of PDL1 Expression in Breast Cancer. Oncotarget 2015, 6, 5449–5464. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Chen, S.; Yang, L.; Li, Y. The Role of PD-1 and PD-L1 in T-Cell Immune Suppression in Patients with Hematological Malignancies. J. Hematol. Oncol. 2013, 6, 74–74. [Google Scholar] [CrossRef]
- Manson, Q.F.; Schrijver, W.A.M.E.; Ter Hoeve, N.D.; Moelans, C.B.; van Diest, P.J. Frequent Discordance in PD-1 and PD-L1 Expression between Primary Breast Tumors and Their Matched Distant Metastases. Clin. Exp. Metastasis 2019, 36, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Homet Moreno, B.; Ribas, A. Anti-Programmed Cell Death Protein-1/Ligand-1 Therapy in Different Cancers. Br. J. Cancer 2015, 112, 1421–1427. [Google Scholar] [CrossRef]
- Sunshine, J.; Taube, J.M. PD-1/PD-L1 Inhibitors. Curr. Opin. Pharm. 2015, 23, 32–38. [Google Scholar] [CrossRef]
- Mavratzas, A.; Seitz, J.; Smetanay, K.; Schneeweiss, A.; Jäger, D.; Fremd, C. Atezolizumab for Use in PD-L1-Positive Unresectable, Locally Advanced or Metastatic Triple-Negative Breast Cancer. Future Oncol. 2019, 16, 4439–4453. [Google Scholar] [CrossRef]
- Schmid, P.; Rugo, H.S.; Adams, S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Henschel, V.; Molinero, L.; Chui, S.Y.; et al. Atezolizumab plus Nab-Paclitaxel as First-Line Treatment for Unresectable, Locally Advanced or Metastatic Triple-Negative Breast Cancer (IMpassion130): Updated Efficacy Results from a Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet Oncol. 2020, 21, 44–59. [Google Scholar] [CrossRef]
- Kwapisz, D. Pembrolizumab and Atezolizumab in Triple-Negative Breast Cancer. Cancer Immunol. Immunother. 2020. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-Mediated Transfer of MRNAs and MicroRNAs Is a Novel Mechanism of Genetic Exchange between Cells. Nat. Cell Biol. 2007, 9, 654. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular Organelles Important in Intercellular Communication. J. Proteomics 2010, 73, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genom. Proteomics Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef]
- Kidd, S.; Spaeth, E.; Dembinski, J.L.; Dietrich, M.; Watson, K.; Klopp, A.; Battula, V.L.; Weil, M.; Andreeff, M.; Marini, F.C. Direct Evidence of Mesenchymal Stem Cell Tropism for Tumor and Wounding Microenvironments Using in Vivo Bioluminescent Imaging. Stem Cells 2009, 27, 2614–2623. [Google Scholar] [CrossRef]
- Zhang, Y.; Daquinag, A.C.; Amaya-Manzanares, F.; Sirin, O.; Tseng, C.; Kolonin, M.G. Stromal Progenitor Cells from Endogenous Adipose Tissue Contribute to Pericytes and Adipocytes That Populate the Tumor Microenvironment. Cancer Res. 2012, 72, 5198. [Google Scholar] [CrossRef]
- Schu, S.; Nosov, M.; O’Flynn, L.; Shaw, G.; Treacy, O.; Barry, F.; Murphy, M.; O’Brien, T.; Ritter, T. Immunogenicity of Allogeneic Mesenchymal Stem Cells. J. Cell Mol. Med. 2012, 16, 2094–2103. [Google Scholar] [CrossRef]
- Wang, X.; Li, J.; Dong, K.; Lin, F.; Long, M.; Ouyang, Y.; Wei, J.; Chen, X.; Weng, Y.; He, T.; et al. Tumor Suppressor MiR-34a Targets PD-L1 and Functions as a Potential Immunotherapeutic Target in Acute Myeloid Leukemia. Cell. Signal. 2015, 27, 443–452. [Google Scholar] [CrossRef]
- Gao, L.; Guo, Q.; Li, X.; Yang, X.; Ni, H.; Wang, T.; Zhao, Q.; Liu, H.; Xing, Y.; Xi, T.; et al. MiR-873/PD-L1 Axis Regulates the Stemness of Breast Cancer Cells. EBioMedicine 2019, 41, 395–407. [Google Scholar] [CrossRef]
- Zhou, Y.; Yamamoto, Y.; Itaya, T.; Xiao, Z.-D.; Ochiya, T. Distinct Mirna Expression Patterns of Extracellular Vesicles Derived From 4 Types of Mesenchymal Stem Cells. J. Stem Cell Res. 2018, 8, 8. [Google Scholar] [CrossRef]
- Crescitelli, R.; Lässer, C.; Szabó, T.G.; Kittel, A.; Eldh, M.; Dianzani, I.; Buzás, E.I.; Lötvall, J. Distinct RNA Profiles in Subpopulations of Extracellular Vesicles: Apoptotic Bodies, Microvesicles and Exosomes. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef] [PubMed]
- Katsuda, T.; Tsuchiya, R.; Kosaka, N.; Yoshioka, Y.; Takagaki, K.; Oki, K.; Takeshita, F.; Sakai, Y.; Kuroda, M.; Ochiya, T. Human Adipose Tissue-Derived Mesenchymal Stem Cells Secrete Functional Neprilysin-Bound Exosomes. Sci. Rep. 2013, 3, 1197. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic Comparison Defines Novel Markers to Characterize Heterogeneous Populations of Extracellular Vesicle Subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.; McKee, C.; Bakshi, S.; Walker, K.; Hakman, E.; Halassy, S.; Svinarich, D.; Dodds, R.; Govind, C.K.; Chaudhry, G.R. Mesenchymal Stem Cells: Cell Therapy and Regeneration Potential. J. Tissue Eng. Regen. Med. 2019, 13, 1738–1755. [Google Scholar] [CrossRef]
- Zhou, Y.; Tian, T.; Zhu, Y.; Jaffar Ali, D.; Hu, F.; Qi, Y.; Sun, B.; Xiao, Z. Exosomes Transfer Among Different Species Cells and Mediating MiRNAs Delivery. J. Cell Biochem. 2017, 118, 4267–4274. [Google Scholar] [CrossRef]
- Khan, A.A.; Betel, D.; Miller, M.L.; Sander, C.; Leslie, C.S.; Marks, D.S. Transfection of Small RNAs Globally Perturbs Gene Regulation by Endogenous MicroRNAs. Nat. Biotechnol. 2009, 27, 549–555. [Google Scholar] [CrossRef]
- Arvey, A.; Larsson, E.; Sander, C.; Leslie, C.S.; Marks, D.S. Target MRNA Abundance Dilutes MicroRNA and SiRNA Activity. Mol. Syst. Biol. 2010, 6, 363. [Google Scholar] [CrossRef]
- Ross, J. MRNA Stability in Mammalian Cells. Microbiol. Rev. 1995, 59, 423–450. [Google Scholar] [CrossRef]
- Larsson, E.; Sander, C.; Marks, D. MRNA Turnover Rate Limits SiRNA and MicroRNA Efficacy. Mol. Syst. Biol. 2010, 6, 433. [Google Scholar] [CrossRef]
- Blank, C.; Gajewski, T.F.; Mackensen, A. Interaction of PD-L1 on Tumor Cells with PD-1 on Tumor-Specific T Cells as a Mechanism of Immune Evasion: Implications for Tumor Immunotherapy. Cancer Immunol. Immunother. 2005, 54, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.; Verhagen, J.; Blaser, K.; Akdis, M.; Akdis, C.A. Mechanisms of Immune Suppression by Interleukin-10 and Transforming Growth Factor-β: The Role of T Regulatory Cells. Immunology 2006, 117, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Nowak, M.; Klink, M.; Glowacka, E.; Sulowska, Z.; Kulig, A.; Szpakowski, M.; Szyllo, K.; Tchorzewski, H. Production of Cytokines During Interaction of Peripheral Blood Mononuclear Cells with Autologous Ovarian Cancer Cells or Benign Ovarian Tumour Cells. Scand. J. Immunol. 2010, 71, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Payne, A.M.; Zorman, J.; Horton, M.; Dubey, S.; ter Meulen, J.; Vora, K.A. Caspase Activation as a Versatile Assay Platform for Detection of Cytotoxic Bacterial Toxins. J. Clin. Microbiol. 2013, 51, 2970–2976. [Google Scholar] [CrossRef] [PubMed]
- Porter, A.G.; Jänicke, R.U. Emerging Roles of Caspase-3 in Apoptosis. Cell Death Differ. 1999, 6, 99–104. [Google Scholar] [CrossRef]
- Chan, F.K.-M.; Moriwaki, K.; De Rosa, M.J. Detection of Necrosis by Release of Lactate Dehydrogenase Activity. Methods Mol. Biol. 2013, 979, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of Cell Viability by the Lactate Dehydrogenase Assay. Cold Spring Harb. Protoc. 2018, 2018, pdb.prot095497. [Google Scholar] [CrossRef] [PubMed]
- Tsang, J.Y.S.; Au, W.-L.; Lo, K.-Y.; Ni, Y.-B.; Hlaing, T.; Hu, J.; Chan, S.-K.; Chan, K.-F.; Cheung, S.-Y.; Tse, G.M. PD-L1 Expression and Tumor Infiltrating PD-1+ Lymphocytes Associated with Outcome in HER2+ Breast Cancer Patients. Breast Cancer Res. Treat. 2017, 162, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Matikas, A.; Zerdes, I.; Lövrot, J.; Richard, F.; Sotiriou, C.; Bergh, J.; Valachis, A.; Foukakis, T. Prognostic Implications of PD-L1 Expression in Breast Cancer: Systematic Review and Meta-Analysis of Immunohistochemistry and Pooled Analysis of Transcriptomic Data. Clin. Cancer Res. 2019, 25, 5717. [Google Scholar] [CrossRef] [PubMed]
- Mittendorf, E.A.; Philips, A.V.; Meric-Bernstam, F.; Qiao, N.; Wu, Y.; Harrington, S.; Su, X.; Wang, Y.; Gonzalez-Angulo, A.M.; Akcakanat, A.; et al. PD-L1 Expression in Triple-Negative Breast Cancer. Cancer Immunol. Res. 2014, 2, 361. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Yagishita, S.; Hagiwara, K.; Yoshioka, Y.; Kosaka, N.; Takeshita, F.; Fujiwara, T.; Tsuta, K.; Nokihara, H.; Tamura, T.; et al. The Clinical Relevance of the MiR-197/CKS1B/STAT3-Mediated PD-L1 Network in Chemoresistant Non-Small-Cell Lung Cancer. Mol. Ther. 2015, 23, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Tao, Z.; Hai, B.; Liang, H.; Shi, Y.; Wang, T.; Song, W.; Chen, Y.; OuYang, J.; Chen, J.; et al. MiR-424(322) Reverses Chemoresistance via T-Cell Immune Response Activation by Blocking the PD-L1 Immune Checkpoint. Nat. Commun. 2016, 7, 11406–11406. [Google Scholar] [CrossRef] [PubMed]
- Reza-Zaldivar, E.E.; Hernández-Sapiéns, M.A.; Minjarez, B.; Gutiérrez-Mercado, Y.K.; Márquez-Aguirre, A.L.; Canales-Aguirre, A.A. Potential Effects of MSC-Derived Exosomes in Neuroplasticity in Alzheimer’s Disease. Front. Cell. Neurosci. 2018, 12, 317. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Wang, S.; Zhao, R.C. Exosomes from Mesenchymal Stem/Stromal Cells: A New Therapeutic Paradigm. Biomark. Res. 2019, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Priebe, W.; Glod, J.; Banerjee, D. Activation of Signal Transducers and Activators of Transcription 3 and Focal Adhesion Kinase by Stromal Cell-Derived Factor 1 Is Required for Migration of Human Mesenchymal Stem Cells in Response to Tumor Cell-Conditioned Medium. Stem Cells 2009, 27, 857–865. [Google Scholar] [CrossRef]
- Rattigan, Y.; Hsu, J.-M.; Mishra, P.J.; Glod, J.; Banerjee, D. Interleukin 6 Mediated Recruitment of Mesenchymal Stem Cells to the Hypoxic Tumor Milieu. Exp. Cell Res. 2010, 316, 3417–3424. [Google Scholar] [CrossRef]
- Senst, C.; Nazari-Shafti, T.; Kruger, S.; Bentrup, K.H.Z.; Dupin, C.L.; Chaffin, A.E.; Srivastav, S.K.; Wörner, P.M.; Abdel-Mageed, A.B.; Alt, E.U.; et al. Prospective Dual Role of Mesenchymal Stem Cells in Breast Tumor Microenvironment. Breast Cancer Res. Treat. 2013, 137, 69–79. [Google Scholar] [CrossRef]
- Xie, C.; Yang, Z.; Suo, Y.; Chen, Q.; Wei, D.; Weng, X.; Gu, Z.; Wei, X. Systemically Infused Mesenchymal Stem Cells Show Different Homing Profiles in Healthy and Tumor Mouse Models. Stem Cells Transl. Med. 2017, 6, 1120–1131. [Google Scholar] [CrossRef]
- Smith, C.L.; Chaichana, K.L.; Lee, Y.M.; Lin, B.; Stanko, K.M.; O’Donnell, T.; Gupta, S.; Shah, S.R.; Wang, J.; Wijesekera, O.; et al. Pre-Exposure of Human Adipose Mesenchymal Stem Cells to Soluble Factors Enhances Their Homing to Brain Cancer. Stem Cells Transl. Med. 2015, 4, 239–251. [Google Scholar] [CrossRef]
- Arvelo, F.; Sojo, F.; Cotte, C. Tumour Progression and Metastasis. Ecancermedicalscience 2016, 10, 617–617. [Google Scholar] [CrossRef]
- Tewari, K.; Nakayama, Y.; Suresh, M. Role of Direct Effects of IFN-γ on T Cells in the Regulation of CD8 T Cell Homeostasis. J. Immunol. 2007, 179, 2115. [Google Scholar] [CrossRef] [PubMed]
- Bhat, P.; Leggatt, G.; Waterhouse, N.; Frazer, I.H. Interferon-γ Derived from Cytotoxic Lymphocytes Directly Enhances Their Motility and Cytotoxicity. Cell Death Dis. 2017, 8, e2836. [Google Scholar] [CrossRef] [PubMed]
- Burmeister, A.R.; Marriott, I. The Interleukin-10 Family of Cytokines and Their Role in the CNS. Front. Cell. Neurosci. 2018, 12, 458. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, N.; Zhou, J.; Ding, C.; Jin, Y.; Cui, X.; Pu, K.; Zhu, Y. Peptide Blocking of PD-1/PD-L1 Interaction for Cancer Immunotherapy. Cancer Immunol. Res. 2018, 6, 178. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Yamamoto, Y.; Takeshita, F.; Yamamoto, T.; Xiao, Z.; Ochiya, T. Delivery of miR-424-5p via Extracellular Vesicles Promotes the Apoptosis of MDA-MB-231 TNBC Cells in the Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 844. https://doi.org/10.3390/ijms22020844
Zhou Y, Yamamoto Y, Takeshita F, Yamamoto T, Xiao Z, Ochiya T. Delivery of miR-424-5p via Extracellular Vesicles Promotes the Apoptosis of MDA-MB-231 TNBC Cells in the Tumor Microenvironment. International Journal of Molecular Sciences. 2021; 22(2):844. https://doi.org/10.3390/ijms22020844
Chicago/Turabian StyleZhou, Yueyuan, Yusuke Yamamoto, Fumitaka Takeshita, Tomofumi Yamamoto, Zhongdang Xiao, and Takahiro Ochiya. 2021. "Delivery of miR-424-5p via Extracellular Vesicles Promotes the Apoptosis of MDA-MB-231 TNBC Cells in the Tumor Microenvironment" International Journal of Molecular Sciences 22, no. 2: 844. https://doi.org/10.3390/ijms22020844
APA StyleZhou, Y., Yamamoto, Y., Takeshita, F., Yamamoto, T., Xiao, Z., & Ochiya, T. (2021). Delivery of miR-424-5p via Extracellular Vesicles Promotes the Apoptosis of MDA-MB-231 TNBC Cells in the Tumor Microenvironment. International Journal of Molecular Sciences, 22(2), 844. https://doi.org/10.3390/ijms22020844




